Abstract
A systematic review on the topic of automatic detection of COVID-19 using audio signals was performed. A total of 48 papers were obtained after screening 659 records identified in the PubMed, IEEE Xplore, Embase, and Google Scholar databases. The reviewed studies employ a mixture of open-access and self-collected datasets. Because COVID-19 has only recently been investigated, there is a limited amount of available data. Most of the data are crowdsourced, which motivated a detailed study of the various pre-processing techniques used by the reviewed studies. Although 13 of the 48 identified papers show promising results, several have been performed with small-scale datasets (<200). Among those papers, convolutional neural networks and support vector machine algorithms were the best-performing methods. The analysis of the extracted features showed that Mel-frequency cepstral coefficients and zero-crossing rate continue to be the most popular choices. Less common alternatives, such as non-linear features, have also been proven to be effective. The reported values for sensitivity range from 65.0% to 99.8% and those for accuracy from 59.0% to 99.8%.
1. Introduction
COVID-19 is an infectious respiratory disease caused by the SARS-CoV-2 virus [1]. Soon after its outbreak, near the end of 2019, the disease became a global pandemic. As of 27 February 2022, confirmed cases have surpassed 433 million, with more than 5.9 million reported deaths worldwide [2]. Several variants of the virus have been identified within two years, and some have spread rapidly across the globe. The SARS-CoV-2 virus has strong mutation capabilities, albeit its variants differ in transmissibility and severity. Factors such as advanced age or the presence of comorbidities seem to elevate the risk of developing severe clinical symptoms regardless of the virus variant. Due to the very high transmissibility of SARS-CoV-2, which has already pushed several national healthcare systems to the limit, a continuously increasing amount of effort has been exerted into reducing and controlling the spread of the virus. Individual and collective measures, such as face-mask mandates, the development of vaccines, social distancing, isolation and quarantine periods, and restrictions placed on mobility and public life, have been employed with different degrees of effectiveness [3,4,5].
A prompt diagnosis of newly confirmed cases is critical to the strategy used to fight COVID-19. Since the early days of the pandemic, the gold standard for COVID-19 diagnosis has been the polymerase chain reaction (PCR) test. However, these tests have deficiencies: non-specific amplifications might arise and the sample amount is very limited. Furthermore, they are expensive, slow, and complicated to perform, and require highly qualified professionals and specialized lab equipment [6]. Antigen tests are an alternative based on the detection of viral proteins located on the surface of the virus [6]. These types of tests are more affordable than PCR tests, can be self-performed, and yield results within 15 to 30 min. Nonetheless, they report a poor sensitivity [7,8], which led the WHO to recommend against their use for patient care [9].
Both PCR and antigen tests have the additional burden of being invasive. To overcome these issues, many experts have tried to develop non-invasive, automated, fast testing methods that can be self-performed and offer reliably good results. These works have been based on the analysis of audio signals because COVID-19 generally attacks both the upper and the lower respiratory tracts, with the lungs being particularly affected [10]. Because these are the main body parts involved in the mechanisms of sound production [11], it is safe to assume that COVID-19 might alter the signals of breathing, coughing, and vocalization. Coughing seems to be a common factor in a majority of COVID-19 patients, no matter the variant of the disease [12]. Hence, many studies have used coughing as their data to perform a diagnosis. Such non-invasive, automated methods were also employed before the outbreak of this pandemic. One of the earliest works on automatic cough detection by Matos et al. dates back to 2006 [13], while Shin et al. employed these types of signals for the first time to determine anomalous health conditions [14]. A large number of studies with a similar purpose have been focused on several other illnesses, including pneumonia, pertussis, and asthma. The generally adopted methodology relies on the extraction of spectral and statistical features from the audio signal, which are then fed into a classification algorithm.
Several papers have been published since the pandemic on this topic. Udugama et al. [15] and Thompson et al. [16] focus their reviews on clinical diagnostic methods. Serrurier et al. [17] presented a work that reviews automatic cough acquisition, detection, and classification methods. However, the included studies were not necessarily related to COVID-19. Schuller et al. [18] analyzed the applicability and limitations of computerized audio tools to contain the crisis due to the SARS-CoV-2 virus. A recent publication [19] discussed the challenges and opportunities of deep learning for cough-based COVID-19 diagnosis from an overall perspective. However, this review performs a detailed step-wise examination of the methodology of existing works, either using machine learning- or deep learning-based methods. Specifically, this work focuses on the audio pre-processing and feature extraction processes, which we believe will allow future researchers to improve the performance and generalization of these methods.
2. Methods
2.1. Study Guidelines
This review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement (PRISMA) [20]. A review protocol was drafted using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocols [21].
2.2. Search Strategy and Study Eligibility
In this work, the following search terms were employed: (automatic (detection OR diagnosis)) AND (cough OR COVID-19) AND (audio OR sound). To ensure the thoroughness of the review, several databases were searched for papers published between 1 December 2019 and 1 January 2022. Gray literature (e.g., government reports or white papers) was not included in this review in an attempt to only include peer-reviewed studies. The search for this review was completed in April 2022.
2.3. Inclusion and Exclusion Criteria
In the first pre-screening phase, duplicate records and papers published in languages other than English, or that were not accessible, were discarded. The screening phase consisted of several steps. First, all the papers that were reviews, case reports, or corrigenda were removed. Then, all the studies that were considered not relevant based on their title were discarded. Finally, the abstracts of the papers were reviewed, the key information was extracted from the main text, and those considered irrelevant were discarded. Reference lists of eligible studies were also hand-searched but no additional studies were included on this basis.
2.4. Data Extraction and Risk of Bias
Each potential study for inclusion underwent full-text screening and was assessed to extract study-specific information and data. For each of the included articles, we extracted information from the below perspectives: the year the paper was published, author(s), used dataset(s), devices, number of subjects, subjects’ gender, pre-processing techniques, feature extraction techniques, and evaluation metrics. The authors (JA and ME) relied on QUADAS-2 to assess the applicability of each study, and the authors reached a consensus when needed.
3. Results
3.1. Study Selection
The paper presents the findings from a systematic review of articles on the topic of “Automatic COVID-19 detection using audio signals”. In this section, we present our findings regarding the selection of studies, the employed datasets, the pre-processing and feature extraction stages, and the choice of classifier algorithm (Table 1). The research papers included in this review were found by searching the following databases: PubMed, IEEE Xplore, Embase, and Google Scholar. By applying the aforementioned search strategy, papers were identified. During the pre-screening phase, 128 papers were discarded, thus leaving for the screening process. The first stage of this process resulted in the exclusion of 80 studies. As a result of the second screening stage, 233 papers were further removed, thus leaving papers for eligibility assessment. During the last stage, 170 papers were considered to be unsuitable for the review. Finally, the remaining papers were selected and included in this review. A flow chart summarizing the selection process is shown in Figure 1.
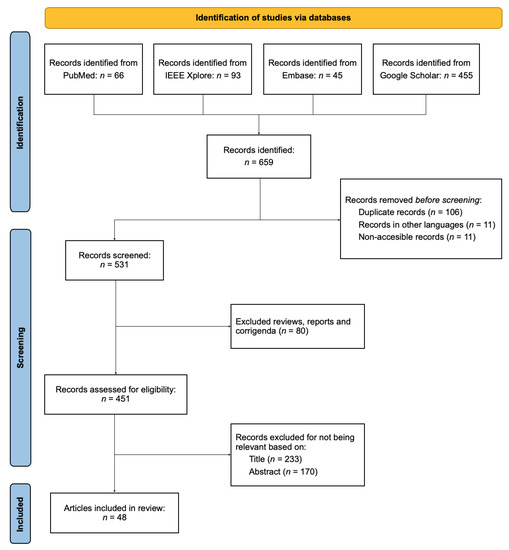
Figure 1.
Flowchart summarizing the search and selection process of the reviewed papers.
The majority of the forty-eight studies reviewed in this paper used already existent datasets, while six papers complemented this with their own data, and nine papers exclusively used self-collected audios. The largest available datasets were collected by the Coughvid Project [22] and the University of Cambridge [23], respectively. The former is open-access, whereas the latter can be made available upon request for research purposes. The Coswara [24] and Virufy [25] datasets were widely used among the reviewed papers. The mid-sized Coswara dataset as well as the Coughvid and Cambridge datasets consist of crowdsourced data. The Virufy dataset has both crowdsourced and clinical data. However, only the latter, which amounts to less than 100 files, is accessible. Other small datasets that are not publicly available include the Sarcos [26] and NoCoCoDa [27] datasets. Furthermore, early pandemic studies complemented their data with cough audio files from non-COVID-related datasets. Figure 2 shows the number of papers that employed each of these datasets. The audio files contain breathing, voiced, and principally cough sounds. Although most reviewed studies use cough sounds, other studies combined these with breathing and voiced sounds or chose to use sounds other than coughs. The number of selected samples varies significantly, ranging from 80 [28] to 16,007 [29]. Note that clinically obtained datasets usually require oversight from an institutional review board, which takes time for the studies to run, data to be collected, and then shared. Hence, they exist less often. On the contrary, the applications developed to collect crowdsourced data include disclaimers informing the user beforehand that their audio recordings will be used for research purposes.
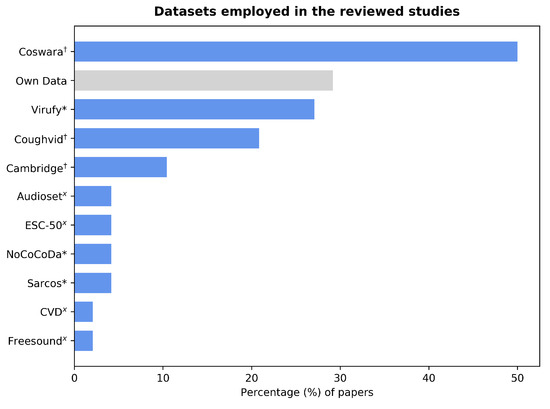
Figure 2.
