Towards Automated Classification of Zooplankton Using Combination of Laser Spectral Techniques and Advanced Chemometrics
Abstract
1. Introduction
2. Materials and Methods
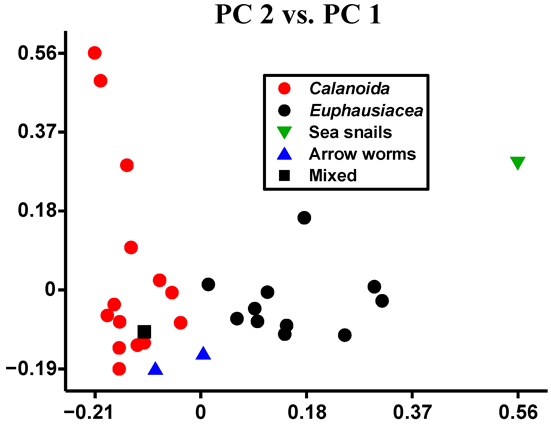
3. Results and Discussion
Critical Analysis and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keith, L.H.; Crummett, W.; Deegan, J.; Libby, R.A.; Taylor, J.K.; Wentler, G. Principles of Environmental Analysis. Anal. Chem. 1983, 55, 2210–2218. [Google Scholar] [CrossRef]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data: An Introduction to Cluster Analysis; Wiley-Interscience: Hoboken, NJ, USA, 2005; ISBN 978-0-471-73578-7. [Google Scholar]
- Gaudiuso, R.; Melikechi, N.; Abdel-Salam, Z.A.; Harith, M.A.; Palleschi, V.; Motto-Ros, V.; Busser, B. Laser-Induced Breakdown Spectroscopy for Human and Animal Health: A Review. Spectrochim. Acta Part B At. Spectrosc. 2019, 152, 123–148. [Google Scholar] [CrossRef]
- Rehse, S.J. A Review of the Use of Laser-Induced Breakdown Spectroscopy for Bacterial Classification, Quantification, and Identification. Spectrochim. Acta Part B At. Spectrosc. 2019, 154, 50–69. [Google Scholar] [CrossRef]
- Sushkov, N.I. Qualitative Classification of Biological Materials. In Laser-Induced Breakdown Spectroscopy in Biological, Forensic and Materials Sciences; Galbács, G., Ed.; Springer International Publishing: Cham, Switzerland, 2022; ISBN 9783031145018. [Google Scholar]
- González, P.; Álvarez, E.; Díez, J.; López-Urrutia, Á.; del Coz, J.J. Validation Methods for Plankton Image Classification Systems. Limnol. Oceanogr. Methods 2017, 15, 221–237. [Google Scholar] [CrossRef]
- Lumini, A.; Nanni, L. Ocean Ecosystems Plankton Classification BT—Recent Advances in Computer Vision: Theories and Applications. In Recent Advances in Computer Vision. Theory and Applications; Hassaballah, M., Hosny, K.M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 261–280. ISBN 978-3-030-03000-1. [Google Scholar]
- Pastore, V.P.; Zimmerman, T.G.; Biswas, S.K.; Bianco, S. Annotation-Free Learning of Plankton for Classification and Anomaly Detection. Sci. Rep. 2020, 10, 12142. [Google Scholar] [CrossRef]
- Kuzminykh, D.; Polykovskiy, D.; Zhebrak, A. Extracting Invariant Features from Images Using an Equivariant Autoencoder. Proc. Mach. Learn. Res. 2018, 95, 438–453. [Google Scholar]
- Wang, C.; Yu, Z.; Zheng, H.; Wang, N.; Zheng, B. CGAN-Plankton: Towards Large-Scale Imbalanced Class Generation and Fine-Grained Classification. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 855–859. [Google Scholar]
- Salvesen, E.; Saad, A.; Stahl, A. Robust Deep Unsupervised Learning Framework to Discover Unseen Plankton Species. In Proceedings of the SPIE, Fourteenth International Conference on Machine Vision (ICMV 2021 Rome, Italy), Hangzhou, China, 4 March 2022; Volume 12084, p. 120840V. [Google Scholar]
- Zorov, N.B.; Popov, A.M.; Zaytsev, S.M.; Labutin, T.A. Qualitative and Quantitative Analysis of Environmental Samples by Laser-Induced Breakdown Spectrometry. Russ. Chem. Rev. 2015, 84, 1021–1050. [Google Scholar] [CrossRef]
- Wang, Q.; Xiangli, W.; Teng, G.; Cui, X.; Wei, K. A Brief Review of Laser-Induced Breakdown Spectroscopy for Human and Animal Soft Tissues: Pathological Diagnosis and Physiological Detection. Appl. Spectrosc. Rev. 2021, 56, 221–241. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, D.; Wang, W.; Chen, F.; Xu, Y.; Nie, J.; Chu, Y.; Guo, L. A Review of Calibration-Free Laser-Induced Breakdown Spectroscopy. TrAC Trends Anal. Chem. 2022, 152, 116618. [Google Scholar] [CrossRef]
