Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention
Abstract
1. Introduction
1.1. Problem Statement
1.2. Related Works
1.2.1. Deep Learning Approach
1.2.2. Machine Learning Approach
1.3. Proposed Method
- -
- Backbone model including DenseNet201, InceptionResNetV2, ResNet50/152, NasNetLarge, NasNetMobile, and MobileNetV2/V3;
- -
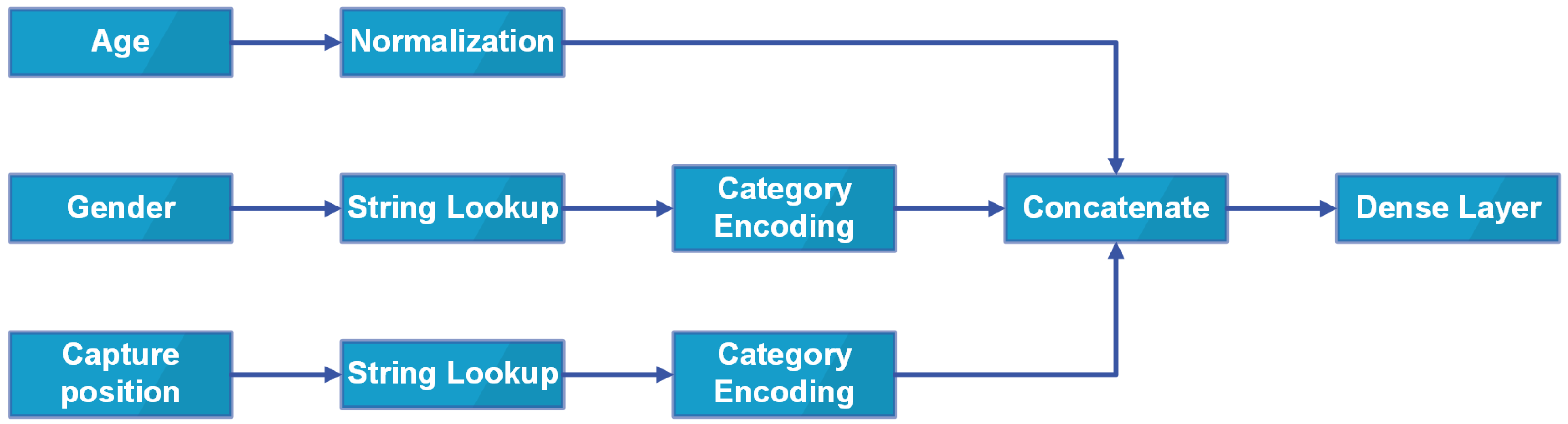
- Using metadata including age, gender, localization as another input of the model;
- -
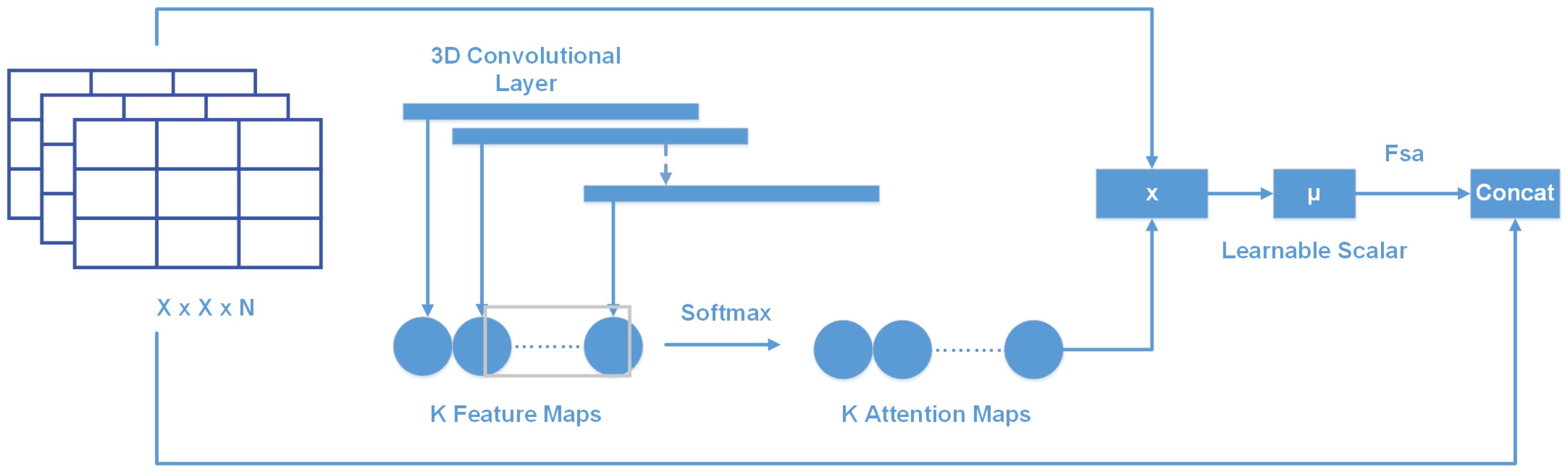
- Using Soft-Attention as a feature extractor of the model;
- -
- A new weight loss function.
2. Materials and Methods
2.1. Materials
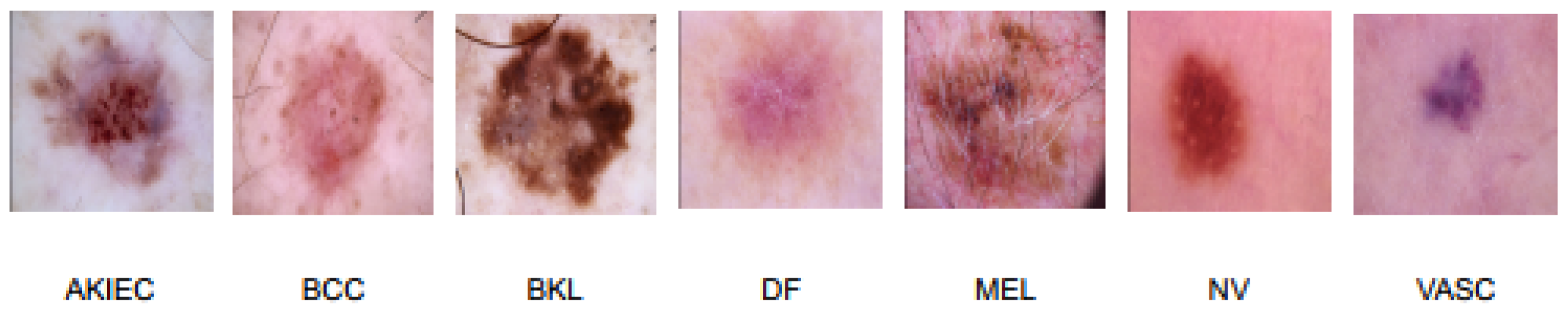
2.1.1. Image Data
2.1.2. Metadata
2.2. Methodology
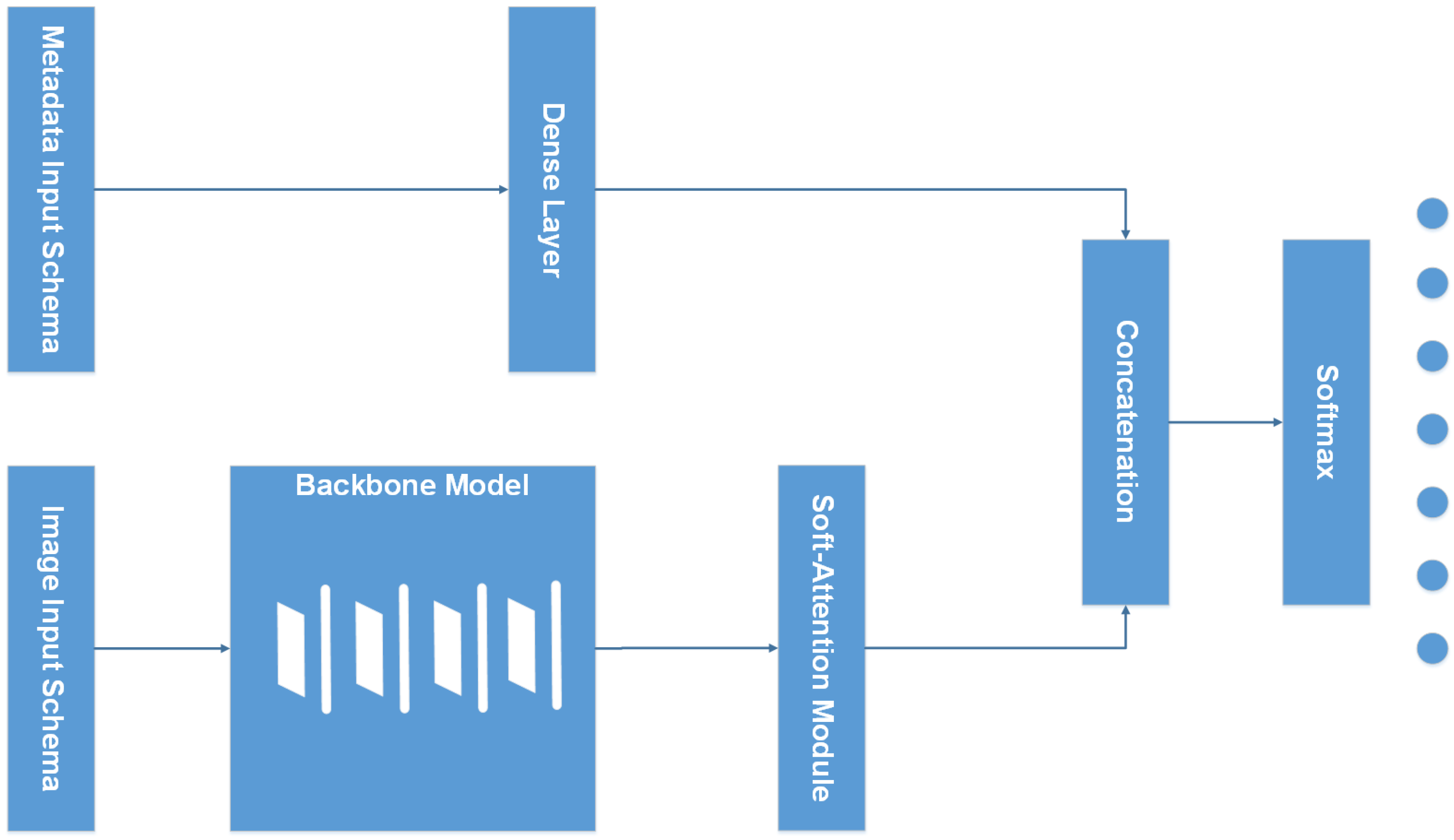
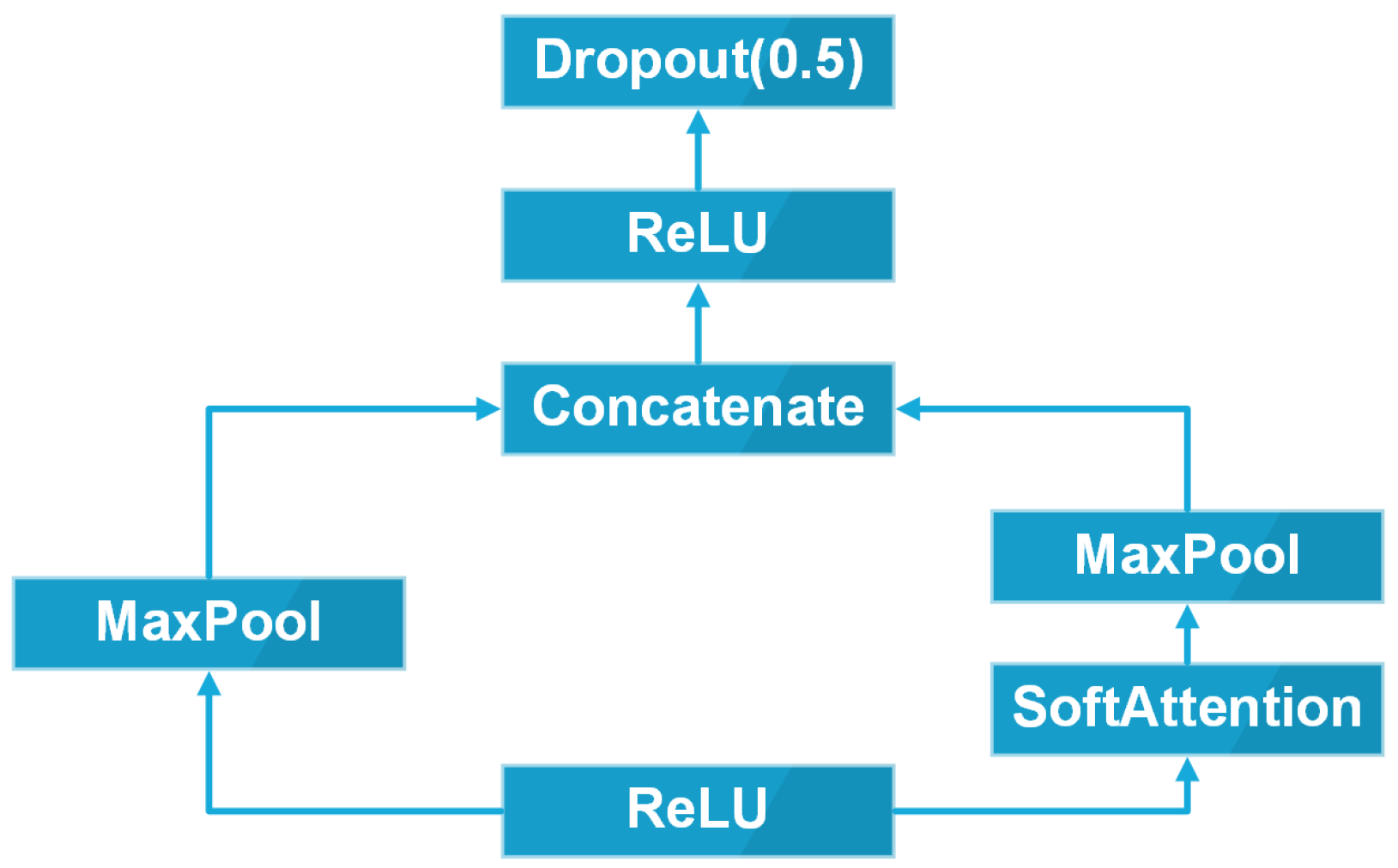
2.2.1. Overall Architecture
2.2.2. Input Schema
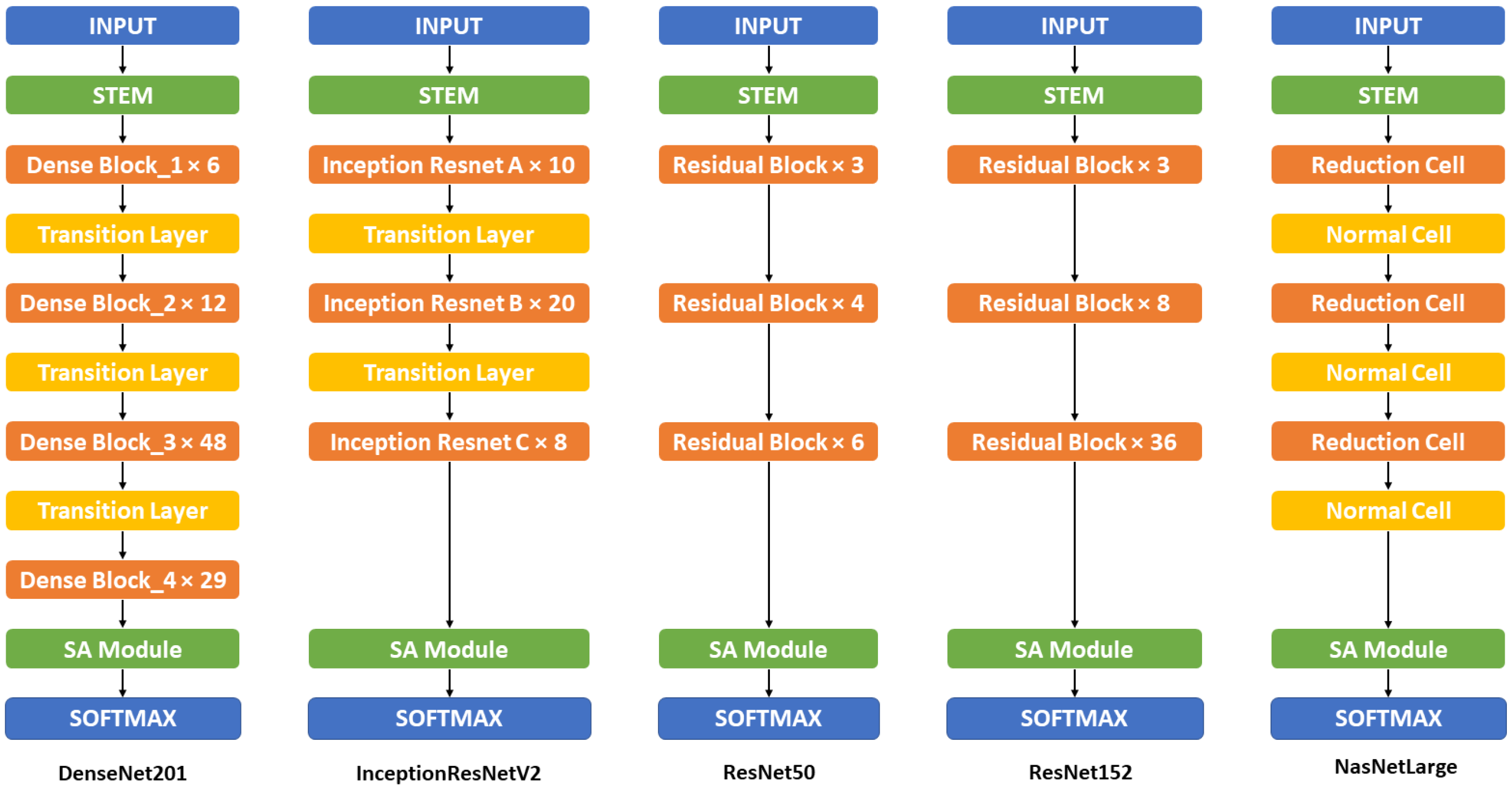
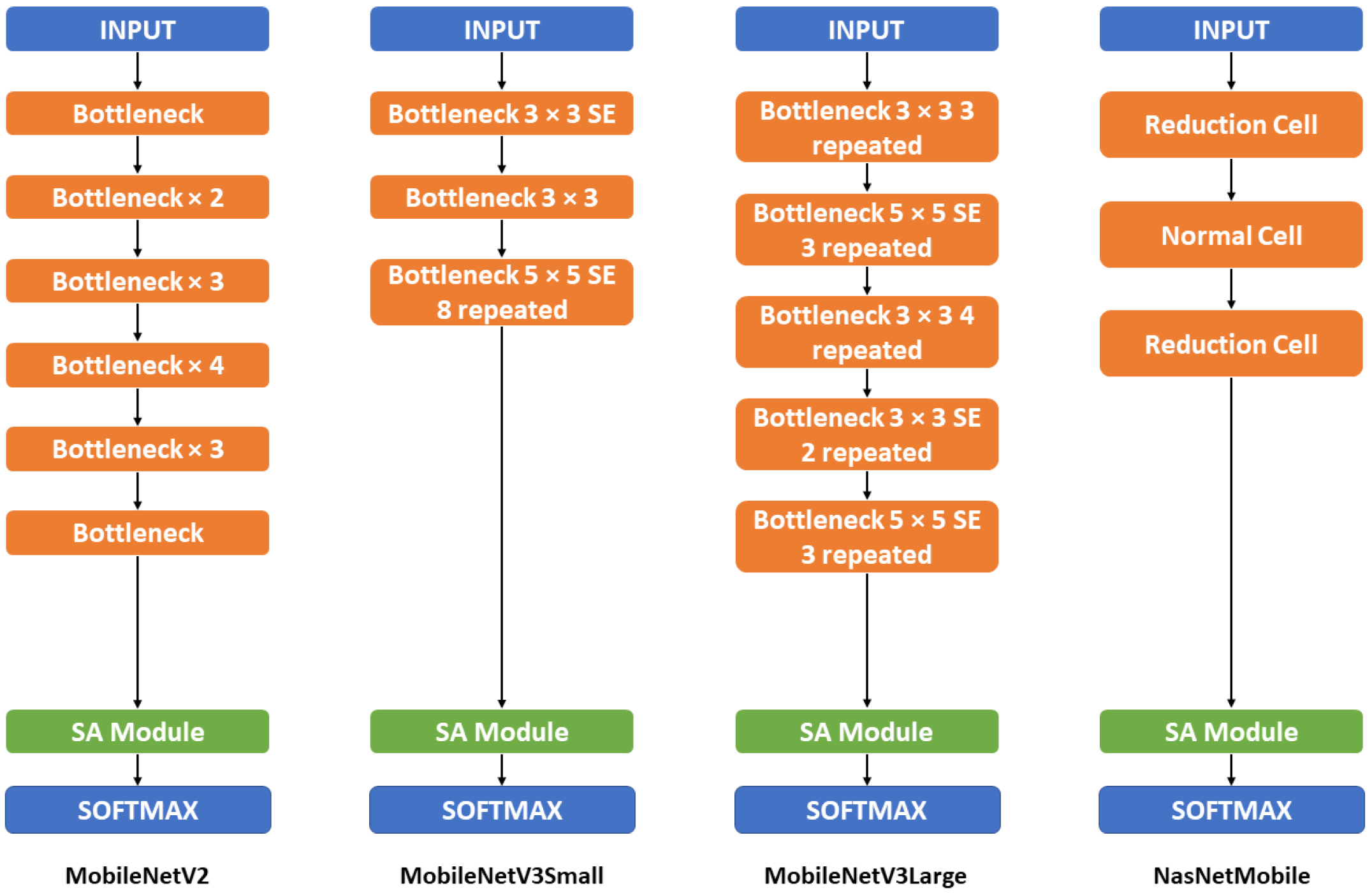
2.2.3. Backbone Model
2.2.4. Soft-Attention Module
2.2.5. Loss Function
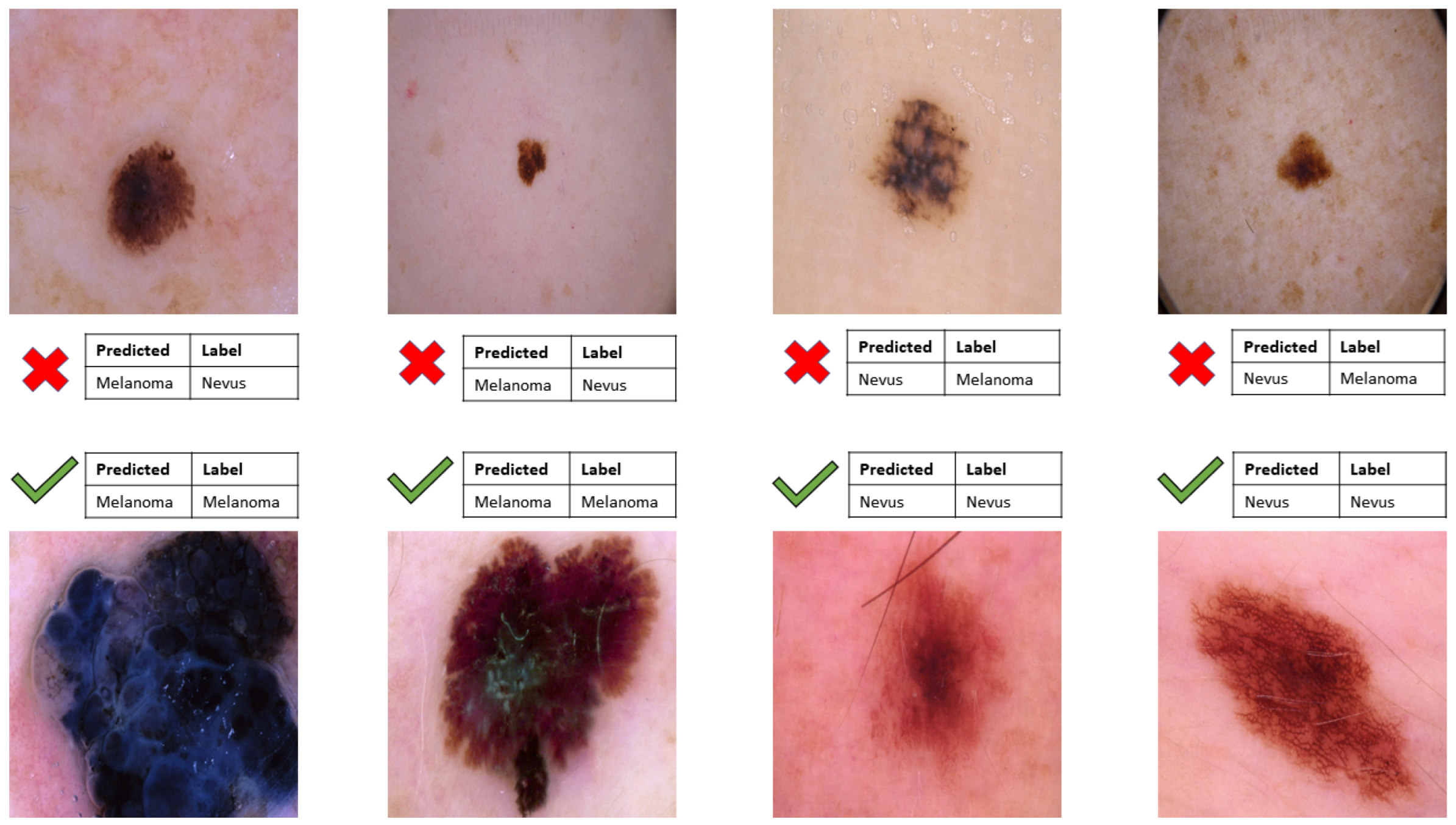
3. Results
3.1. Experimental Setup
3.1.1. Training
- -
- Rotation range: rotate the image in an angle range of 180.
- -
- Width and height shift range: Shift the image horizontally and vertically in a range of 0.1, respectively.
- -
- Zoom range: Zoom in or zoom out the image in a range of 0.1 to create new image.
- -
- Horizontal and vertical flipping: Flipping the image horizontally and vertically to create a new image.
3.1.2. Tools
3.1.3. Evaluation Metrics
3.2. Discussion
| Model | ACC (AD) | ACC (MD) |
|---|---|---|
| InceptionResNetV2 | 0.79 | 0.90 |
| DenseNet201 | 0.84 | 0.89 |
| ResNet50 | 0.76 | 0.70 |
| ResNet152 | 0.81 | 0.57 |
| NasNetLarge | 0.56 | 0.84 |
| MobileNetV2 | 0.83 | 0.81 |
| MobileNetV3Small | 0.83 | 0.78 |
| MobileNetV3Large | 0.85 | 0.86 |
| NasNetMobile | 0.84 | 0.86 |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAD | Computer-aided diagnosis |
| AI | Artificial Intelligence |
| AKIEC | Actinic keratoses and intraepithelial carcinoma or Bowen’s disease |
| BCC | Basal Cell Carcinoma |
| BKL | Benign Keratosis-like Lesions |
| DF | Dermatofibroma |
| MEL | Melanoma |
| NV | Melanocytic Nevi |
| VASC | Vascular Lesions |
| HISTO | Histopathology |
| FOLLOWUP | Follow-up examination |
| CONSENSUS | Expert Consensus |
| CONFOCAL | Confocal Microscopy |