Bar graph showing the percentage of papers that used each of the identified datasets. Of them, 29% employed their own self-collected data, which have not been formally turned into a dataset. Those marked with “x” are non-COVID datasets that originated before the start of the pandemic; those marked with “*” include clinically collected data and those marked with “†” consist of crowdsourced data.
3.2. Pre-Processing Methods
Pre-processing is generally considered a crucial step in any process involving signal analysis. Employing low-quality data could induce inaccurate results. Hence, to ensure that the analysis and classification results are robust and accurate, pre-processing the raw data is required [30]. While most papers employed several pre-processing techniques, three papers were found [31,32,33], which did not report using any pre-processing steps. Moreover, two other papers [34,35] explicitly specified working with the unprocessed signal. Note that this review does not consider data augmentation techniques to be signal pre-processing, even if some papers include it in their pre-processing sections. Figure 3 presents a summary of the pre-processing techniques used in the reviewed studies.
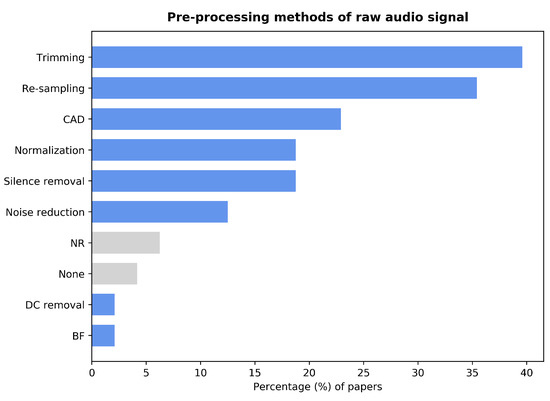
Figure 3.
Bar graph showing the percentage of papers that employ each of the pre-processing techniques. Trimming: manual segmentation of the raw audio signal. Re-sampling: reduction of the sampling rate of the raw audio signal. CAD: algorithm-based Cough Automatic Detection method. Normalization: amplitude normalization of the raw audio signal. BF: application of a low-pass Butterworth filter. Silence removal: algorithm-based method to remove silence parts of the raw audio signal. DC removal: subtraction of the DC part of the raw audio signal. None: papers that report using the raw audio signal for feature extraction. NR: papers that did not report any pre-processing.
Cough Automatic Detection (CAD) is an algorithm-based method to detect whether one or more coughs are present in an audio signal. This method was used by 23% of the studies and it is particularly relevant for crowdsourced data. Since audio files recorded in an uncontrolled environment could include unwanted or irrelevant sounds, it could lead to faulty models. The creators of the Coughvid dataset provided each file with a cough probability score based on the result of a self-developed Extreme Gradient Boosting cough classifier [22] and created a cough segmentation algorithm [36]. The combination of these two steps as CAD was used by nine out of the 10 studies that employed the Coughvid dataset. A few studies used other classifiers to detect the presence of cough. Zhang et al. [37] employed a self-designed Convolutional Neural Network (CNN). Tena et al. [38] made use of the TensorFlow-based YAMNet deep neural network. Kamble et al. [39] built a Recurrent Neural Network (RNN) with a long short-term memory architecture. Feng et al. [40] achieved this with a simple k-Nearest Neighbors (k-NN) algorithm. In most cases, these cough detection methods were applied to self-collected data since other datasets do not require this step. In the Coswara and Cambridge datasets, the presence of cough in each audio file has been annotated manually [23,24]; the available recordings from the Virufy dataset were collected in a controlled environment, thus ensuring 100% the presence of cough [25].
Other straightforward pre-processing techniques include re-sampling, amplitude normalization, and manual trimming of the audio signals. These were employed by 35%, 19%, and 39% of the studies, respectively. The trimming procedure was used in two ways: either for silence removal or to segment analyzable sounds into chunks of fixed length. Some papers used a non-manual silence removal process. Feng et al. [40] used a Support Vector Machine (SVM) model to classify the frames of a recording between silence and sound based on the energy of the signal. Pahar et al. [26,41] removed the silences using a simple energy detector, although they did not provide any more details. A similar approach was used by Milani et al. [42], again without specifying the details of the process. Grant et al. [43], Nellore et al. [44], and Sangle and Gaikwad [45] used the signal’s energy as a reference. However, these studies effectively removed the silence parts by hard thresholding. Rao et al. [46] chose a simpler way; they considered any part of the recording below an amplitude threshold to be silent and limited the non-silence parts to a fixed-length duration. Gupta et al. [47] applied a low-pass fourth-order Butterworth filter with a cut-off frequency of 6 kHz (cf. [48]). Kahn et al. [49] removed the DC component of the signal. Unfortunately, they did not further clarify how this was carried out. Noise reduction is another pre-processing technique employed by 12% of the reviewed studies. Tawfik et al. [31] used a wavelet-based de-noising algorithm from the scikit-image library. Zhang et al. [37] chose a spectral gating algorithm based on a hard threshold applied to the Root Mean Square (RMS) of the original signal. Verde et al. [50] used a band-stop filter, as shown in [51]. Three other papers [52,53,54] mentioned noise reduction techniques without specifying how it was performed.
3.3. Feature Extraction
A total of 54 extracted feature types were found, which can be classified into four categories: frequency-space (F-type), time-space (T-type), frequency-time-space (FT-type), and statistical (S-type) features. The first three categories refer to features extracted from a frequency, time, and frequency-time representation of the audio signal. The fourth category consists of the features gained from statistical considerations of the corresponding independent variable. The usage of the different features and feature types in the reviewed papers is shown in Figure 4a–d.
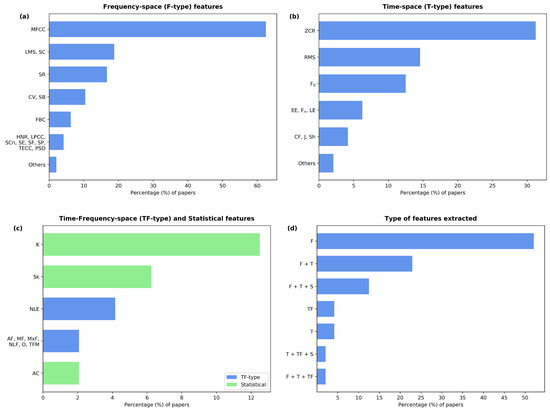
Figure 4.
Percentage of papers that extracted each listed feature grouped according to feature types: F-type (a), T-type (b), and TF-type and S-type (c). Additionally, the percentage of papers using each of the given combinations of feature types is shown (d). Legend ⇒ Mel-Frequency Cepstral Coefficients (MFCC), Log-Mel Spectrogram (LMS), Chroma Vector (CV), Spectral Centroid (SC), Spectral Roll-off (SR), Spectral Bandwidth (SB), Filter Bank Coefficients (FBC), Harmonics-to-Noise Ratio (HNR), Linear-Predictive Coding Coefficients (LPCC), Spectral Contrast (SCn), Spectral Entropy (SE), Spectral Flux (SF), Spectral flatness (SP), Teager Energy Cepstral Coefficients (TECC), Power Spectrum Density (PSD), Zero Crossing Rate (ZCR), Root-Mean-Square (RMS), fundamental Frequency (F), Entropy of the Energy (EE), formant Frequencies (F), Log Energy (LE), Crest Factor (CF), Jitter (J), Shimmer (Sh), Kurtosis (K), Skewness (Sk), Non-Linear Entropies (NLE), Average, Mean and Maximum Frequency (AF, MF, and MxF), Non-Linear Features (NLF), Onset (O), Time-Frequency Moment (TFM), and AutoCorrelation (AC).
The most commonly employed feature was the Mel-Frequency Cepstral Coefficient (MFCC), which is an F-type feature. MFCC features were used in 62% of the studies, although the number of extracted coefficients varied extensively among the papers. Five of the studies [32,37,40,50,54] did not report the number of coefficients that were extracted, and two papers [29,55] mentioned the use of over 60 coefficients. In most cases, 12 or 13 coefficients were used. Other F-type features employed by several papers (10–20%) were the spectral centroid, the spectral bandwidth, the spectral roll-off, the Log-Mel Spectrogram (LMS), and the Chroma Vector (CV). Similar to the MFCC, the CV is a multi-coefficient feature. Two papers [31,52] did not report the number of CV coefficients, while the rest extracted 12 of them. The spectral entropy, the power spectrum density, and the filter bank coefficients were present in 6% of the studies. Other F-type features include the spectral flux, the harmonics-to-noise ratio, the spectral flatness, the spectral contrast, the Teager energy cepstral coefficients, and the linear predictive coding coefficients. These were found in 4% of the papers. Finally, the rest of the F-type features, only observed in 2% of the papers, were spectral spread, relative spectra perceptual linear prediction, Harmonic Ratio (HR), spectral energy, spectral information, noise-to-harmonics ratio, cepstral peak prominence, tonal centroid, non-negative matrix factorization coefficients, eigenvalue ratios, and gamma-tone cepstral coefficients. The formal definitions of these features or their algorithms can easily be found in the signal processing literature. For a formal definition of HR, see [56].
The most frequently evaluated T-type feature was the zero-crossing rate, which was found in 31% of the studies. The fundamental frequency and the RMS were found in 12% and 14% of the papers, respectively. The entropies of the energy, the log-energy, and the formant frequencies were also considered in 6% of the cases. The following T-type features were found in approximately 4% of the studies: the signal’s energy, symbolic recurrence quantification analysis, crest factor, jitter, shimmer, Hjorth descriptors, number and degree of voice breaks, Higuchi and Katz fractal dimension, and maximal phonation time.