- Gunawan, R.; Imran, A.; Ahmed, I.; Liu, Y.; Chu, Y.; Guo, L.; Yang, M.; Lau, C. FROZEN! Intracellular Multi-Electrolyte Analysis Measures Millimolar Lithium in Mammalian Cells. Analyst 2021, 146, 5186–5197. [Google Scholar] [CrossRef]
- Luarte, D.; Myakalwar, A.K.; Velásquez, M.; Álvarez, J.; Sandoval, C.; Fuentes, R.; Yañez, J.; Sbarbaro, D. Combining Prior Knowledge with Input Selection Algorithms for Quantitative Analysis Using Neural Networks in Laser Induced Breakdown Spectroscopy. Anal. Methods 2021, 13, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Teng, G.; Wang, Q.; Cui, X.; Wei, K.; Xiangli, W.; Chen, G. Feature-Level Fusion of Laser-Induced Breakdown Spectroscopy and Raman Spectroscopy for Improving Support Vector Machine in Clinical Bacteria Identification. J. Raman Spectrosc. 2021, 52, 805–814. [Google Scholar] [CrossRef]
- Khan, M.N.; Wang, Q.; Idrees, B.S.; Teng, G.; Xiangli, W.; Cui, X.; Wei, K. Evaluation of Human Melanoma and Normal Formalin Paraffin-Fixed Samples Using Raman and LIBS Fused Data. Lasers Med. Sci. 2022, 37, 2489–2499. [Google Scholar] [CrossRef] [PubMed]
- Eum, C.; Jang, D.; Kim, J.; Choi, S.; Cha, K.; Chung, H. Improving the Accuracy of Spectroscopic Identification of Geographical Origins of Agricultural Samples through Cooperative Combination of Near-Infrared and Laser-Induced Breakdown Spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2018, 149, 281–287. [Google Scholar] [CrossRef]
- Mishra, P.; Roger, J.M.; Marini, F.; Biancolillo, A.; Rutledge, D.N. Parallel Pre-Processing through Orthogonalization (PORTO) and Its Application to near-Infrared Spectroscopy. Chemom. Intell. Lab. Syst. 2021, 212, 104190. [Google Scholar] [CrossRef]
- Vandeginste, B.G.M.; Massart, D.L.; Buydens, L.M.C.; De Jong, S.; Lewi, P.J.; Smeyers-Verbeke, J. Chapter 33—Supervised Pattern Recognition. In Handbook of Chemometrics and Qualimetrics: Part B; Vandeginste, B.G.M., Massart, D.L., Buydens, L.M.C., De Jong, S., Lewi, P.J., Smeyers-Verbeke, J.B.T.-D.H., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; Volume 20, pp. 207–241. ISBN 0922-3487. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Routledge: New York, NY, USA, 1984. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Vandeginste, B.G.M.; Massart, D.L.; Buydens, L.M.C.; De Jong, S.; Lewi, P.J.; Smeyers-Verbeke, J. Chapter 35—Relations between Measurement Tables. In Handbook of Chemometrics and Qualimetrics: Part B; Vandeginste, B.G.M., Massart, D.L., Buydens, L.M.C., De Jong, S., Lewi, P.J., Smeyers-Verbeke, J.B.T.-D.H., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; Volume 20, pp. 307–347. ISBN 0922-3487. [Google Scholar]
- Putnam, R.A.; Mohaidat, Q.I.; Daabous, A.; Rehse, S.J. A Comparison of Multivariate Analysis Techniques and Variable Selection Strategies in a Laser-Induced Breakdown Spectroscopy Bacterial Classification. Spectrochim. Acta Part B At. Spectrosc. 2013, 87, 161–167. [Google Scholar] [CrossRef]
- Rehse, S.J.; Diedrich, J.; Palchaudhuri, S. Identification and Discrimination of Pseudomonas Aeruginosa Bacteria Grown in Blood and Bile by Laser-Induced Breakdown Spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2007, 62, 1169–1176. [Google Scholar] [CrossRef]
- Malenfant, D.J.; Gillies, D.J.; Rehse, S.J. Bacterial Suspensions Deposited on Microbiological Filter Material for Rapid Laser-Induced Breakdown Spectroscopy Identification. Appl. Spectrosc. 2016, 70, 485–493. [Google Scholar] [CrossRef]