| RGB | Red Green Blue |
| BGR | Blue Green Red |
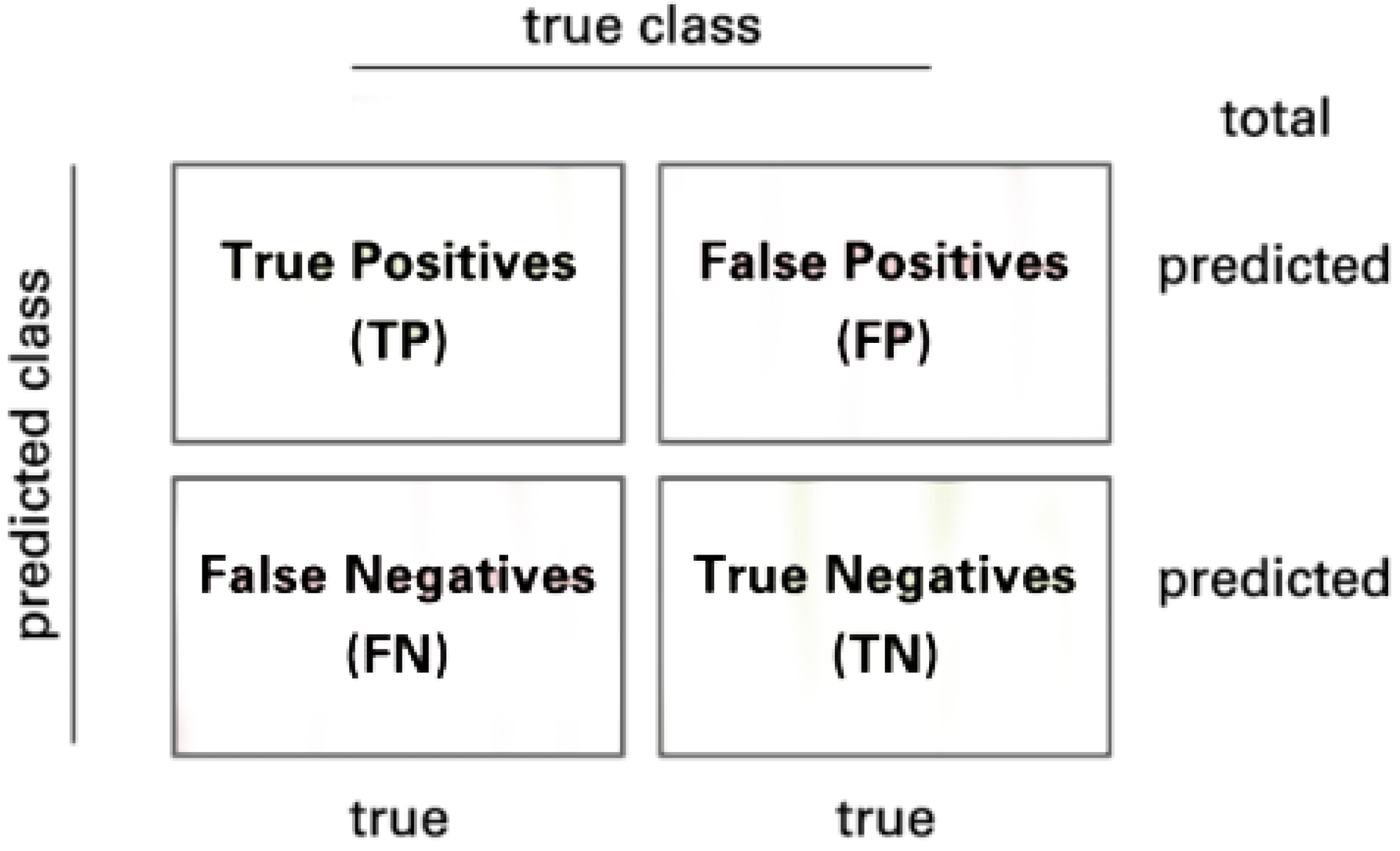
| TP | True Positives |
| FN | False Negatives |
| TN | True Negatives |
| FP | False Positives |
| Sens | Sensitivity |
| Spec | Specificity |
| AUC | Area Under the Curve |
| ROC | Receiver Operating Curve |
Appendix A. Detailed Model Structure
| DenseNet-201 | DenseNet-201 + SA | Inception-ResNetV2 | Inception-ResNetV2 + SA | ResNet-50 | ResNet-50 + SA | ResNet-152 | ResNet-152 + SA | NasNet-Large | NasNet-Large + SA |
|---|---|---|---|---|---|---|---|---|---|
| Conv2D | Conv2D | STEM | STEM | Conv2D | Conv2D | Conv2D | Conv2D | Conv2D | Conv2D |
| Pooling | Pooling | Pooling | Pooling | Pooling | Pooling | Pooling | Pooling | ||
| DenseBlock × 6 | DenseBlock × 6 | Inception ResNet A × 10 | Inception ResNet A × 10 | Residual Block × 3 | Residual Block × 3 | Residual Block × 3 | Residual Block × 3 | Reduction Cell × 2 | Reduction Cell × 2 |
| Conv2D | Conv2D | Reduction A | Reduction A | Normal Cell × N | Normal Cell × N | ||||
| Average pool | Average pool | ||||||||
| DenseBlock × 12 | DenseBlock × 12 | Inception ResNet B × 20 | Inception ResNet B × 20 | Residual Block × 4 | Residual Block × 4 | Residual Block × 8 | Residual Block × 8 | Reduction Cell | Reduction Cell |
| Conv2D | Conv2D | Reduction B | Reduction B | Normal Cell × N | Normal Cell × N | ||||
| Average pool | Average pool | ||||||||
| DenseBlock × 48 | DenseBlock × 12 | Inception ResNet C × 5 | Inception ResNet C × 5 | Residual Block × 6 | Residual Block × 6 | Residual Block × 36 | Residual Block × 36 | Reduction Cell | Reduction Cell |
| Conv2D | Conv2D | Normal Cell × N | Normal Cell × N-2 | ||||||
| Average pool | Average pool | ||||||||
| DenseBlock × 29 | DenseBlock × 29 | Residual Block × 3 | Residual Block × 3 | ||||||
| DenseBlock × 3 | SA Module | SA Module | SA Module | SA Module | SA Module | ||||
| GAP | Average pool | GAP | GAP | ||||||
| FC 1000D | Dropout (0.8) | FC 1000D | FC 1000D | ||||||
| SoftMax | SoftMax | SoftMax | SoftMax | SoftMax | SoftMax | SoftMax | SoftMax | SoftMax | SoftMax |
Appendix B. Detailed Mobile-based Model Structure
| MobileNetV2 | MobileNetV2 + SA | MobileNetV3 Small | MobileNetV3 Small + SA | MobileNetV3 Large | MobileNetV3 Large + SA | NasNet Mobile | NasNetMobile + SA |
|---|---|---|---|---|---|---|---|