Moreover, the S-type features skewness, kurtosis, and autocorrelation were considered by 6%, 12%, and 2% of the papers, respectively. Regarding the FT-type features, the use of non-linear entropies was found in 4% of the studies, while the mean, maximum, and average frequency (cf. [38]), the time-frequency moment, the onset, and some non-linear features (cf. [35]) were found in 2% of the studies.
3.4. Classification Algorithms
We identified 13 types of classification algorithms for COVID-19 diagnosis, which were used either independently, i.e., without combining them with other algorithms, or as an ensemble. They belong to two categories: neural network-based and supervised machine learning algorithms. The results are summarized in Figure 5. CNNs were used independently by 29% of the studies, making them the preferred choice amongst the neural network-based algorithms. Other types of networks include RNNs and Feedforward Neural Networks (FNN), which were used independently by 8% and 6% of the reviewed papers, respectively. Some of the studies integrated these three types into ensembles. Alkhodari and Khandoker [57] and Shen et al. [55] combined a CNN with an RNN, while Sanjeev et al. [52] and Fakhry et al. [58] used an FNN together with a CNN. Son and Lee [59] employed a Dense Neural Network (DNN) in combination with a CNN. Finally, Saha et al. [34] utilized a dense convolutional network, as introduced by Huang et al. [60].
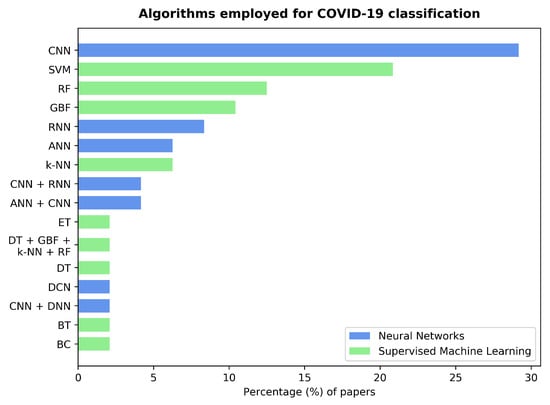
Figure 5.
Bar graph showing the percentage of papers employing different classifier algorithms or ensembles. The colors of the bars highlight the type of algorithm, with 54% of the papers using neural network-based algorithms and 48% of them using supervised machine learning. Note that some studies report results with several algorithms (see Table 1); thus, the sum of these percentages exceeds 100%. Legend⇒ Convolutional Neural Network (CNN), Support Vector Machine (SVM), Random Forests (RF), Gradient Boosting Framework (GBF), Recurrent Neural Network (RNN), Feedforward Neural Network (FNN), k-Nearest Neighbors (k-NN), Extremely randomized Trees (ET), Decision Tree (DT), Dense Convolutional Network (DCN), Dense Neural Network (DNN), Bagged Tree (BT), and Binary Classifier (BC).
Supervised machine learning algorithms were used independently in most cases. SVM, Random Forests (RF), Gradient Boosting Framework (GBF), and k-NN algorithms were observed in 21%, 12%, 10%, and 6% of the studies, respectively. Khan et al. [49] implemented a Decision Tree (DT), while Milani et al. [42] chose a bagged tree, i.e., a DT with a bootstrapping aggregation meta-algorithm (cf. [61]). Chowdhury et al. [62] used an extremely randomized trees classification algorithm. A different approach was taken by Pancaldi et al. [63], who employed a score-based binary classifier. The only instance of a supervised machine learning algorithm ensemble was found in [47], where Gupta et al. implemented a combination of GBF, DT, RF, and k-NN classifiers.
3.5. Metrics
For the analysis of the performance of the reviewed papers, the following evaluation metrics were used: specificity, sensitivity, F-score, accuracy, and Area Under the Curve (AUC). As shown in Figure 6a, only 10% of the studies reported all five values, while most of them included three or four metrics (38% and 31% of the papers, respectively). The works by Kamble et al. [64], Grant et al. [43], and Chaudhari et al. [65] only reported the AUC value. The most-reported metric was sensitivity followed by accuracy (Figure 6b); these appeared in 83% and 73% of the papers, respectively. The AUC value (62%) and specificity (60%) were reported similarly often; the F-score was only mentioned in 48% of the studies.
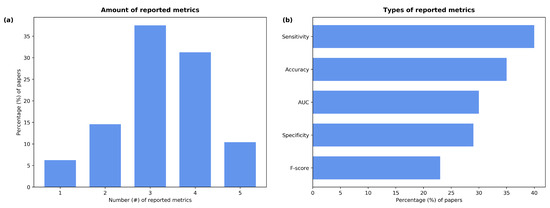
Figure 6.
Percentage of papers that (a) reported the given number of metrics and (b) used each one of them.

Table 1.
Summary of the reviewed papers. Non-reported data are given as “nr”. Datasets: T = type of collected data (B: breath, C: cough, V: voice), number of selected samples. Pre-processing: CAD = Cough Activity Detection. Features: Extracted features (number of features, only reported if the given feature is not a one-element category); numbers of features given as indicate the use of the fundamental coefficients together with the corresponding and values. Classifier: employed classification algorithm. Metrics: Specificity (SP), Sensitivity (SE), F-score (F1), Accuracy (ACC), and Area Under the Curve (AUC), given in percentages.
Table 1.
Summary of the reviewed papers. Non-reported data are given as “nr”. Datasets: T = type of collected data (B: breath, C: cough, V: voice), number of selected samples. Pre-processing: CAD = Cough Activity Detection. Features: Extracted features (number of features, only reported if the given feature is not a one-element category); numbers of features given as indicate the use of the fundamental coefficients together with the corresponding and values. Classifier: employed classification algorithm. Metrics: Specificity (SP), Sensitivity (SE), F-score (F1), Accuracy (ACC), and Area Under the Curve (AUC), given in percentages.
| Date | Ref. | Datasets [T] | Device (SR) | Pre-Processing | Features | Classifier | SP (%) | SE (%) | F1 (%) | ACC (%) | AUC (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2022 | Son and Lee [59] | Coughvid [C] (6092) | nr (48 kHz) | CAD, Re-sampling | MFCC (13), SP, LMS, SCn, SB, SC, SR, CV (12) | CNN + DNN | 94.0 | 93.0 | nr | 94.0 | nr |
| 2022 | Tawfik et al. [31] | Coswara, Virufy [C] (1171, 121) | nr (nr) | Noise reduction, Trimming, CAD | MFCC (20), O, CV (nr), SC, SR, ZCR, SB, CQT | CNN | 99.7 | 99.6 | 98.4 | 98.5 | nr |
| 2022 | Alkhodari and Khandoker [57] | Coswara [B] (480) | Smartphone (48 kHz) | Re-sampling | MFCC (13), K, SE, ZCR, EE, Sk, HFD, KFD | CNN + RNN | 93.0 | 93.7 | 93.3 | 93.3 | 88.0 |
| 2022 | Kamble et al. [64] | Coswara [C] (1436) | nr (44.1 kHz) | nr | TECC (63×3) | GBF | nr | nr | nr | nr | 86.6 |
| 2022 | Gupta et al. [47] | Coughvid [C] (2500) | nr (nr) | CAD, Normalization, BF, Re-sampling | MFCC (13), RMS, SR, SC, ZCR, CF | DT + RF + k-NN + GBF | nr | nr | 79.9 | 79.9 | 79.7 |
| 2022 | Haritaoglu et al. [29] | Coswara, Virufy, Coughvid, Own data [C] (Total: 16007) | nr (nr) | CAD, Re-sampling | MFCC (64), LMS | SVM, CNN | SVM: 51.0, CNN: 74.0 | SVM: 89.0, CNN: 77.0 | nr | SVM: 59.0, CNN: 75.0 | SVM: 80.7, CNN: 80.2 |
| 2022 | Islam et al. [56] | Virufy [C] | nr (nr) | Trimming | SC, SE, SF, SR, MFCC (13), CV (12), HR | CNN | 100.0 | 95.0 | 97.4 | 97.5 | nr |
| 2022 | Pancaldi et al. [63] | Own data [B] | Stethoscope (4 kHz) | Trimming | LPCC (8) | BC | 81.8 | 70.6 | nr | 75.0 | nr |
| 2021 | Milani et al. [42] | Coswara [V] (nr) | nr (nr) | Silence removal | GTCC (nr) | k-NN, BT | nr | nr | 90.9 | 90.0 | nr |
| 2021 | Nellore et al. [44] | Coswara [C] (1273) | nr (nr) | Silence removal | FBC (128×3), F , F (4), RMS | FNN | 34.3 | 85.6 | nr | nr | 65.7 |