- Cisewski, J.; Snyder, E.; Hannig, J.; Oudejans, L. Support Vector Machine Classification of Suspect Powders Using Laser-Induced Breakdown Spectroscopy (LIBS) Spectral Data. J. Chemom. 2012, 26, 143–149. [Google Scholar] [CrossRef]
- Burges, C.J.C. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Metzinger, A.; Rajkó, R.; Galbács, G. Discrimination of Paper and Print Types Based on Their Laser Induced Breakdown Spectra. Spectrochim. Acta Part B At. Spectrosc. 2014, 94, 48–57. [Google Scholar] [CrossRef]
- Pořízka, P.; Klus, J.; Képeš, E.; Prochazka, D.; Hahn, D.W.; Kaiser, J. On the Utilization of Principal Component Analysis in Laser-Induced Breakdown Spectroscopy Data Analysis, a Review. Spectrochim. Acta Part B At. Spectrosc. 2018, 148, 65–82. [Google Scholar] [CrossRef]
- Choi, S.; Cichocki, A.; Park, H.-M.; Lee, S.-Y. Blind Source Separation and Independent Component Analysis: A Review. Neural Inf. Process. Lett. Rev. 2005, 6, 1–57. [Google Scholar]
- Lee, D.D.; Seung, H.S. Learning the Parts of Objects by Non-Negative Matrix Factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Monakhova, Y.B.; Rutledge, D.N. Independent Components Analysis (ICA) at the “Cocktail-Party” in Analytical Chemistry. Talanta 2020, 208, 120451. [Google Scholar] [CrossRef]
- Cichocki, A.; Zdunek, R.; Phan, A.H.; Amari, S. Nonnegative Matrix and Tensor Factorizations: Applications to Exploratory Multi-Way Data Analysis and Blind Source Separation; Wiley Publishing: Chichester, UK, 2009; ISBN 0470746661. [Google Scholar]
- Tavakkoli, E.; Rajkó, R.; Abdollahi, H. Duality Based Direct Resolution of Unique Profiles Using Zero Concentration Region Information. Talanta 2018, 184, 557–564. [Google Scholar] [CrossRef]
- Hérault, J.; Jutten, C.; Ans, B. Détection de Grandeurs Primitives Dans Un Message Composite Par Une Architecture de Calcul Neuromimétique En Apprentissage Non Supervisé. In Dixième Colloque sur le Traitement du Signal et ses Applications, Actes du Xème Colloque; GRETSI: Nice, France, 1985; pp. 1017–1022. [Google Scholar]
- Kassouf, A.; Rakwe, M.E.; Chebib, H.; Ducruet, V.; Rutledge, D.N.; Maalouly, J.; Matn, J. El Independent Components Analysis Coupled with 3D-Front-Face Fluorescence Spectroscopy to Study the Interaction between Plastic Food Packaging and Olive Oil. Anal. Chim. Acta 2014, 839, 14–25. [Google Scholar] [CrossRef]
- Meksiarun, P.; Ishigaki, M.; Huck-Pezzei, V.A.C.; Huck, C.W.; Wongravee, K.; Sato, H.; Ozaki, Y. Comparison of Multivariate Analysis Methods for Extracting the Paraffin Component from the Paraffin-Embedded Cancer Tissue Spectra for Raman Imaging. Sci. Rep. 2017, 7, 44890. [Google Scholar] [CrossRef]
- Hyvärinen, A. Independent Component Analysis: Recent Advances. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2013, 371, 20110534. [Google Scholar] [CrossRef]
- Hyvärinen, A. Survey on Independent Component Analysis. Neural Comput. Surv. 1999, 2, 94–128. [Google Scholar]
- Khlaifi, A. Estimation Des Sources de Pollution Par Modélisation Inverse. Thèse présentée pour l’obtention du Doctorat de l’, Université Paris XII, Créteil, France, 2007; p. 366. [Google Scholar]
- Werheit, P.; Fricke-Begemann, C.; Gesing, M.; Noll, R. Fast Single Piece Identification with a 3D Scanning LIBS for Aluminium Cast and Wrought Alloys Recycling. J. Anal. At. Spectrom. 2011, 26, 2166–2174. [Google Scholar] [CrossRef]