| Conv2D | Conv2D | Conv2D | Conv2D | Conv2D | Conv2D | Normal Cell | Normal Cell |
| bottleneck | bottleneck | bottleneck SE | bottleneck SE | bottleneck 3 repeated | bottleneck 3 repeated | Reduction Cell | Reduction Cell |
| bottleneck 2 repeated | bottleneck 2 repeated | bottleneck | bottleneck | bottleneck SE 3 repeated | bottleneck SE 3 repeated | Normal Cell | Normal Cell |
| bottleneck 3 repeated | bottleneck 3 repeated | bottleneck SE 8 repeated | bottleneck SE 8 repeated | bottleneck 4 repeated | bottleneck 4 repeated | Reduction Cell | Reduction Cell |
| bottleneck 4 repeated | bottleneck 4 repeated | bottleneck SE 2 repeated | bottleneck SE 2 repeated | Normal Cell | |||
| bottleneck 3 repeated | bottleneck 3 repeated | bottleneck SE 3 repeated | bottleneck SE 3 repeated | ||||
| bottleneck 3 repeated | bottleneck | ||||||
| bottleneck | |||||||
| Conv2D | Conv2D SE | Conv2D SE | Conv2D | Conv2D | |||
| AP | Pool | Pool | Pool | Pool | |||
| Conv2D | SA Module | Conv2D 2 repeated | SA Module | Conv2D 2 repeated | SA Module | SA Module | |
| Softmax | Softmax | Softmax | Softmax | Softmax | Softmax | Softmax | Softmax |
Appendix C. Detailed Model Performance
Appendix C.1. F1-Score Model Performance
| Model | akiec | bcc | bkl | df | mel | nv | vasc | Mean |
|---|---|---|---|---|---|---|---|---|
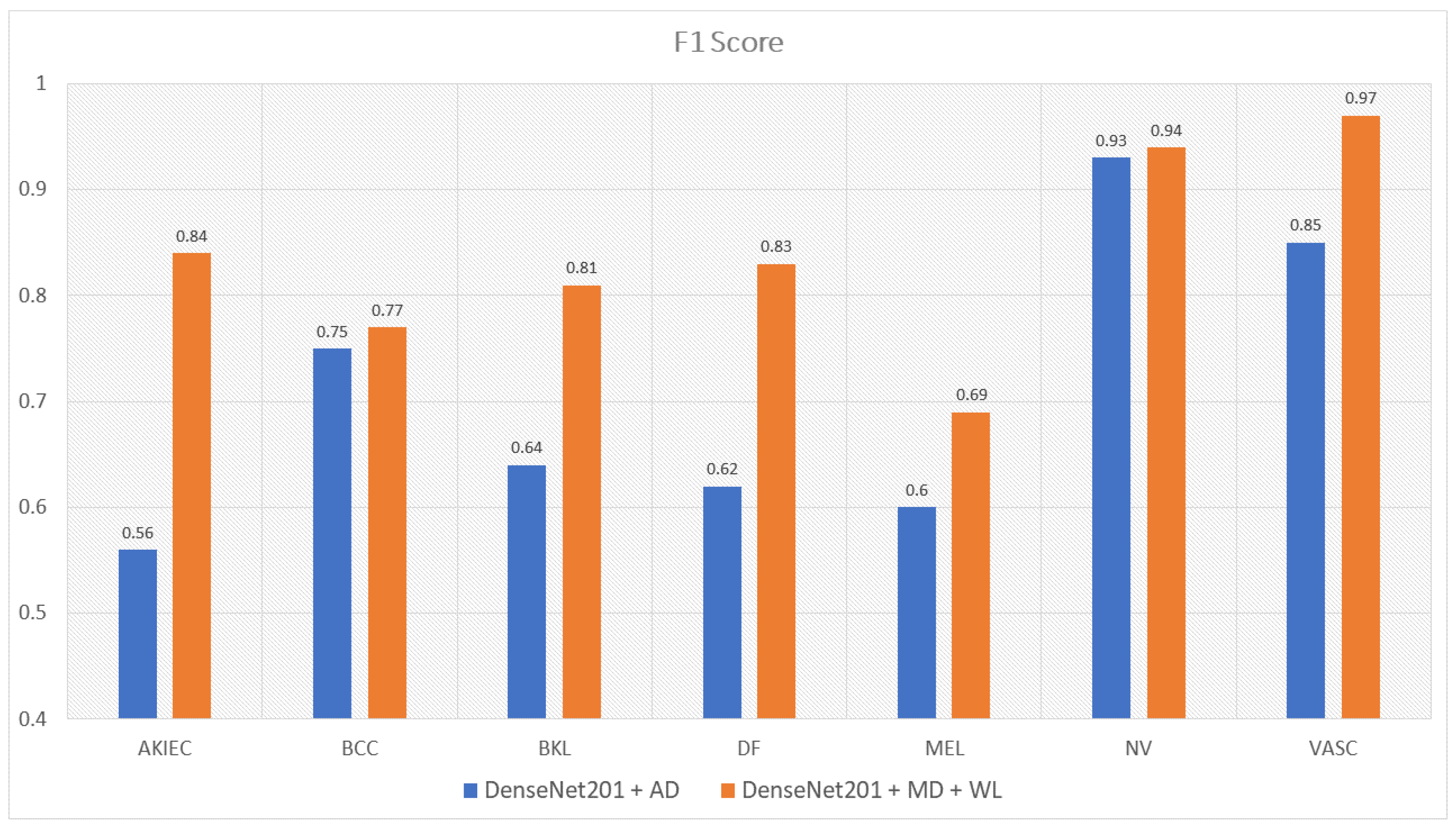
| DenseNet201 with Augmented Data | 0.56 | 0.75 | 0.64 | 0.62 | 0.60 | 0.93 | 0.85 | 0.70 |
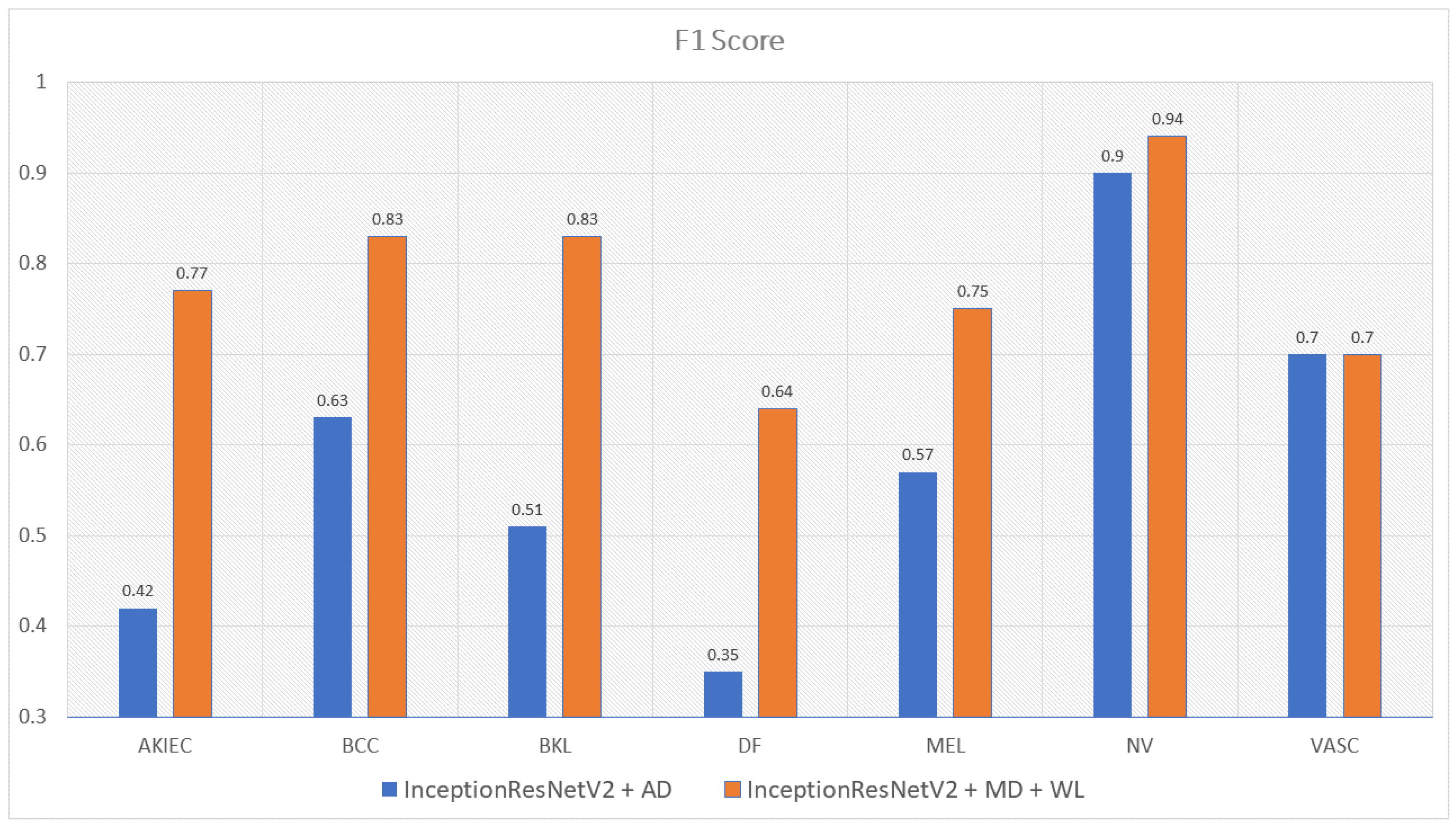
| InceptionResNetV2 with Augmented Data | 0.42 | 0.63 | 0.51 | 0.35 | 0.57 | 0.9 | 0.7 | 0.58 |
| Resnet50 with Augmented Data | 0.39 | 0.59 | 0.42 | 0.6 | 0.42 | 0.88 | 0.79 | 0.58 |
| VGG16 with Augmented Data | 0.35 | 0.62 | 0.42 | 0.32 | 0.47 | 0.89 | 0.77 | 0.54 |
| DenseNet201 with Metadata and WeightLoss | 0.84 | 0.77 | 0.81 | 0.83 | 0.69 | 0.94 | 0.97 | 0.83 |
| InceptionResNetV2 with Metadata and WeightLoss | 0.77 | 0.83 | 0.83 | 0.64 | 0.75 | 0.94 | 0.7 | 0.81 |
| Resnet50 with Metadata and WeightLoss | 0.49 | 0.59 | 0.55 | 0.36 | 0.45 | 0.83 | 0.8 | 0.58 |
| Resnet152 with Metadata and WeightLoss | 0.42 | 0.38 | 0.41 | 0.15 | 0.4 | 0.75 | 0.75 | 0.46 |
| NasNetLarge with Metadata and WeightLoss | 0.79 | 0.79 | 0.8 | 0.74 | 0.65 | 0.92 | 0.92 | 0.80 |
| MobileNetV2 with Metadata and WeightLoss | 0.68 | 0.79 | 0.66 | 0.78 | 0.54 | 0.9 | 0.9 | 0.75 |