| 2021 | Pahar et al. [41] | Coswara, Cambridge, Sarcos [C] , 517, | nr (Cos.: 44.1 kHz, Cam.: 16 kHz, Sar.: 44.1 kHz) | Re-sampling, Silence removal | ZCR, FBC, K, MFCC (13×3) | CNN | 92.7 | 95.7 | nr | 94.0 | 95.9 |
| 2021 | Rahman and Lestari [66] | Coswara, Coughvid [C] (5120, 2601) | nr (nr) | Cou.: CAD, Cos.: None | NMFC | SVM | 55.6 | 90.0 | nr | nr | 73.3 |
| 2021 | Saha et al. [34] | Coswara [C] (426) | nr (44.1 kHz) | None | LMS | DCN | 98.8 | 99.5 | nr | 99.5 | 98.9 |
| 2021 | Sangle and Gaikwad [45] | Coswara [C] (825) | nr (44.1 kHz) | Normalization, Silence removal | MFCC (13×3), ZCR, K, Sk | CNN | 99.2 | 92.3 | 96.0 | 96.0 | 96.0 |
| 2021 | Sanjeev et al. [52] | Own data [C] (1350) | nr (nr) | Noise reduction, Silence removal | MFCC (20), SC, CV (nr), RMS, ZCR, SB, SR | FNN + CNN | nr | 82.0 | 83.0 | 85.0 | 94.0 |
| 2021 | Shkanov et al. [67] | Own data [C] (1876) | nr (nr) | Trimming | EVR (2) | SVM, RF | nr | nr | SVM: 93.0, RF: 94.0 | SVM: 93.0, RF: 94.0 | nr |
| 2021 | Wang et al. [68] | Own data [B] (104) | Stethoscope (nr) | Trimming | LMS | CNN | 93.1 | 90.0 | nr | 91.3 | nr |
| 2021 | Solak [35] | Virufy [C] (194) | nr (48 kHz) | None | NLF (9) | SVM | 96.8 | 93.1 | nr | 95.8 | 93.2 |
| 2021 | Zhang et al. [37] | Own data, Coswara, Virufy [C] (321, nr, nr) | nr (nr) | Noise reduction, CAD, Trimming | MFCC (nr) | CNN | 95.8 | 95.3 | 96.1 | 95.8 | 98.1 |
| 2021 | Chowdhury et al. [62] | Cambridge, Coswara, NoCoCoDa, Virufy [C] (525, 1319, 73, 121) | nr (nr) | Re-sampling | MFCC (40), TC (6), LMS, CV (12), SCn (7) | ET, GBF | nr | ET: 74.0, GBF: 80.0 | nr | nr | ET: 83.0, GBF: 82.0 |
| 2021 | Rao et al. [46] | Coughvid, Coswara [C] (nr) | nr (nr) | Silence removal, Re-sampling | LMS | CNN | 77.9 | 80.0 | nr | nr | 82.3 |
| 2021 | Shen et al. [55] | Coswara, Own data [C] (610, 707) | nr (nr) | Re-sampling, Trimming | bw-MFCC (128) | CNN + RNN | 93.3 | 81.8 | nr | nr | 96.1 |
| 2021 | Tena et al. [38] | Own data, Coswara, Cambridge, Virufy [C] (813 total) | nr (nr) | CAD, Re-sampling, Normalization | SEn, TFM, MxF, MF, AF, NLE (3), SI, K | RF | 85.1 | 86.0 | 85.6 | 85.5 | 89.6 |
| 2021 | Vahedian-Azimi et al. [32] | Own data [V] (748) | Recorder (44.1 kHz) | nr | F , J, Sh, HNR, NHR, AC, CPP, MFCC (nr), MPT, NVB, DVB | FNN | nr | 91.6 | 90.6 | 89.7 | nr |
| 2021 | Erdoğan and Narin [53] | Virufy [C] (1187) | nr (nr) | Normalization, Noise reduction | NLE (6) | SVM | 97.3 | 99.5 | 98.6 | 98.4 | nr |
| 2021 | Grant et al. [43] | Coswara [V] (1199) | nr (44.1 kHz) | Silence removal, Trimming, Normalization | MFCC (20×3), RASTA-PLP (20) | RF | nr | nr | nr | nr | 79.4 |
| 2021 | Irawati and Zakaria [69] | Coswara, Virufy [C] (150, 121) | nr (Cos.: 44.1 kHz, Vir.: 48 kHz) | Trimming | MFCC (20), SB, RMS, ZCR | GBF | nr | nr | Cos.: 87.0, Vir.: 82.0 | Cos.: 86.0, Vir.: 86.2 | nr |
| 2021 | Khan et al. [49] | Own data [C] (1579) | Stethoscope (8 kHz) | DC removal, Normalization, Trimming | HD (3) | k-NN, DT | k-NN: 97.8, DT: 92.0 | k-NN: 99.8, DT: 93.9 | nr | k-NN: 99.8, DT: 94.4 | nr |
| 2021 | Melek-Manshouri [70] | Virufy [C] | nr (nr) | Trimming | MFCC (13) | SVM | 91.7 | 98.6 | nr | 95.9 | nr |
| 2021 | Melek [71] | NoCoCoDa, Virufy [C] (59, 121) | nr (NoC.: 44.1 kHz, Vir.: 48 kHz) | Trimming | MFCC (19) | k-NN | nr | 97.2 | 98.0 | 98.3 | 98.6 |
| 2021 | Nessiem et al. [72] | Cambridge [C, B], (1035) | nr (16 kHz) | Re-sampling | LMS | CNN | 62.8 | 77.6 | nr | 67.7 | 77.6 |
| 2021 | Verde et al. [50] | Coswara [V] (166) | nr (nr) | Noise reduction | MFCC (nr), F , J, Sh, SC, SR | RF | 70.6 | 94.1 | 84.2 | 82.4 | 90.1 |
| 2021 | Banerjee and Nilhani [73] | Coswara, Coughvid [C] (233, 640) | nr (nr) | CAD, Trimming | LMS | CNN | 99.7 | 67.2 | 79.3 | 96.0 | nr |
| 2021 | Kamble et al. [39] | Coswara [C] (1253) | nr (44.1 kHz) | CAD | TECC (40×3) | GBF | 53.6 | 80.5 | nr | nr | 76.3 |
| 2021 | Maor et al. [74] | Own data [V] (434) | nr (nr) | Re-sampling | LMS | RF, SVM | 53.0 | 85.0 | nr | nr | 72.0 |
| 2021 | Pahar et al. [26] | Coswara, Sarcos [C] (1171, 44) | nr (44.1 kHz) | Silence removal, Normalization | MFCC (13), ZRC, K, LE | CNN, RNN | CNN: 98.0, RNN: 96.0 | CNN: 93.0, RNN: 91.0 | nr | CNN: 95.3, RNN: 92.9 | CNN: 97.6, RNN: 93.8 |
| 2021 | Vrindavanam et al. [75] | Freesound, Coswara, Cambridge [C] (150 total) | nr (44.1 kHz) | Re-sampling | MFCC (12), ZCR, F , RMS, CF, SB, PSD, SC, SR, LE, SP, LPCC, F (4) | SVM, RF | nr | SVM: 81.2, RF: 73.8 | SVM: 78.4, RF: 78.0 | SVM: 83.9, RF: 85.2 | nr |
| 2021 | Fakhry et al. [58] | Coughvid [C] (1880) | nr (44.1 kHz) | CAD, Re-sampling | MFCC (13), LMS | CNN + FNN | 99.2 | 85.0 | nr | nr | 91.0 |
| 2021 | Han et al. [76] | Own data [V] | Smartphone, Computer (nr) | Trimming, Re-sampling, Normalization | F , MFCC (12), RMS, HNR, ZCR | SVM | 82.0 | 68.0 | nr | nr | 79.0 |
| 2021 | Khriji et al. [54] | Audioset, ESC-50 [C, B] (nr) | nr (nr) | Noise reduction | PSD, FBC, MFCC (nr) | RNN | nr | 78.8 | 79.0 | 80.3 | nr |
| 2021 | Pal and Sankarasubbu [77] | Own data [C], (150) | nr (nr) | Re-sampling, Normalization, Trimming | MFCC (12), LE, EE, ZCR, Sk, F (4), F , K | FNN | 90.3 | 90.1 | 90.6 | 90.8 | nr |
| 2021 | Feng et al. [40] | Coswara, Virufy [C] (633, 25) | nr (nr) | Silence removal, CAD | MFCC (nr), E, EE, ZCR, SE, SC, SS, SF | RNN | nr | nr | nr | Cos.: 90.0, Vir.: 81.2 | Cos.: 92.8, Vir.: 79.0 |
| 2021 | Lella and Pja [78] | Cambridge [C, B, V] | Smartphone, Computer (16 kHz) | Trimming | MFCC (159×3) | CNN | nr | nr | 93.5 | 92.3 | nr |
| 2021 | Brown et al. [23] | Cambridge [C, B] (491) | Smartphone, Computer (nr) | Trimming, Re-sampling | MFCC (13×3), SC, RMS, SR, ZCR | SVM | nr | 72.0 | nr | nr | 82.0 |
| 2021 | Mouawad et al. [33] | CVD [C] (1927) | nr (22.05 kHz) | nr | s-RQA (nr) | GBF | nr | 65.0 | 62.0 | 97.0 | 84.0 |
| 2020 | Hassan et al. [28] | Own Data [C] | Microphone (nr) | Trimming | SC, SR, ZCR, MFCC (nr×3) | RNN | nr | 96.4 | 97.9 | 97.0 | 97.4 |
| 2020 | Chaudhari et al. [65] | Virufy, Coughvid, Coswara [C] (1442, 941, 105) | Smartphone (nr) | Re-sampling | MFCC (39), LMS | CNN | nr | nr | nr | nr | 77.1 |
| 2020 | Bansal et al. [79] | Audioset, ESC-50 [C] total) | nr (44.1 kHz) | Trimming | MFCC (40), SC, SR, ZCR | CNN | nr | 81.0 | 69.6 | 70.6 | nr |
4. Discussion
The development of a rapid, self-performable, automatized detection method can be a helpful tool for diagnosing and controlling the spread of diseases. In the case of highly contagious pandemic diseases, this is particularly important. In this systematic review, we selected and analyzed studies that addressed this issue analogously: first, the data from either crowdsourced or clinically obtained datasets underwent different signal processing techniques; then, a set of features was extracted and fed into a classification algorithm (see Figure 7), which characterized the data as positive or negative for COVID-19. In some instances, demographic information or clinical data are also used as input for the classifier.
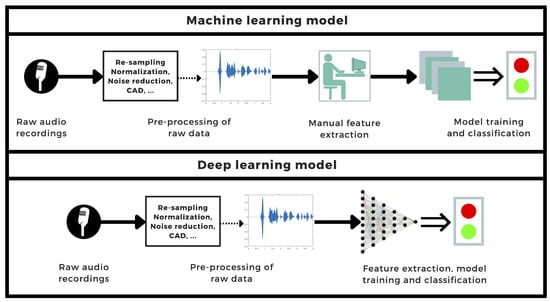
Figure 7.