- Lobus, N.V.; Drits, A.V.; Flint, M. V Accumulation of Chemical Elements in the Dominant Species of Copepods in the Ob Estuary and the Adjacent Shelf of the Kara Sea. Oceanology 2018, 58, 405–415. [Google Scholar] [CrossRef]
- Lobus, N.V.; Arashkevich, E.G.; Flerova, E.A. Major, Trace, and Rare-Earth Elements in the Zooplankton of the Laptev Sea in Relation to Community Composition. Environ. Sci. Pollut. Res. 2019, 26, 23044–23060. [Google Scholar] [CrossRef]
- Lobus, N. V Elemental Composition of Zooplankton in the Kara Sea and the Bays on the Eastern Side of Novaya Zemlya. Oceanology 2016, 56, 809–818. [Google Scholar] [CrossRef]
- Freese, D.; Niehoff, B.; Søreide, J.E.; Sartoris, F.J. Seasonal Patterns in Extracellular Ion Concentrations and PH of the Arctic Copepod Calanus Glacialis. Limnol. Oceanogr. 2015, 60, 2121–2129. [Google Scholar] [CrossRef]
- Martin, M.Z.; Labbé, N.; André, N.; Harris, R.; Ebinger, M.; Wullschleger, S.D.; Vass, A.A. High Resolution Applications of Laser-Induced Breakdown Spectroscopy for Environmental and Forensic Applications. Spectrochim. Acta Part B At. Spectrosc. 2007, 62, 1426–1432. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Kourti, T.; MacGregor, J.F. Analysis of Multiblock and Hierarchical PCA and PLS Models. J. Chemom. 1998, 12, 301–321. [Google Scholar] [CrossRef]
- Mishra, P.; Roger, J.M.; Jouan-Rimbaud-Bouveresse, D.; Biancolillo, A.; Marini, F.; Nordon, A.; Rutledge, D.N. Recent Trends in Multi-Block Data Analysis in Chemometrics for Multi-Source Data Integration. TrAC Trends Anal. Chem. 2021, 137, 116206. [Google Scholar] [CrossRef]
- Cariou, V.; Jouan-Rimbaud Bouveresse, D.; Qannari, E.M.; Rutledge, D.N. ComDim Methods for the Analysis of Multiblock Data in a Data Fusion Perspective. In Data Handling in Science and Technology; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 31, pp. 179–204. [Google Scholar]
- Mazerolles, G.; Devaux, M.F.; Dufour, E.; Qannari, E.M.; Courcoux, P. Chemometric Methods for the Coupling of Spectroscopic Techniques and for the Extraction of the Relevant Information Contained in the Spectral Data Tables. Chemom. Intell. Lab. Syst. 2002, 63, 57–68. [Google Scholar] [CrossRef]
- Cariou, V.; Qannari, E.M.; Rutledge, D.N.; Vigneau, E. ComDim: From Multiblock Data Analysis to Path Modeling. Food Qual. Prefer. 2018, 67, 27–34. [Google Scholar] [CrossRef]
- Qannari, E.M.; Wakeling, I.; MacFie, H.J.H. A Hierarchy of Models for Analysing Sensory Data. Food Qual. Prefer. 1995, 6, 309–314. [Google Scholar] [CrossRef]
- Qannari, E.M.; Wakeling, I.; Courcoux, P.; MacFie, H.J.H. Defining the Underlying Sensory Dimensions. Food Qual. Prefer. 2000, 11, 151–154. [Google Scholar] [CrossRef]
- Makimori, G.Y.F.; Bona, E. Commercial Instant Coffee Classification Using an Electronic Nose in Tandem with the ComDim-LDA Approach. Food Anal. Methods 2019, 12, 1067–1076. [Google Scholar] [CrossRef]
- Vieira, T.F.; Makimori, G.Y.F.; dos Santos Scholz, M.B.; Zielinski, A.A.F.; Bona, E. Chemometric Approach Using ComDim and PLS-DA for Discrimination and Classification of Commercial Yerba Mate (Ilex Paraguariensis St. Hil.). Food Anal. Methods 2020, 13, 97–107. [Google Scholar] [CrossRef]
- Ríos-Reina, R.; Callejón, R.M.; Savorani, F.; Amigo, J.M.; Cocchi, M. Data Fusion Approaches in Spectroscopic Characterization and Classification of PDO Wine Vinegars. Talanta 2019, 198, 560–572. [Google Scholar] [CrossRef]