| MobileNetV3Large with Metadata and WeightLoss | 0.72 | 0.76 | 0.75 | 0.92 | 0.58 | 0.92 | 0.92 | 0.79 |
| MobileNetV3Small with Metadata and WeightLoss | 0.6 | 0.72 | 0.61 | 0.75 | 0.47 | 0.89 | 0.89 | 0.70 |
| NasNetMobile with Metadata and WeightLoss | 0.76 | 0.74 | 0.78 | 0.73 | 0.63 | 0.93 | 0.93 | 0.78 |
Appendix C.2. Recall Model Performance
| Model | akiec | bcc | bkl | df | mel | nv | vasc | Mean |
|---|---|---|---|---|---|---|---|---|
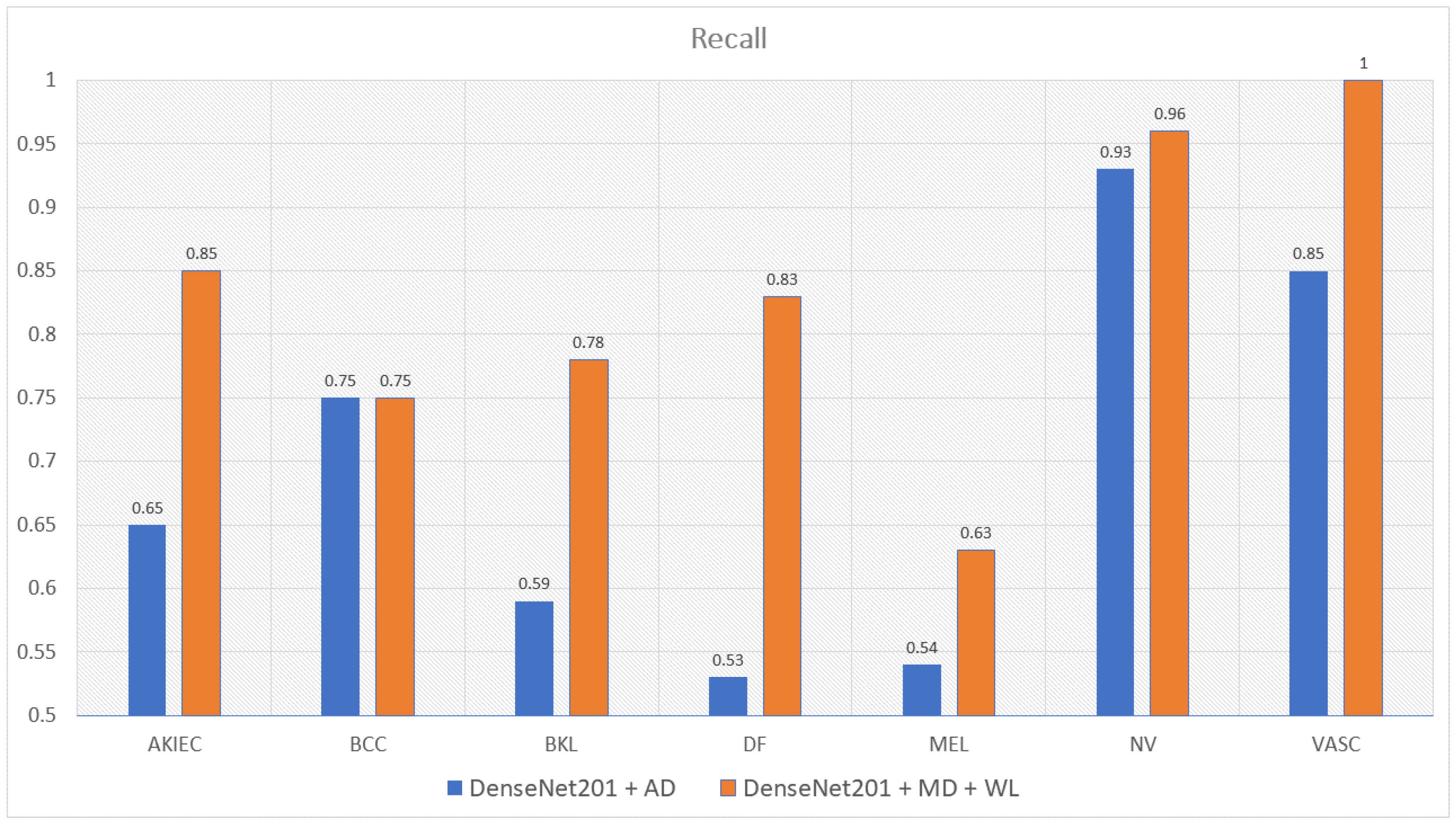
| DenseNet201 with Augmented Data | 0.65 | 0.75 | 0.59 | 0.53 | 0.54 | 0.93 | 0.85 | 0.69 |
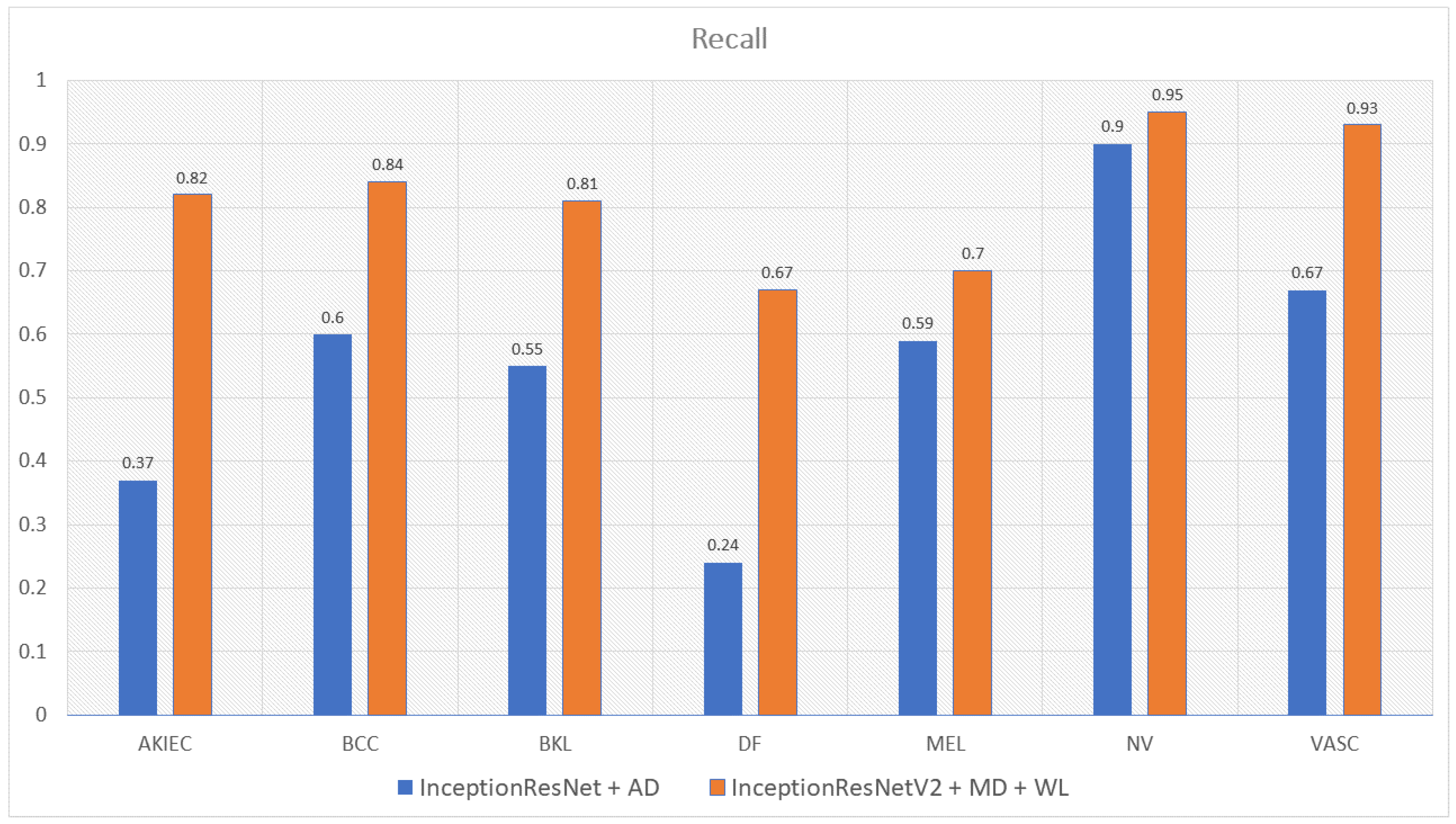
| InceptionResNetV2 with Augmented Data | 0.37 | 0.60 | 0.55 | 0.24 | 0.59 | 0.9 | 0.67 | 0.56 |
| Resnet50 with Augmented Data | 0.33 | 0.56 | 0.38 | 0.53 | 0.40 | 0.92 | 0.81 | 0.56 |
| VGG16 with Augmented Data | 0.31 | 0.66 | 0.37 | 0.24 | 0.40 | 0.94 | 0.71 | 0.51 |
| DenseNet201 with Metadata and WeightLoss | 0.85 | 0.75 | 0.78 | 0.83 | 0.63 | 0.96 | 1 | 0.82 |
| InceptionResNetV2 with Metadata and WeightLoss | 0.82 | 0.84 | 0.81 | 0.67 | 0.7 | 0.95 | 0.93 | 0.81 |
| Resnet50 with Metadata and WeightLoss | 0.67 | 0.63 | 0.54 | 0.83 | 0.63 | 0.74 | 0.86 | 0.70 |
| Resnet152 with Metadata and WeightLoss | 0.51 | 0.49 | 0.35 | 0.76 | 0.47 | 0.63 | 0.48 | 0.52 |
| NasNetLarge with Metadata and WeightLoss | 0.73 | 0.71 | 0.83 | 0.92 | 0.59 | 0.9 | 0.93 | 0.81 |
| MobileNetV2 with Metadata and WeightLoss | 0.7 | 0.86 | 0.72 | 0.75 | 0.58 | 0.86 | 1 | 0.78 |
| MobileNetV3Large with Metadata and WeightLoss | 0.72 | 0.76 | 0.75 | 0.92 | 0.58 | 0.92 | 0.92 | 0.80 |
| MobileNetV3Small with Metadata and WeightLoss | 0.76 | 0.84 | 0.68 | 1 | 0.52 | 0.82 | 0.93 | 0.79 |
| NasNetMobile with Metadata and WeightLoss | 0.82 | 0.73 | 0.83 | 0.92 | 0.53 | 0.93 | 0.93 | 0.81 |
Appendix C.3. Detailed Mobile Model Performance
| Model | [8] | [9] Small | [9] Large | [13] Mobile |
|---|---|---|---|---|
| Accuracy (avg) | 0.81 | 0.78 | 0.86 | 0.86 |
| Balanced Accuracy (avg) | 0.86 | 0.87 | 0.87 | 0.88 |
| Precision (avg) | 0.71 | 0.63 | 0.75 | 0.73 |
| F1-score (avg) | 0.75 | 0.70 | 0.79 | 0.78 |
| Sensitivity (avg) | 0.78 | 0.79 | 0.80 | 0.81 |
| Specificity (avg) | 0.95 | 0.95 | 0.95 | 0.96 |
| AUC (avg) | 0.96 | 0.95 | 0.96 | 0.97 |
References