General workflow diagram of common machine learning- and deep learning-based COVID-19 detection processes using audio signals.
Most of the employed datasets primarily consist of cough sounds. This type of sound is more suitable for developing a generalizable method. Breathing sounds often need to be recorded with a stethoscope to ensure good quality, which requires a clinical recording environment. Vocalized sounds are less general; some instructions or a detailed description of the kind of sounds that must be generated are required. Additionally, cough sounds have the advantage of being anharmonic. The features of two arbitrarily produced coughs can be easily compared [80], whereas a vocalized sound (e.g., a sustained vowel) is affected by the anatomy, thus making it necessary to differentiate between ages or genders. Furthermore, the features obtained from a recorded speech or phrase could be language-biased due to the associated defining phonetics. Nonetheless, some of the reviewed studies that do not employ cough sounds have reported promising results.
The amount of original data impacts the statistical significance of a study. In the past, data augmentation techniques have been used successfully for developing cough detection algorithms [81]. They have been most often applied to enlarge the available datasets or to balance the majority and minority classes. It has been shown that augmenting a balanced dataset to 4–5 times its original size can improve classification accuracy by more than 10% [82]. We consider that this technique needs to be approached cautiously. While a classifier should perform better when trained with more data, typical data augmentation algorithms create new data points by slightly variating the original data. Such a technique could result in the introduction of some bias. Some studies have investigated the performance of data augmentation in cough detection or image recognition, i.e., when the original pre-processed file is given as input to the classifier. To the best of our knowledge, there is a lack of detailed studies on whether data augmentation results in a positive impact or a negative bias in classification based on extracted features.
The choice of extracted features is essential to ensure effective classifier performance. Both the shortage of meaningful features and the abundance of irrelevant ones can contribute to the deterioration of classification efficiency. For example, in the case of MFCC, the most common choice was between 12 and 13 coefficients. The reasons for this choice might be twofold. While it has been shown that using eight to 14 coefficients can suffice for speech analysis [83], reducing the large number of initially extracted MFCC is often desired. For this purpose, the Karhunen–Loeve transform, which in speech analysis can be approximated by a direct cosine transform [84], is a frequent choice [85]. Several of the proposed methods only employ a subset of the extracted features. Some use a dimensionality reduction algorithm to select an optimal feature subset. We could not find two papers with the same choices regarding feature extraction and selection as well as classification algorithms. Moreover, these choices are not thoroughly justified, and the validation methods employed also varied. Therefore, and based on the results of this review, it is difficult to conduct a quantitative analysis of which features should be extracted, which feature selection algorithm works best, or which classifier is more effective.
Equally important is the quality of the extracted features, which is closely related to the quality of the audio data. Unless data are collected very carefully in a controlled environment, some pre-processing is needed to ensure good quality. Thus, it is safe to assume that self-performed recordings will be noisy and unclean. Noise reduction techniques were infrequently considered in the reviewed studies. This is a surprising fact, given that the majority of the used audio files came from crowdsourced datasets, which had not been curated in regard to noise. Noisy signals could bias the extracted features and obstruct the silence removal and CAD processes.
A coughing sound typically consists of an explosive, an intermediate, and a voiced phase [86]. While some coughs may be multiple (i.e., the next explosive phase starts before the last voiced phase ends), in most cases, the separation between individual coughs is clear. To analyze the sound signal, we also consider full cycles (i.e., all three phases) to be more appropriate than segments of fixed length. Extracting the individual coughs via manual trimming or annotation can be extremely time-consuming when working with large datasets. Additionally, it is not suitable for an automatized, self-performable detection method. The usage of an algorithm-based technique solves this issue, but it can be problematic for crowdsourced data, which do not undergo any screening of the sound types in the recording. By embedding cough segmentation into a CAD algorithm, both problems can be tackled at once.
Re-sampling is a technique that aims to improve computational speed by reducing the number of frames and thus the number of data points of a given audio file. A detailed comparison of the sampling rates and metric values in the reviewed studies did not show any correlation between these factors. In general, the choice of sampling rate should provide a good enough improvement in computational speed without a sensible loss of information. Although most of the studies that considered re-sampling agree with this choice, Alkhodari and Khandoker [57] achieved good results despite reducing the sampling rate from 48 kHz to 4 kHz.
The reported metrics are the most direct way of evaluating the performance of a proposed method. These values are greatly affected by the type and amount of available data and by the pre-processing steps, feature extraction, and classification algorithm that are used. As a consequence of such diversity, comparing the metric results between papers is rather ineffective. Nevertheless, to offer a qualitative evaluation of the performance of the reviewed studies, here, we will discuss the best-performing papers. The entire diagnosis process includes several steps. Thus, there is a large number of variables involved. Each of the metrics describes a different aspect of the performance of a method. Therefore, the quality of a study improves when several metrics are reported since a high value on one metric does not directly imply high values in other ones. In this way, we considered those papers with three or more reported evaluation metrics of 90% or higher each to be amongst the best performing. Note that other criteria could be used to determine the best-performing models. However, evaluating the study’s performance based on these metrics has to be consistently used in all papers.
Son and Lee [59] proposed concatenating the outputs of a CNN, fed with a handcrafted feature set, and a DNN that receives the LMS as input. While they did not use any cross-validation method, they used 70% of the data for training, 15% for validation, and 15% for testing. Although they accessed several datasets, they only reported the results of their proposed method with the Coughvid dataset. Furthermore, they worked with a heavily unbalanced dataset containing 10 healthy subjects for each COVID-19-positive subject (10N to 1P). A similar approach was taken by Tawfik et al. [31]. They mixed two different datasets but only employed a slightly unbalanced subset (2N to 1P). The evaluation was done using a 10-fold cross-validation technique. These two differences might explain why they obtained better results than Son and Lee [59]. Cross-validation improves the robustness of the method, while the use of balanced datasets for the training phase is particularly important, even if it does not play a crucial role in the testing phase [87].
Islam et al. [56] reported excellent results. However, that study used a balanced but very small dataset, which was clinically collected, i.e., noise-free and annotated. They used a 70/20/10 split of their data and used five-fold cross-validation. Their pre-processing techniques were tailored for this type of data, thus making them less generalizable. A more detailed explanation of their feature extraction process would be desired. Islam et al. [56]) compared features extracted from COVID-19 positive subjects and healthy subjects. However, silent frames, which were not removed before the feature extraction, can be easily identified. That might have biased the results of their work. Two other studies employed the same dataset, and while they took different approaches, they achieved similar results. Melek-Manshouri [70] extracted MFCC from the pre-processed signal and selected the best ones using a sequential forward search method, whose resulting output was fed into an SVM with an RBF kernel. The evaluation was performed with a leave-one-out cross-validation technique. Melek [71] employed a slightly larger but still small dataset and used a Euclidean k-NN classifier with leave-one-out cross-validation to determine the hyperparameters for feature extraction and evaluate their proposed method. In the case of the k-NN classifier, feature selection using a sequential forward search technique did not improve the results. However, the study disregarded noisy recordings. Such a treatment is not generalizable since only recordings collected in a controlled environment, such as a clinic or hospital, can be assuredly noise-free.
Saha et al. [34] performed data augmentation by pairing the original signals with random white noise and time-stretching the resulting audio. Consequently, the amount of initial data was enlarged 40-fold. Although data augmentation can help improve the classifier performance, as discussed above, Saha et al. [34] failed to address the main issue: their dataset was deeply unbalanced (10P to 1N). The evaluation was done with a 70/10/20 data split but without cross-validation. Other studies using data augmentation techniques are those by Sangle and Gaikwad [45] and Solak [35]. In both cases, oversampling was employed to balance the datasets. The former did an 80/20 split for evaluation without cross-validation. They also tried different sets of handcrafted features. However, their results seemed inconclusive since the accuracy and F-score values for the various combinations suggested random behavior. The dataset employed by the latter was still very limited and the evaluation techniques were not reported. Interestingly, Solak [35] worked with a set of handcrafted, nonlinear features motivated by the nonlinear nature of cough signals.
Erdoğan and Narin [53] also employed non-linear features. They had access to the complete Virufy dataset, including a large corpus of crowdsourced data. Unfortunately, since these data are not open-access, the replicability of this study is strongly hindered. On a positive note, the dataset was highly balanced, with almost the same number of annotated recordings from COVID-19 subjects and healthy subjects. They used the ReliefF algorithm to select the best among the extracted features. The evaluation was performed using an SVM classifier with a linear kernel and five-fold cross-validation.
Pahar et al. [26] explored two crowdsourced datasets: the large Coswara dataset and the small Sarcos dataset. After balancing via the synthetic minority over-sampling technique [88], the former was used to train both a CNN and an RNN model. The RNN model, in combination with leave-p-out cross-validation and sequential forward search techniques, was first applied to select the best subset of extracted handcrafted features. Both classifiers were subsequently tested while employing the same cross-validation technique. The CNN model had the best results when using the Coswara dataset for evaluation, whereas the RNN performed best with the Sarcos dataset. Moreover, the selected hyperparameters were different in both cases. Some of the results suggest that their proposed methods might not be generalizable. For example, when they applied the CNN classifier with its corresponding hyperparameters to the Sarcos dataset, the accuracy and AUC values decreased by 20%, and the specificity was 40% lower. The same authors took a different approach in another study [41]. First, they pre-trained a CNN model with several not-labeled datasets, which were created to develop cough detection or tuberculosis diagnosis algorithms. This technique is called transfer learning and it serves the same purpose as data augmentation. Although it allows the neural network to be trained with more original data, it might introduce some bias to the extracted features. For evaluation, the data was split (80% for training and 20% for testing), and nested cross-validation was applied.