- Gibbons, E.; Léveillé, R.; Berlo, K. Data Fusion of Laser-Induced Breakdown and Raman Spectroscopies: Enhancing Clay Mineral Identification. Spectrochim. Acta Part B At. Spectrosc. 2020, 170, 105905. [Google Scholar] [CrossRef]
- Gyftokostas, N.; Nanou, E.; Stefas, D.; Kokkinos, V.; Bouras, C.; Couris, S. Classification of Greek Olive Oils from Different Regions by Machine Learning-Aided Laser-Induced Breakdown Spectroscopy and Absorption Spectroscopy. Molecules 2021, 26, 1241. [Google Scholar] [CrossRef]
- Breitwieser, M.; Vigneau, E.; Viricel, A.; Becquet, V.; Lacroix, C.; Erb, M.; Huet, V.; Churlaud, C.; Le Floch, S.; Guillot, B.; et al. What Is the Relationship between the Bioaccumulation of Chemical Contaminants in the Variegated Scallop Mimachlamys Varia and Its Health Status? A Study Carried out on the French Atlantic Coast Using the Path ComDim Model. Sci. Total Environ. 2018, 640, 662–670. [Google Scholar] [CrossRef]
- Sushkov, N.I.; Galbács, G.; Fintor, K.; Lobus, N.V.; Labutin, T.A. A Novel Approach for Discovering Correlations between Elemental and Molecular Composition Using Laser-Based Spectroscopic Techniques. Analyst 2022, 147, 3248–3257. [Google Scholar] [CrossRef]
- Zaytsev, S.M.; Popov, A.M.; Labutin, T.A. Stationary Model of Laser-Induced Plasma: Critical Evaluation and Applications. Spectrochim. Acta Part B At. Spectrosc. 2019, 158, 105632. [Google Scholar] [CrossRef]
- Brunet, J.-P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and Molecular Pattern Discovery Using Matrix Factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Buciu, I. Non-Negative Matrix Factorization, a New Tool for Feature Extraction: Theory and Applications. Int. J. Comput. Commun. Control 2008, 3, 67–74. [Google Scholar]
- Pearse, R.W.B.; Gaydon, A.G. The Identification of Molecular Spectra; Chapman & Hall: London, UK, 1963. [Google Scholar]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference Database of Raman Spectra of Biological Molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Lin-Vien, D.; Colthup, N.B.; Fateley, W.G.; Grasselli, J.G. The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules, 1st ed.; Academic Press: Cambridge, MA, USA, 1991. [Google Scholar]
- Dias, P.A.; Dunkel, T.; Fajado, D.A.S.; Gallegos, E. de L.; Denecke, M.; Wiedemann, P.; Schneider, F.K.; Suhr, H. Image Processing for Identification and Quantification of Filamentous Bacteria in in Situ Acquired Images. Biomed. Eng. Online 2016, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, E.J.M.; Kok, E.N.D.; Koning, S.G.B.d.; Sanders, J.; Aalbers, A.G.J.; Kok, N.F.M.; Beets, G.L.; Flohil, C.C.; Bruin, S.C.; Kuhlmann, K.F.D.; et al. Hyperspectral Imaging for Tissue Classification, a Way toward Smart Laparoscopic Colorectal Surgery. J. Biomed. Opt. 2019, 24, 16002. [Google Scholar] [CrossRef]
- Nielsen, J.H.; Pedersen, C.; Kiørboe, T.; Nikolajsen, T.; Brydegaard, M.; Rodrigo, P.J. Investigation of autofluorescence in zooplankton for use in classification of larval salmon lice. Appl. Opt. 2019, 26, 7022–7027. [Google Scholar] [CrossRef]
- Limbeck, A.; Brunnbauer, L.; Lohninger, H.; Pořízka, P.; Modlitbová, P.; Kaiser, J.; Janovszky, P.; Kéri, A.; Galbács, G. Methodology and Applications of Elemental Mapping by Laser Induced Breakdown Spectroscopy. Anal. Chim. Acta 2021, 1147, 72–98. [Google Scholar] [CrossRef]
- ThermoFisherScientific Website. Available online: https://www.thermofisher.com/order/catalog/product/IQLAADGABFFAHCMAPB (accessed on 17 October 2022).