- Datta, S.K.; Shaikh, M.A.; Srihari, S.N.; Gao, M. Soft-Attention Improves Skin Cancer Classification Performance. In Interpretability of Machine Intelligence in Medical Image Computing, and Topological Data Analysis and Its Applications for Medical Data; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial Intelligence-Based Image Classification for Diagnosis of Skin Cancer: Challenges and Opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef]
- Poduval, P.; Loya, H.; Sethi, A. Functional Space Variational Inference for Uncertainty Estimation in Computer Aided Diagnosis. arXiv 2020, arXiv:2005.11797. [Google Scholar]
- Gao, H.; Zhuang, L.; Kilian, Q. Weinberger: Densely Connected Convolutional Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the AAAI Conference, New Orleans, LO, USA, 2–7 February 2018. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the IEEE Conference on Computer Vision andPattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Garg, R.; Maheshwari, S.; Shukla, A. Decision Support System for Detection and Classification of Skin Cancer using CNN. In Innovations in Computational Intelligence and Computer Vision; Springer: Singapore, 2019. [Google Scholar]
- Rezvantalab, A.; Safigholi, H.; Karimijeshni, S. Dermatologist Level Dermoscopy Skin Cancer Classification Using Different Deep Learning Convolutional Neural Networks Algorithms. arXiv 2021, arXiv:1810.10348. [Google Scholar]
- Nadipineni, H. Method to Classify Skin Lesions using Dermoscopic images. arXiv 2020, arXiv:2008.09418. [Google Scholar]
- Yao, P.; Shen, S.; Xu, M.; Liu, P.; Zhang, F.; Xing, J.; Shao, P.; Kaffenberger, B.; Xu, R.X. Single Model Deep Learning on Imbalanced Small Datasets for Skin Lesion Classification. IEEE Trans. Med. Imaging 2022, 41, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Young, K.; Booth, G.; Simpson, B.; Dutton, R.; Shrapnel, S. Dermatologist Level Dermoscopy Deep neural network or dermatologist? Nature 2021, 542, 115–118. [Google Scholar]
- Xing, X.; Hou, Y.; Li, H.; Yuan, Y.; Li, H.; Meng, M.Q.H. Categorical Relation-Preserving Contrastive Knowledge Distillation for Medical Image Classification. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Mahbod, A.; Tsch, L.P.; Langs, G.; Ecker, R.; Ellinger, I. The Effects of Skin Lesion Segmentation on the Performance of Dermatoscopic Image Classification. Comput. Methods Programs Biomed. 2020, 197, 105725. [Google Scholar] [CrossRef]