Zhang et al. [37] worked with a small, self-collected dataset complemented with crowdsourced data. Unfortunately, they failed to report the amount of data used from each crowdsourced corpus. For their self-collected data, they applied a CAD pre-processing method. That method needs manual annotation, thus hindering the generalizability of their model. Furthermore, they used MFCCs as features but they did not mention the number of extracted coefficients. The entire dataset with an 80/10/10 split was used for training and testing their CNN-based CAD algorithm. The evaluation was performed with a five-fold cross-validation technique, although they only considered a small data subset.
The studies by Wang et al. [68], Pal and Sankarasubbu [77], and Hassan et al. [28] used very small datasets and failed to report whether a cross-validation technique was used. In Wang et al. [68], the data consisted of breath sounds collected with a stethoscope. That study applied data augmentation in the same way as Saha et al. [34], but did not mention how many new data points were created. Hassan et al. [28] used the smallest dataset of all the reviewed papers. Despite the imbalance (3N to 1P), neither data augmentation nor balancing techniques were applied.
The work by Khan et al. [49] differs from others in that they employed self-collected cough sounds from lung auscultations. They also trimmed the signal in frequency space, only considering the parts with frequencies up to 2 kHz. While this type of data can be high quality, it fails to address the desired self-performability criteria. Additionally, it leaves unanswered the question of whether the same method would offer as good results using other data types. They used Hjorth descriptors as features, which was not observed in the other reviewed papers. They evaluated their method with both a k-NN (k = 10) and a DT classifier, but they failed to report the validation technique they employed. In this case, the classification was three-fold and included COVID-19 positive, dry cough, and wet cough.
The reviewed studies work towards the development of an automatized, self- performable COVID-19 detection method using audio signals. We consider that cough signals are optimal for this purpose. They can be easily produced and recorded and are less biased by demographic factors. To improve the generalizability of the results, it is essential to have large amounts of data. Despite showing good performance metrics, several of the reviewed studies were performed with small 200) datasets. Techniques such as data augmentation and transfer learning can help increase the amount of available data, but we strongly encourage merging all the available open-access datasets. Therefore, the development of a comprehensive pre-processing algorithm that allows combining data collected in different environments is considered to be decisive. In particular, noise reduction and segmentation techniques can improve the quality of the raw data. For non-annotated datasets, we recommend the use of the CAD algorithm developed by the authors of the Coughvid dataset. When applied together with a cough segmentation algorithm, it yields good results, as shown by Son and Lee [59] and Fakhry et al. [58]. Re-sampling could also be beneficial, especially when employing large datasets. A sampling rate between 16 and 22 kHz is regarded as an optimal choice that enhances computational speed without an extensive loss of information. Particular emphasis should be put into the feature extraction process. We believe that a detailed study of each individual feature and its potential to characterize an audio signal as COVID-19 positive or negative would lead to more informed feature choices. In addition to the common feature types used in audio signal analysis, alternative options, as shown by several in several of the reviewed papers, can offer promising results as well. A better understanding of an optimal set of features for the particular case of COVID-19 characterization could also create the opportunity to conduct a more detailed study of different classification algorithms.
Based on the results of this review, we believe that the following recommendations could lead to better and, in particular, more generalizable results:
- Database creators should aim to collect diverse data, including a wide range of ages, genders, ethnicities, etc. These datasets need also be publicly available since combining several databases would allow researchers to have a larger data corpus and reduce the possible biases introduced by demographic factors.
- The pre-processing methods need to be automated and be as non-data-specific as possible. It is essential that researchers consider carefully whether re-sampling techniques improve computational speed enough to compensate for the information loss. An efficient CAD algorithm is critical since manual trimming and labeling become highly impractical when working with large datasets. Data augmentation should be avoided in favor of precise cough segmentation techniques and the creation of large datasets.
- The impact of each feature needs to be investigated individually, thus increasing the efficiency of manual feature extraction processes, allowing the adaptation of the neural network architectures, and improving the generalizability of the methods. Moreover, by fixing a subset of features that does not depend on the available dataset, researchers could design studies to test the performance of several machine learning and deep learning models as the only dependent variable.
5. Conclusions
Over the last two decades, the application of neural networks and machine learning algorithms for cough detection, cough discrimination, and the diagnosis of respiratory diseases, such as pneumonia or asthma, via the analysis of audio signals has notably progressed. Since the outbreak of the COVID-19 pandemic, all efforts have been focused on implementing and improving these methods to diagnose this disease. This systematic review has shown that state-of-the-art studies offer promising results. However, an important issue is the generalizability of the proposed methods. The scientific community should aim to develop a pre-screening tool that could be used instead of the usual test methods (PCR and antigen tests). Such a solution would be fast, accurate, and self-performable (without the necessity of specialized equipment or knowledge) and would significantly reduce the environmental impact associated with mass antigen and PCR tests. The first step is the creation of large open-access datasets of audio recordings that would allow many scientists to contribute to that objective. In combination with this, the development of general, non-dataset-specific pre-processing algorithms and a more detailed study of how both conventional and uncommon acoustical and statistical features can differentiate between healthy and infected subjects should be targeted. In particular, we suggest the individual study of every feature and the development of a quantifying method to establish the adequacy of each one for distinguishing between healthy and COVID-19-positive subjects. Although this might seem similar to dimensionality reduction methods, such as principal component analysis, an individual study of each feature would minimize the correlation in the feature selection step, thus strongly contributing to the reduction of error sources as well as to the generalization of the method.
Author Contributions
M.E. designed and led the study. J.G.A., M.E. and C.M. conceived the study. One author (J.G.A.) conducted the literature search, and two authors (J.G.A. and M.E.) independently screened the titles and abstracts for potentially eligible studies. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there are no competing interest.
List of Acronyms
| AC | Autocorrelation | LPCC | Linear predictive coding coefficients | |
| AF | Average frequency | LMS | Log-energy | |
| BC | Binary classifier | MF | Mean frequency | |
| BF | Butterworth filter | MFCC | Mel-frequency cepstral coefficients | |
| BT | Bagged tree | MPT | Maximal phonation time | |
| CF | Crest factor | MxF | Maximum frequency | |
| CV | Chroma vector | NHR | Noise-to-harmonic ratio | |
| CAD | Cough automatic detection | NLE | Non-linear entropies | |
| CNN | Convolutional neural network | NLF | Non-linear features | |
| CQT | Constant-Q transform | NMFC | Non-negative matrix factorization coefficients | |
| CPP | Cepstral peak coefficients | NVB | Number of voice breaks | |
| DCN | Dense convolutional network | O | Onset | |
| DT | Decision tree | PCR | Polymerase chain reaction | |
| DVB | Degree of voice breaks | PSD | Power spectrum density | |
| E | Energy of the signal | RASTA-PLP | Relative spectra perceptual linear prediction | |
| EE | Entropy of the energy | RF | Random forest | |
| ET | Extremely randomized trees | RMS | Root mean square | |
| EVR | Eigenvalue ratios | RNN | Recurrent neural network | |
| FBC | Filter bank coefficients | SB | Spectral bandwidth | |
| FNN | Feedforward neural network | SC | Spectral centroid | |
| F | Fundamental frequency | SCn | Spectral contrast | |
| F | Formant frequencies | SE | Spectral entropy | |
| GBF | Gradient boosting framework | SI | Spectral information | |
| GTCC | Gamma-tone cepstral coefficients | SF | Spectral flux | |
| HD | Hjorth descriptors | Sh | Shimmer | |
| HFD | Higuchi fractal dimension | Sk | Skewness | |
| HNR | Harmonic-to-noise ratio | SP | Spectral flatness | |
| HR | Harmonic ratio | SR | Spectral roll-off | |
| J | Jitter | s-RQA | Symbolic recurrence quantification analysis | |
| K | Kurtosis | SVM | Support vector machine | |
| KFD | Katz fractal dimension | TC | Tonal centroid | |
| k-NN | k-Nearest neighbors | TECC | Teager energy cepstral coefficients | |
| LE | Log-energy | TFM | Time-frequency moment | |
| LMS | Log-Mel spectrogram | ZCR | Zero crossing rate |
References
- World Health Organization (WHO). Naming the Coronavirus Disease (COVID-19) and the Virus That Causes It. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 22 March 2022).
- World Health Organization (WHO). COVID-19 Weekly Epidemiological Update. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—1-march-2022 (accessed on 22 March 2022).
- Mahmoudi, J.; Xiong, C. How social distancing, mobility, and preventive policies affect COVID-19 outcomes: Big data-driven evidence from the District of Columbia-Maryland-Virginia (DMV) megaregion. PLoS ONE 2022, 17, e0263820. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Zu, J.; Fairley, C.K.; Pagán, J.A.; An, L.; Du, Z.; Guo, Y.; Rong, L.; Xiao, Y.; Zhuang, G.; et al. Projected COVID-19 epidemic in the United States in the context of the effectiveness of a potential vaccine and implications for social distancing and face mask use. Vaccine 2021, 39, 2295–2302. [Google Scholar] [CrossRef] [PubMed]
- Kerr, C.C.; Mistry, D.; Stuart, R.M.; Rosenfeld, K.; Hart, G.R.; Núñez, R.C.; Cohen, J.A.; Selvaraj, P.; Abeysuriya, R.G.; Jastrzębski, M.; et al. Controlling COVID-19 via test-trace-quarantine. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kyosei, Y.; Yamura, S.; Namba, M.; Yoshimura, T.; Watabe, S.; Ito, E. Antigen tests for COVID-19. Biophys. Physicobiol. 2021, 18, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Magnard, C.; Valette, M.; Aymard, M.; Lina, B. Comparison of two nested PCR, cell culture, and antigen detection for the diagnosis of upper respiratory tract infections due to influenza viruses. J. Med. Virol. 1999, 59, 215–220. [Google Scholar] [CrossRef]
- Drexler, J.F.; Helmer, A.; Kirberg, H.; Reber, U.; Panning, M.; Müller, M.; Höfling, K.; Matz, B.; Drosten, C.; Eis-Hübinger, A.M. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg. Infect. Dis. 2009, 15, 1662. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Advice on the Use of Point-of-Care Immunodiagnostic Tests for COVID-19: Scientific Brief, 8 April 2020. Technical Report, World Health Organization. 2020. Available online: https://apps.who.int/iris/handle/10665/331713 (accessed on 1 March 2022).