- Zhao, Z.; Chen, L.; Liu, F.; Zhou, F.; Peng, J.; Sun, M. Fast Classification of Geographical Origins of Honey Based on Laser-Induced Breakdown Spectroscopy and Multivariate Analysis. Sensors 2020, 20, 1878. [Google Scholar] [CrossRef]
- Yang, Y.; Hao, X.; Zhang, L.; Ren, L. Application of Scikit and Keras Libraries for the Classification of Iron Ore Data Acquired by Laser-Induced Breakdown Spectroscopy (LIBS). Sensors 2020, 20, 1393. [Google Scholar] [CrossRef]
- Bilge, G.; Velioglu, H.M.; Sezer, B.; Eseller, K.E.; Boyaci, I.H. Identification of Meat Species by Using Laser-Induced Breakdown Spectroscopy. Meat Sci. 2016, 119, 118–122. [Google Scholar] [CrossRef]

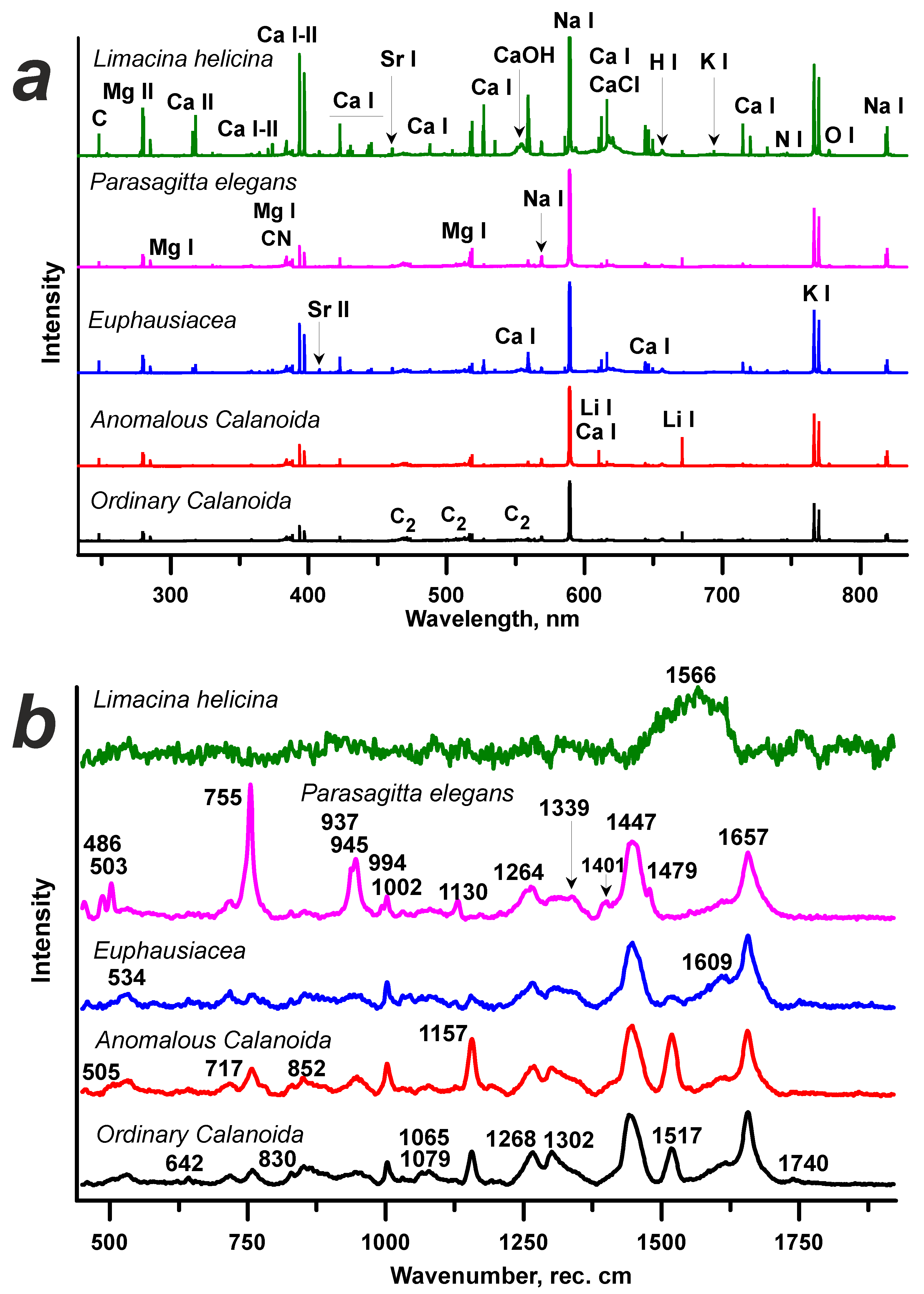

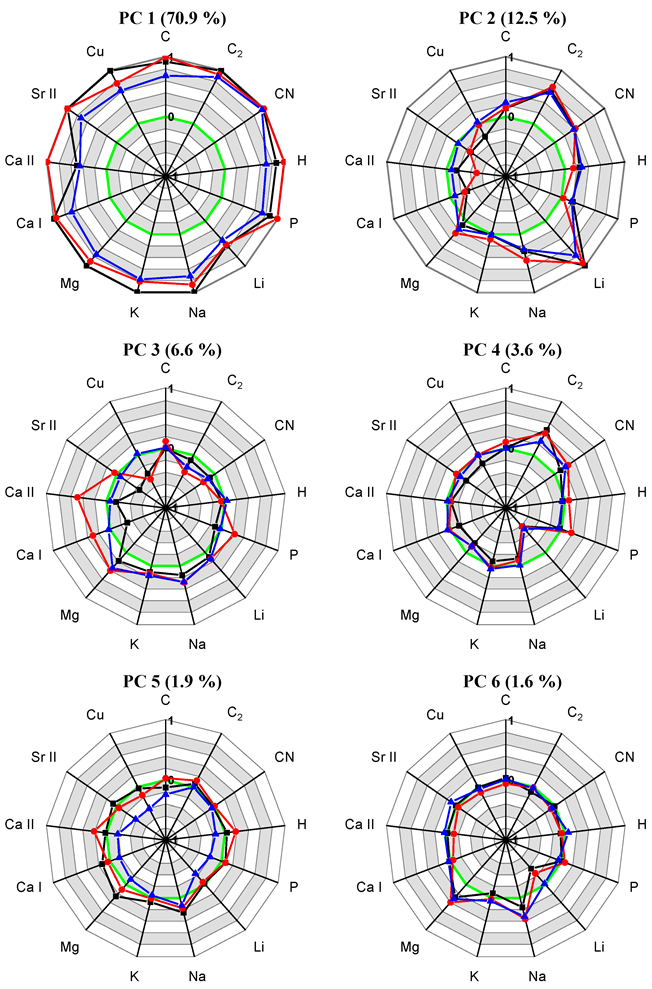

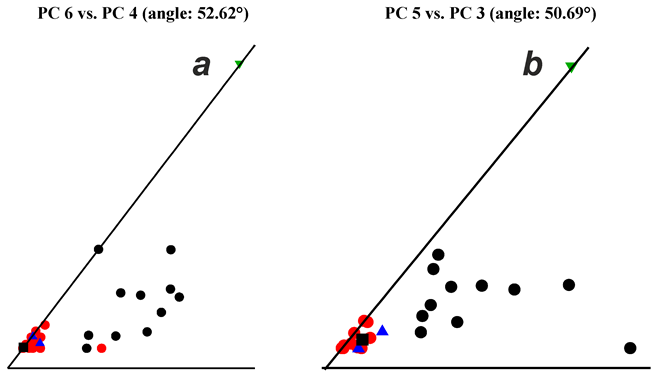
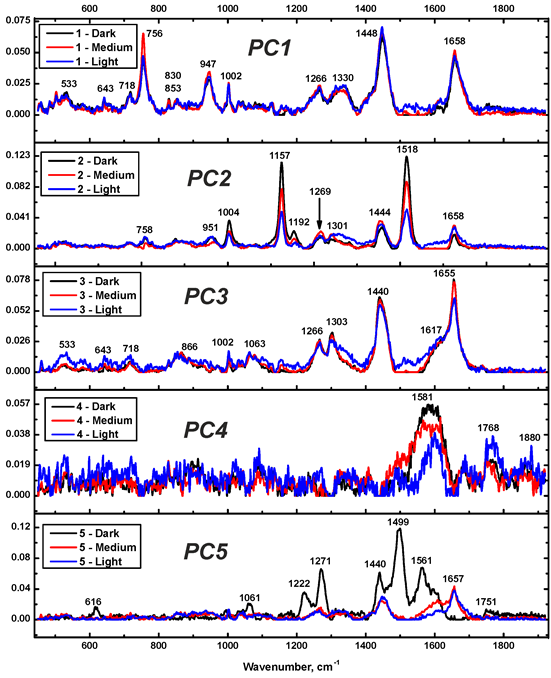
| No. | Method, Dataset | Plane | Sil, % | Sil (all dim.), % |
|---|---|---|---|---|