- Lee, Y.C.; Jung, S.H.; Won, H.H. WonDerM: Skin Lesion Classification with Fine-tuned Neural Networks. arXiv 2019, arXiv:1808.03426. [Google Scholar]
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin Lesion Classification Using Ensembles of Multi-Resolution EfficientNets with Meta Data. MethodsX 2020, 7, 100864. [Google Scholar] [CrossRef]
- Alberti, M.; Botros, A.; Schutz, N.; Ingold, R.; Liwicki, M.; Seuret, M. Trainable Spectrally Initializable Matrix Transformations in Convolutional Neural Networks. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021. [Google Scholar]
- Abayomi-Alli, O.O.; Damasevicius, R.; Misra, S.; Maskeliunas, R.; Abayomi-Alli, A. Malignant skin melanoma detection using image augmentation by oversamplingin nonlinear lower-dimensional embedding manifold. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2600–2614. [Google Scholar] [CrossRef]
- Nawaz, M.; Nazir, T.; Masood, M.; Ali, F.; Khan, M.A.; Tariq, U.; Sahar, N. Robertas Damaševicius Melanoma segmentation: A framework of improved DenseNet77 and UNET convolutional neural network. Int. J. Imaging Syst. Technol. 2022. [Google Scholar] [CrossRef]
- Kadry, S.; Taniar, D.; Damaševičius, R.; Rajinikanth, V.; Lawal, I.A. Extraction of abnormal skin lesion from dermoscopy image using VGG-SegNet. In Proceedings of the 2021 Seventh International conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25–27 March 2021. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2016, arXiv:1409.1556. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- DeVries, T.; Taylor, G.W. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Li, X.; Lu, Y.; Desrosiers, C.; Liu, X. Out-of-Distribution Detection for Skin Lesion Images with Deep Isolation Forest. In International Workshop on Machine Learning in Medical Imaging; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Tsch, L.P.; Rosendahl, C.; Kittler, H. The HAM10000 data set, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 1–9. [Google Scholar]
- Fekri-Ershad, S.; Saberi, M.; Tajeripour, F. An innovative skin detection approach using color based image retrieval technique. arXiv 2012, arXiv:1207.1551. [Google Scholar] [CrossRef]