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Zhang, Z. Mechanics of human voice production and control. J. Acoust. Soc. Am. 2016, 140, 2614–2635. [Google Scholar] [CrossRef]
- Gandhi, R.T.; Lynch, J.B.; del Rio, C. Mild or moderate Covid-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef]
- Matos, S.; Birring, S.; Pavord, I.; Evans, H. Detection of cough signals in continuous audio recordings using hidden Markov models. IEEE Trans. Biomed. Eng. 2006, 53, 1078–1083. [Google Scholar] [CrossRef]
- Shin, S.H.; Hashimoto, T.; Hatano, S. Automatic detection system for cough sounds as a symptom of abnormal health condition. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 486–493. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C. Diagnosing COVID-19: The disease and tools for detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Thompson, D.; Lei, Y. Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sens. Actuators Rep. 2020, 2, 100017. [Google Scholar] [CrossRef]
- Serrurier, A.; Neuschaefer-Rube, C.; Röhrig, R. Past and trends in cough sound acquisition, automatic detection and automatic classification: A comparative review. Sensors 2022, 22, 2896. [Google Scholar] [CrossRef]
- Schuller, B.W.; Schuller, D.M.; Qian, K.; Liu, J.; Zheng, H.; Li, X. Covid-19 and computer audition: An overview on what speech & sound analysis could contribute in the SARS-CoV-2 corona crisis. Front. Digit. Health 2021, 3. [Google Scholar] [CrossRef]
- Ghrabli, S.; Elgendi, M.; Menon, C. Challenges and opportunities of deep learning for cough-based COVID-19 diagnosis: A scoping review. Diagnostics 2022, 12, 2142. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1–9. [Google Scholar] [CrossRef]
- Orlandic, L.; Teijeiro, T.; Atienza, D. The COUGHVID crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Sci. Data 2021, 8, 1–10. [Google Scholar] [CrossRef]
- Brown, C.; Chauhan, J.; Grammenos, A.; Han, J.; Hasthanasombat, A.; Spathis, D.; Xia, T.; Cicuta, P.; Mascolo, C. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. arXiv 2021, arXiv:2006.05919. [Google Scholar]
- Sharma, N.; Krishnan, P.; Kumar, R.; Ramoji, S.; Chetupalli, S.R.; Nirmala, R.; Ghosh, P.K.; Ganapathy, S. Coswara—A database of breathing, cough, and voice sounds for COVID-19 diagnosis. In Proceedings of the Interspeech 2020, ISCA, Shanghai, China, 25–29 October 2020. [Google Scholar] [CrossRef]
- Virufy. Virufy—About us. Available online: https://virufy.org/en/about/#about-us (accessed on 30 March 2022).
- Pahar, M.; Klopper, M.; Warren, R.; Niesler, T. COVID-19 cough classification using machine learning and global smartphone recordings. Comput. Biol. Med. 2021, 135, 104572. [Google Scholar] [CrossRef] [PubMed]
- Cohen-McFarlane, M.; Goubran, R.; Knoefel, F. Novel coronavirus cough database: NoCoCoDa. IEEE Access 2020, 8, 154087–154094. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Shahin, I.; Alsabek, M.B. COVID-19 detection system using recurrent neural networks. In Proceedings of the 2020 International Conference On Communications, Computing, Cybersecurity, and Informatics (CCCI), Sharjah, United Arab Emirates, 3–5 November 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Haritaoglu, E.D.; Rasmussen, N.; Xiao, J.; Chaudhari, G.; Rajput, A.; Govindan, P.; Yamaura, M.; Gomezjurado, L.; Khanzada, A.; Pilanci, M.; et al. Using deep learning with large aggregated datasets for COVID-19 classification from cough. arXiv 2022, arXiv:2201.01669. [Google Scholar]
- Verma, N.K.; Salour, A. Intelligent Condition Based Monitoring: For Turbines, Compressors, and Other Rotating Machines; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Tawfik, M.; Nimbhore, S.; Al-Zidi, N.M.; Ahmed, Z.A.; Almadani, A.M. Multi-features extraction for automating COVID-19 detection from cough sound using deep neural networks. In Proceedings of the 2022 4th International Conference on Smart Systems and Inventive Technology (ICSSIT), Tirunelveli, India, 20–22 January 2022; pp. 944–950. [Google Scholar] [CrossRef]
- Vahedian-Azimi, A.; Keramatfar, A.; Asiaee, M.; Atashi, S.S.; Nourbakhsh, M. Do you have COVID-19? An artificial intelligence-based screening tool for COVID-19 using acoustic parameters. J. Acoust. Soc. Am. 2021, 150, 1945–1953. [Google Scholar] [CrossRef]
- Mouawad, P.; Dubnov, T.; Dubnov, S. Robust detection of COVID-19 in cough sounds. SN Comput. Sci. 2021, 2, 1–13. [Google Scholar] [CrossRef]
- Saha, S.; Bhadra, R.; Kar, S. An audio signal-based COVID-19 detection methodology using modified DenseNet121. In Proceedings of the 2021 IEEE 18th India Council International Conference (INDICON), Guwahati, India, 19–21 December 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Solak, F.Z. Identification of COVID-19 from cough Sounds using non-linear analysis and machine learning. Avrupa Bilim Teknol. Derg. 2021, 710–716. [Google Scholar] [CrossRef]
- Orlandic, L.; Teijeiro, T.; Atienza, D. Cough Segmentation Algorithm. Available online: https://c4science.ch/diffusion/10770/browse/master/src/ (accessed on 15 August 2022).
- Zhang, X.; Pettinati, M.; Jalali, A.; Rajput, K.S.; Selvaraj, N. Novel COVID-19 screening using cough recordings of a mobile patient monitoring system. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual Conference, 31 October–4 November 2021; pp. 2353–2357. [Google Scholar] [CrossRef]
- Tena, A.; Clarià, F.; Solsona, F. Automated detection of COVID-19 cough. Biomed. Signal Process. Control 2021, 71, 103175. [Google Scholar] [CrossRef]
- Kamble, M.R.; Gonzalez-Lopez, J.A.; Grau, T.; Espin, J.M.; Cascioli, L.; Huang, Y.; Gomez-Alanis, A.; Patino, J.; Font, R.; Peinado, A.M.; et al. PANACEA cough sound-based diagnosis of COVID-19 for the DiCOVA 2021 Challenge. arXiv 2021, arXiv:2106.04423. [Google Scholar]
- Feng, K.; He, F.; Steinmann, J.; Demirkiran, I. Deep-learning based approach to identify COVID-19. In Proceedings of the SoutheastCon 2021, Atlanta, GA, USA, 10–13 March 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Pahar, M.; Klopper, M.; Warren, R.; Niesler, T. COVID-19 detection in cough, breath and speech using deep transfer learning and bottleneck features. Comput. Biol. Med. 2021, 141, 105153. [Google Scholar] [CrossRef]
- Milani, M.M.; Ramashini, M.; Krishani, M. Speech signal analysis of COVID-19 patients via machine learning approach. In Proceedings of the 2021 International Conference on Decision Aid Sciences and Application (DASA), Sakheer, Bahrain, 7–8 December 2021; pp. 314–319. [Google Scholar] [CrossRef]
- Grant, D.; McLane, I.; West, J. Rapid and scalable COVID-19 screening using speech, breath, and cough recordings. In Proceedings of the 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), Athens, Greece, 27–30 July 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Nellore, B.T.; Sreeram, G.; Dhawan, K.; Reddy, P.B. Evaluating speech production-based acoustic features for COVID-19 classification using cough signals. In Proceedings of the 2021 IEEE 18th India Council International Conference (INDICON), Guwahati, India, 19–21 December 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Sangle, S.; Gaikwad, C. COVID-19 detection using spectral and statistical features of cough and breath sounds. In Proceedings of the 2021 International Conference on Decision Aid Sciences and Application (DASA), Sakheer, Bahrain, 7–8 December 2021; pp. 182–186. [Google Scholar] [CrossRef]
- Rao, S.; Narayanaswamy, V.; Esposito, M.; Thiagarajan, J.; Spanias, A. Deep learning with hyper-parameter tuning for COVID-19 cough detection. In Proceedings of the 2021 12th International Conference on Information, Intelligence, Systems & Applications (IISA), Chania Crete, Greece, 12–14 July 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Gupta, R.; Krishna, T.A.; Adeeb, M. Cough sound based COVID-19 detection with stacked ensemble model. In Proceedings of the 2022 4th International Conference on Smart Systems and Inventive Technology (ICSSIT), Tirunelveli, India, 20–22 January 2022; pp. 1391–1395. [Google Scholar] [CrossRef]
- AlHinai, N. Chapter 1—Introduction to biomedical signal processing and artificial intelligence. In Biomedical Signal Processing and Artificial Intelligence in Healthcare; Developments in Biomedical Engineering and Bioelectronics, Zgallai, W., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–28. [Google Scholar] [CrossRef]
- Khan, M.U.; Ali, S.; Habib, K.; Khan, H.; Tariq, F.; Bibi, S. A novel intelligent model for COVID-19 detection using cough auscultations and Hjorth descriptors. In Proceedings of the ICAME21, International Conference on Advances in Mechanical Engineering, Islamabad, Pakistan, 25 August 2021; pp. 1–10. [Google Scholar]