| Crustaceans–Arrow worms–Sea snails discrimination | ||||
| 1 | PCA—Raman only, full (5/97) | 3-1 | 10 | –10 |
| 2 | PCA—Raman only, short (5/89) | 5-1 | 65 | 39 |
| 3 | NMF—Raman only, full (5/84) | 4-2 | 11.5 | –6 |
| 4 | NMF—Raman only, short (5/75) | 4-1 | 47 | 34 |
| Calanoida–Euphausiacea discrimination | ||||
| 5 | PCA—LIBS only, full (5/99) | 5-1 | 34 | 17 |
| 6 | PCA—LIBS only, short (6/97) | 2-1 | 62 | 22 |
| 7 | NMF—LIBS only, full (5/88) | 5-2 | 56 | 6 |
| 8 | NMF—LIBS only, short (6/83) | 6-4 | 60 | 37 |
| 9 | SUM-PCA—LIBS + Raman (5/91) | 2-1 | 67 | 21 |
| 10 | NMF—LIBS + Raman (5/72) | 5-3 | 63 | 28 |
| 11 | ComDimPCA—LIBS + Raman (3/99.06) | 2-1 | 39 | 30 |
| 12 | ComDimICA—LIBS + Raman (3/96.89) | 3-2 | 36 | 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sushkov, N.I.; Galbács, G.; Janovszky, P.; Lobus, N.V.; Labutin, T.A. Towards Automated Classification of Zooplankton Using Combination of Laser Spectral Techniques and Advanced Chemometrics. Sensors 2022, 22, 8234. https://doi.org/10.3390/s22218234
Sushkov NI, Galbács G, Janovszky P, Lobus NV, Labutin TA. Towards Automated Classification of Zooplankton Using Combination of Laser Spectral Techniques and Advanced Chemometrics. Sensors. 2022; 22(21):8234. https://doi.org/10.3390/s22218234
Chicago/Turabian StyleSushkov, Nikolai I., Gábor Galbács, Patrick Janovszky, Nikolay V. Lobus, and Timur A. Labutin. 2022. "Towards Automated Classification of Zooplankton Using Combination of Laser Spectral Techniques and Advanced Chemometrics" Sensors 22, no. 21: 8234. https://doi.org/10.3390/s22218234
APA StyleSushkov, N. I., Galbács, G., Janovszky, P., Lobus, N. V., & Labutin, T. A. (2022). Towards Automated Classification of Zooplankton Using Combination of Laser Spectral Techniques and Advanced Chemometrics. Sensors, 22(21), 8234. https://doi.org/10.3390/s22218234