- Fred, A. Agarap Deep Learning using Rectified Linear Units (ReLU). arXiv 2019, arXiv:1803.08375. [Google Scholar]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, Attend and Tell: Neural Image Caption Generation with Visual Attention. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015; 2015. [Google Scholar]
- Shaikh, M.A.; Duan, T.; Chauhan, M.; Srihari, S.N. Attention based writer independent verification. In Proceedings of the 2020 17th International Conference on Frontiers in Handwriting Recognition, Dortmund, Germany, 8–10 September 2020. [Google Scholar]
- Tomita, N.; Abdollahi, B.; Wei, J.; Ren, B.; Suriawinata, A.; Hassanpour, S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw. 2020, 2, e1914645. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Wookey, S. The Real-World-Weight Cross-Entropy Loss Function: Modeling the Costs of Mislabeling. IEEE Access 2020, 8, 4806–4813. [Google Scholar] [CrossRef]
- King, G.; Zeng, L. Logistic Regression in Rare Events Data. Political Anal. 2001, 9, 137–163. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
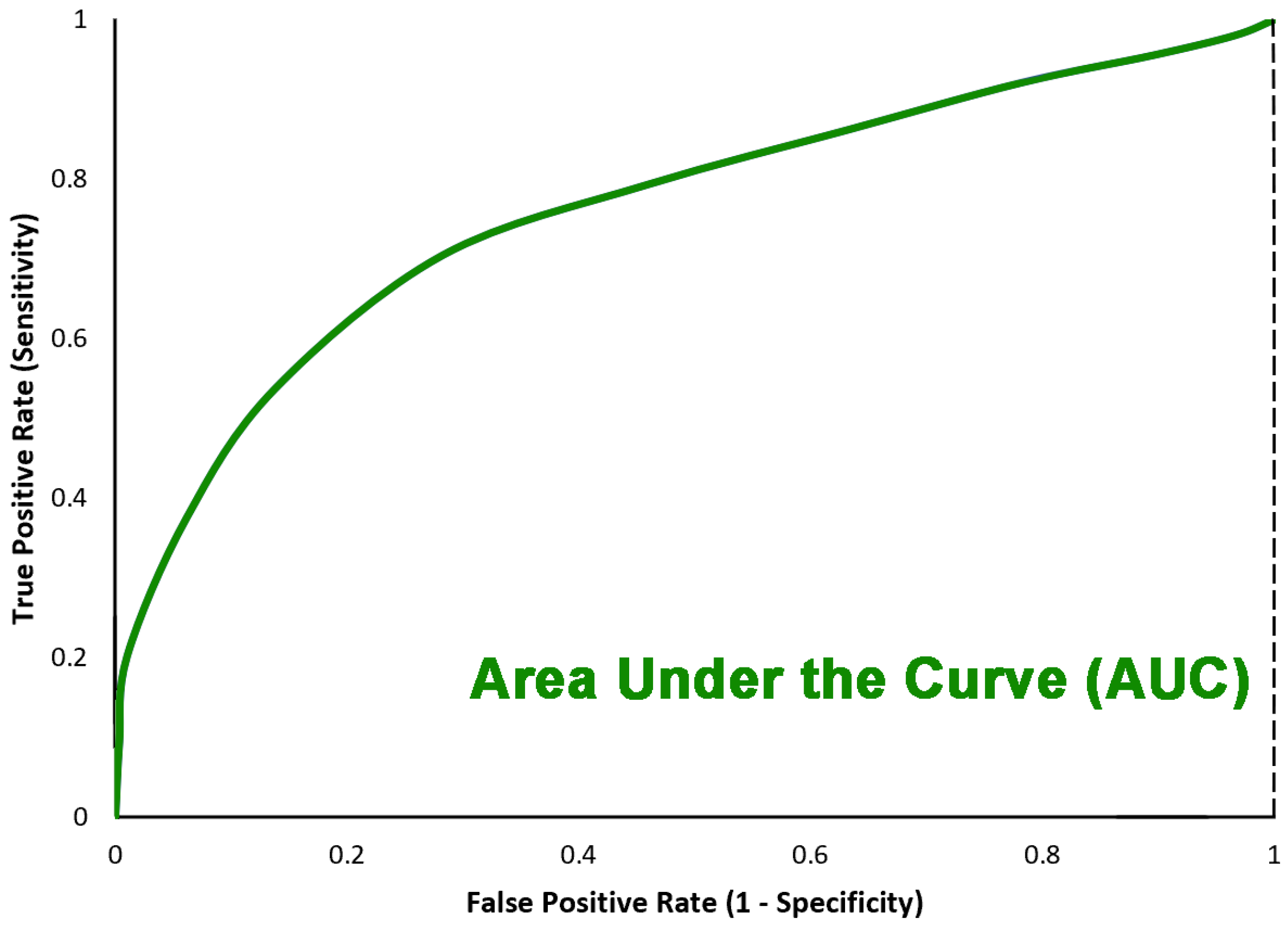
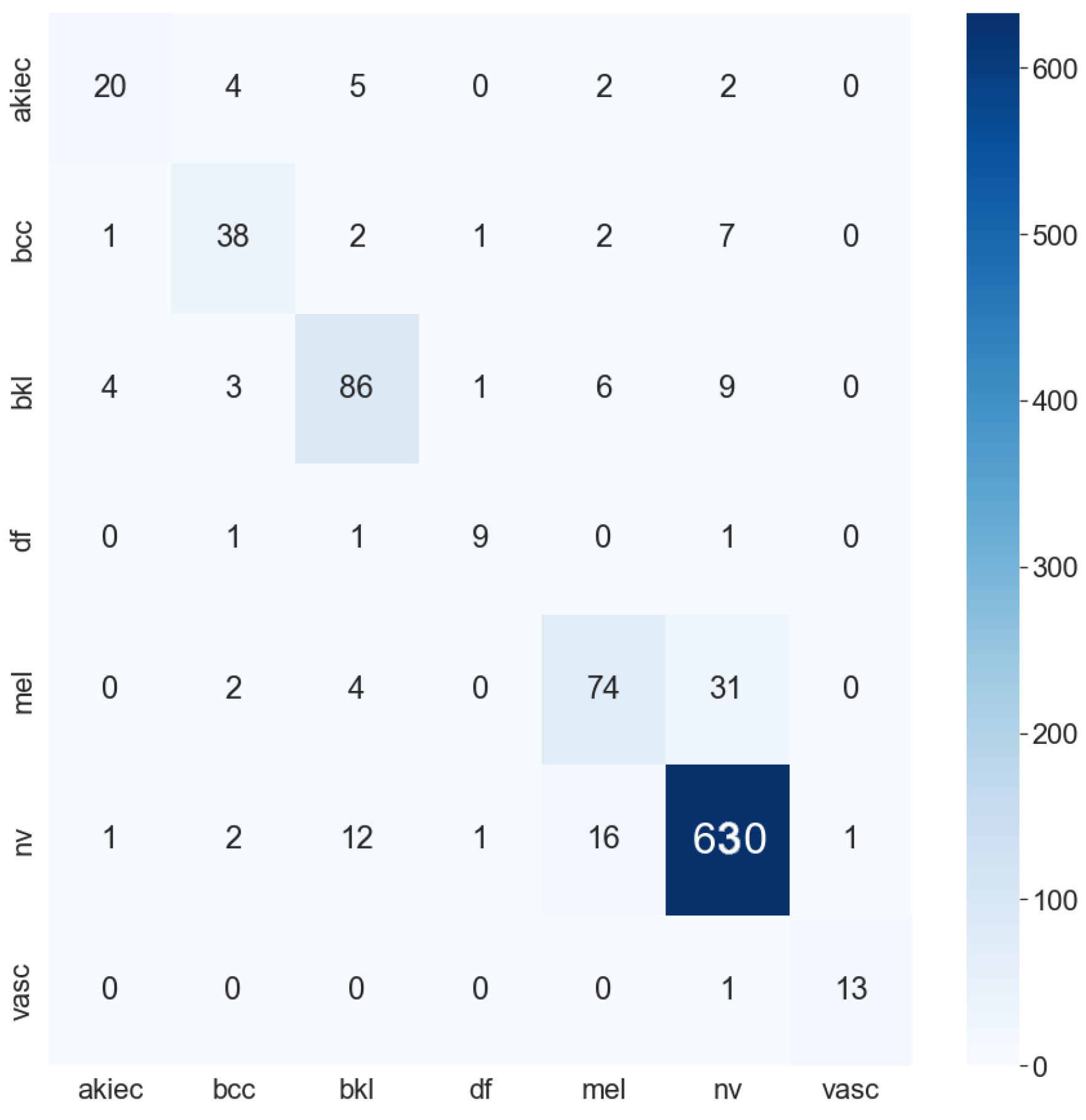
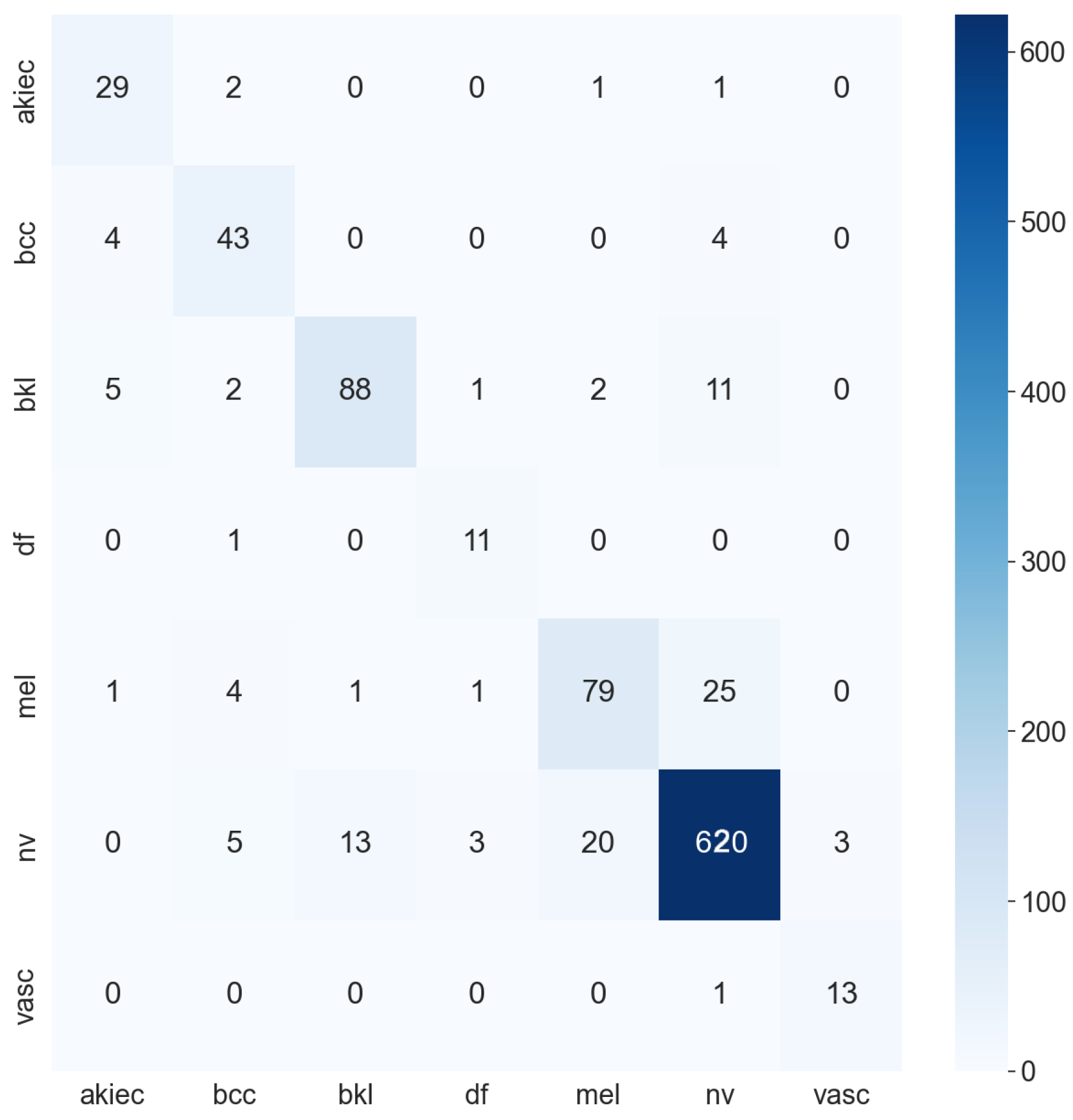
| Work | Deep Learning | Machine Learning | Data Augmentation | Feature Extractor | Data Set | Result |
|---|---|---|---|---|---|---|
| [1] | Classify | x | HAM10000 | 0.93 (ACC) | ||
| [14] | Classify | Classify | x | x | HAM10000 | 0.9 (ACC) |
| [15] | Classify | Classify | x | HAM10000, PH2 | ||
| [16] | Classify | x | HAM10000 | 0.88 (ACC) | ||
| [17] | Classify | x | HAM10000 | 0.86 (ACC) | ||
| [18] | Classify | x | x | HAM10000, BCN-20000, MSK | 0.85 (ACC) | |
| [19] | Classify | x | HAM10000 | 0.85 (ACC) | ||
| [20] | Classify | x | HAM10000 | 0.92 (AUC) | ||
| [21] | Classify | x | HAM10000 | 0.92 (AUC) | ||
| [22] | Classify | x | HAM10000 | 0.74 (recall) | ||
| [23] | Classify | x | x | HAM10000 | ||
| [24] | Classify | x | HAM10000 | 0.92 (ACC) | ||
| [25] | Seg | HAM10000 | 0.99 (ACC) | |||
| [26] | Seg | HAM10000 | 0.97 (ACC) |
| Class | AKIEC | BCC | BKL | DF | MEL | NV | VASC | Total |
|---|---|---|---|---|---|---|---|---|
| No. Sample | 327 | 514 | 1099 | 115 | 1113 | 6705 | 142 | 10,015 |
| ID | Age | Gender | Local |
|---|---|---|---|
| ISIC-00001 | 15 | Male | back |
| ISIC-00002 | 85 | Female | elbow |
| Model | Size (MB) | No. Trainable Parameters | Depth |
|---|---|---|---|
| Resnet50 | 98 | 25,583,592 | 107 |