- Verde, L.; De Pietro, G.; Sannino, G. Artificial intelligence techniques for the non-invasive detection of COVID-19 through the analysis of voice signals. Arab. J. Sci. Eng. 2021, 1–11. [Google Scholar] [CrossRef]
- Meiniar, W.; Afrida, F.A.; Irmasari, A.; Mukti, A.; Astharini, D. Human voice filtering with band-stop filter design in MATLAB. In Proceedings of the 2017 International Conference on Broadband Communication, Wireless Sensors and Powering (BCWSP), Jakarta, Indonesia, 21–23 November 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Sanjeev, S.; Balne, C.C.S.; Reddy, T.J.; Reddy, G.P. Deep learning-based mixed data approach for COVID-19 detection. In Proceedings of the 2021 IEEE 18th India Council International Conference (INDICON), Guwahati, India, 19–21 December 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Erdoğan, Y.E.; Narin, A. COVID-19 detection with traditional and deep features on cough acoustic signals. Comput. Biol. Med. 2021, 136, 104765. [Google Scholar] [CrossRef]
- Khriji, L.; Ammari, A.; Messaoud, S.; Bouaafia, S.; Maraoui, A.; Machhout, M. COVID-19 recognition based on patient’s coughing and breathing patterns analysis: Deep learning approach. In Proceedings of the 2021 29th Conference of Open Innovations Association (FRUCT), Tampere, Finland, 12–14 May 2021; pp. 185–191. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, X.; Wang, W.; Huang, Z.; Zhang, P.; Yan, Y. Cough-based COVID-19 detection with multi-band long-short term memory and convolutional neural networks. In Proceedings of the 2nd International Symposium on Artificial Intelligence for Medicine Sciences, Beijing, China, 29–31 October 2021; pp. 209–215. [Google Scholar] [CrossRef]
- Islam, R.; Abdel-Raheem, E.; Tarique, M. A study of using cough sounds and deep neural networks for the early detection of COVID-19. Biomed. Eng. Adv. 2022, 100025. [Google Scholar] [CrossRef]
- Alkhodari, M.; Khandoker, A.H. Detection of COVID-19 in smartphone-based breathing recordings: A pre-screening deep learning tool. PLoS ONE 2022, 17, e0262448. [Google Scholar] [CrossRef]
- Fakhry, A.; Jiang, X.; Xiao, J.; Chaudhari, G.; Han, A.; Khanzada, A. Virufy: A multi-branch deep learning network for automated detection of COVID-19. arXiv 2021, arXiv:2103.01806. [Google Scholar]
- Son, M.J.; Lee, S.P. COVID-19 diagnosis from crowdsourced cough sound data. Appl. Sci. 2022, 12, 1795. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Chowdhury, N.K.; Kabir, M.A.; Rahman, M. An Ensemble-based Multi-Criteria Decision Making Method for COVID-19 Cough Classification. arXiv 2021, arXiv:2110.00508. [Google Scholar]
- Pancaldi, F.; Pezzuto, G.S.; Cassone, G.; Morelli, M.; Manfredi, A.; D’Arienzo, M.; Vacchi, C.; Savorani, F.; Vinci, G.; Barsotti, F.; et al. VECTOR: An algorithm for the detection of COVID-19 pneumonia from velcro-like lung sounds. Comput. Biol. Med. 2022, 105220. [Google Scholar] [CrossRef]
- Kamble, M.R.; Patino, J.; Zuluaga, M.A.; Todisco, M. Exploring auditory acoustic features for the diagnosis of the COVID-19. arXiv 2022, arXiv:2201.09110. [Google Scholar]
- Chaudhari, G.; Jiang, X.; Fakhry, A.; Han, A.; Xiao, J.; Shen, S.; Khanzada, A. Virufy: Global applicability of crowdsourced and clinical datasets for AI detection of COVID-19 from cough. arXiv 2020, arXiv:2011.13320. [Google Scholar]
- Rahman, D.A.; Lestari, D.P. COVID-19 classification using cough sounds. In Proceedings of the 2021 8th International Conference on Advanced Informatics: Concepts, Theory and Applications (ICAICTA), Bandung, Indonesia, 29–30 September 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Shkanov, B.; Zolotova, A.; Alexandrov, M.; Koshulko, O. Express diagnosis of COVID-19 on cough audiograms with machine learning algorithms from Scikit-learn library and GMDH Shell tool. In Proceedings of the 2021 IEEE 16th International Conference on Computer Sciences and Information Technologies (CSIT), Lviv, Ukraine, 22–25 September 2021; pp. 410–414. [Google Scholar] [CrossRef]
- Wang, J.; Chen, P.; Zhang, C.; Kang, Y. Corona virus disease 2019 respiratory cycle detection based on convolutional neural network. In Proceedings of the 2021 IEEE Biomedical Circuits and Systems Conference (BioCAS), Berlin, Germany, 7–9 October 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Irawati, M.E.; Zakaria, H. Classification model for COVID-19 detection through recording of cough using XGboost classifier algorithm. In Proceedings of the 2021 International Symposium on Electronics and Smart Devices (ISESD), Bandung, Indonesia, 29–30 June 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Melek Manshouri, N. Identifying COVID-19 by using spectral analysis of cough recordings: A distinctive classification study. Cogn. Neurodynamics 2021, 16, 239–253. [Google Scholar] [CrossRef]
- Melek, M. Diagnosis of COVID-19 and non-COVID-19 patients by classifying only a single cough sound. Neural Comput. Appl. 2021, 33, 17621–17632. [Google Scholar] [CrossRef]
- Nessiem, M.A.; Mohamed, M.M.; Coppock, H.; Gaskell, A.; Schuller, B.W. Detecting COVID-19 from breathing and coughing sounds using deep neural networks. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 183–188. [Google Scholar] [CrossRef]
- Banerjee, A.; Nilhani, A. A residual network based deep learning model for detection of COVID-19 from cough sounds. arXiv 2021, arXiv:2106.02348. [Google Scholar]
- Maor, E.; Tsur, N.; Barkai, G.; Meister, I.; Makmel, S.; Friedman, E.; Aronovich, D.; Mevorach, D.; Lerman, A.; Zimlichman, E.; et al. Noninvasive vocal biomarker is associated with severe acute respiratory syndrome coronavirus 2 infection. Mayo Clin. Proc. Innov. Qual. Outcomes 2021, 5, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Vrindavanam, J.; Srinath, R.; Shankar, H.H.; Nagesh, G. Machine learning based COVID-19 cough classification Models-A comparative analysis. In Proceedings of the 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 8–10 April 2021; pp. 420–426. [Google Scholar] [CrossRef]
- Han, J.; Brown, C.; Chauhan, J.; Grammenos, A.; Hasthanasombat, A.; Spathis, D.; Xia, T.; Cicuta, P.; Mascolo, C. Exploring automatic COVID-19 diagnosis via voice and symptoms from crowdsourced data. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 8328–8332. [Google Scholar] [CrossRef]
- Pal, A.; Sankarasubbu, M. Pay attention to the cough: Early diagnosis of COVID-19 using interpretable symptoms embeddings with cough sound signal processing. In Proceedings of the 36th Annual ACM Symposium on Applied Computing, Virtual Conference, 22–26 March 2021; pp. 620–628. [Google Scholar] [CrossRef]
- Lella, K.K.; Pja, A. Automatic COVID-19 disease diagnosis using 1D convolutional neural network and augmentation with human respiratory sound based on parameters: Cough, breath, and voice. AIMS Public Health 2021, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Bansal, V.; Pahwa, G.; Kannan, N. Cough classification for COVID-19 based on audio MFCC features using convolutional neural networks. In Proceedings of the 2020 IEEE International Conference on Computing, Power and Communication Technologies (GUCON), Greater Noida, India, 2–4 October 2020; pp. 604–608. [Google Scholar] [CrossRef]
- Lortie, C.L.; Thibeault, M.; Guitton, M.J.; Tremblay, P. Effects of age on the amplitude, frequency and perceived quality of voice. Age 2015, 37, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Salamon, J.; Bello, J.P. Deep convolutional neural networks and data augmentation for environmental sound classification. IEEE Signal Process. Lett. 2017, 24, 279–283. [Google Scholar] [CrossRef]
- Aquino, N.R.; Gutoski, M.; Hattori, L.T.; Lopes, H.S. The effect of data augmentation on the performance of Convolutional Neural Networks. Braz. Soc. Comput. Intell. 2017. [Google Scholar] [CrossRef]
- O’Shaughnessy, D. Speech Communication: Human and Machine; Wiley-IEEE Press: Hoboken, NJ, USA, 1999; ISBN 978-0-780-33449-6. [Google Scholar]
- Merhav, N.; Lee, C.H. On the asymptotic statistical behavior of empirical cepstral coefficients. IEEE Trans. Signal Process. 1993, 41, 1990–1993. [Google Scholar] [CrossRef]
- Logan, B. Mel frequency cepstral coefficients for music modeling. In Proceedings of the International Symposium on Music Information Retrieval, Plymouth, MA, USA, 23–25 October 2000. [Google Scholar]
- Korpáš, J.; Sadloňová, J.; Vrabec, M. Analysis of the cough sound: An overview. Pulm. Pharmacol. 1996, 9, 261–268. [Google Scholar] [CrossRef]
- Wei, Q.; Dunbrack, R.L., Jr. The role of balanced training and testing data sets for binary classifiers in bioinformatics. PLoS ONE 2013, 8, e67863. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).