| Resnet152 | 232 | 60,268,520 | 311 |
| DenseNet201 | 80 | 20,013,928 | 402 |
| InceptionResNetV2 | 215 | 55,813,192 | 449 |
| MobileNetV2 | 14 | 3,504,872 | 105 |
| MobileNetV3Small | Unknown | 2,542,856 | 88 |
| MobileNetV3Large | Unknown | 5,483,032 | 118 |
| NasnetMobile | 23 | 5,289,978 | 308 |
| NasnetLarge | 343 | 88,753,150 | 533 |
| Model | MobileNetV3Large | DenseNet201 | InceptionResnetV2 |
|---|---|---|---|
| No. Trainable Parameters | 5,490,039 | 17,382,935 | 47,599,671 |
| Depth | 118 | 402 | 449 |
| Accuracy | 0.86 | 0.89 | 0.90 |
| Training Time (seconds/epoch) | 116 | 1000 | 3500 |
| Infer Time (seconds) | 0.13 | 1.16 | 4.08 |
| Model | AUC (AD) | AUC (MD) |
|---|---|---|
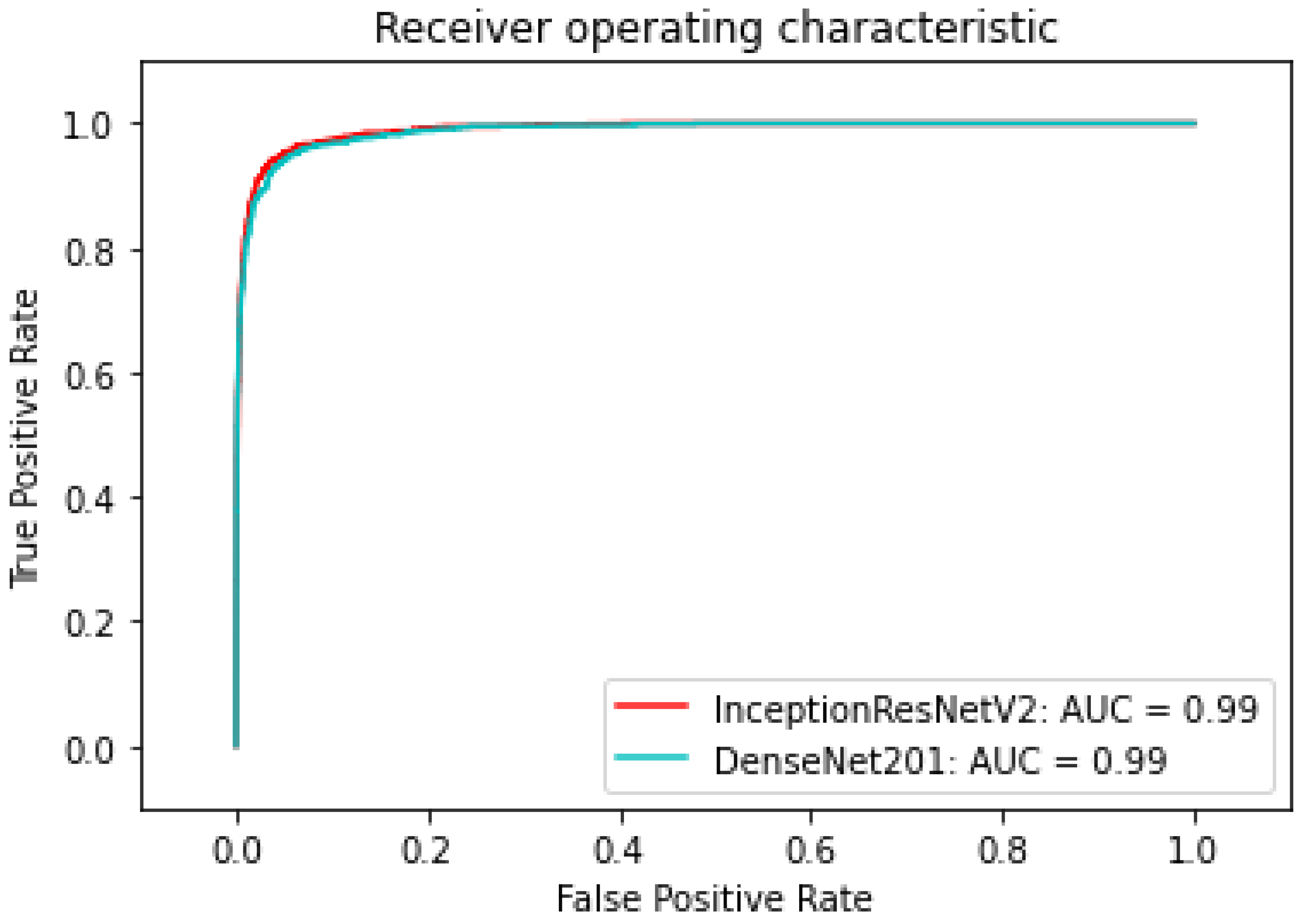
| InceptionResNetV2 | 0.971 | 0.99 |
| DenseNet201 | 0.93 | 0.99 |
| ResNet50 | 0.95 | 0.93 |
| ResNet152 | 0.97 | 0.87 |
| NasNetLarge | 0.74 | 0.96 |
| MobileNetV2 | 0.95 | 0.97 |
| MobileNetV3Small | 0.67 | 0.96 |
| MobileNetV3Large | 0.96 | 0.97 |
| NasNetMobile | 0.96 | 0.97 |
| Model | No Weight | Original Loss Accuracy | New Loss Accuracy |
|---|---|---|---|
| InceptionResNetV2 | 0.74 | 0.79 | 0.90 |
| DenseNet201 | 0.81 | 0.84 | 0.89 |
| MobileNetV3Large | 0.79 | 0.80 | 0.86 |
| Approach | Accuracy | Precision | F1-score | Recall | AUC |
|---|---|---|---|---|---|
| InceptionResNetV2 [1] | 0.93 | 0.89 | 0.75 | 0.71 | 0.97 |
| [14] | - | 0.88 | 0.77 | 0.74 | - |
| [16] | 0.88 | - | - | - | - |
| [17] | 0.86 | - | - | - | - |
| GradCam and Kernel SHAP [18] | 0.88 | - | - | - | - |
| Student and Teacher [19] | 0.85 | 0.76 | 0.76 | - | - |
| Proposed Method | 0.9 | 0.86 | 0.86 | 0.81 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.D.; Bui, N.D.; Do, H.K. Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention. Sensors 2022, 22, 7530. https://doi.org/10.3390/s22197530
Nguyen VD, Bui ND, Do HK. Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention. Sensors. 2022; 22(19):7530. https://doi.org/10.3390/s22197530
Chicago/Turabian StyleNguyen, Viet Dung, Ngoc Dung Bui, and Hoang Khoi Do. 2022. "Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention" Sensors 22, no. 19: 7530. https://doi.org/10.3390/s22197530
APA StyleNguyen, V. D., Bui, N. D., & Do, H. K. (2022). Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention. Sensors, 22(19), 7530. https://doi.org/10.3390/s22197530





