Hollow-Core Fiber-Based Biosensor: A Platform for Lab-in-Fiber Optical Biosensors for DNA Detection
Abstract
:1. Introduction
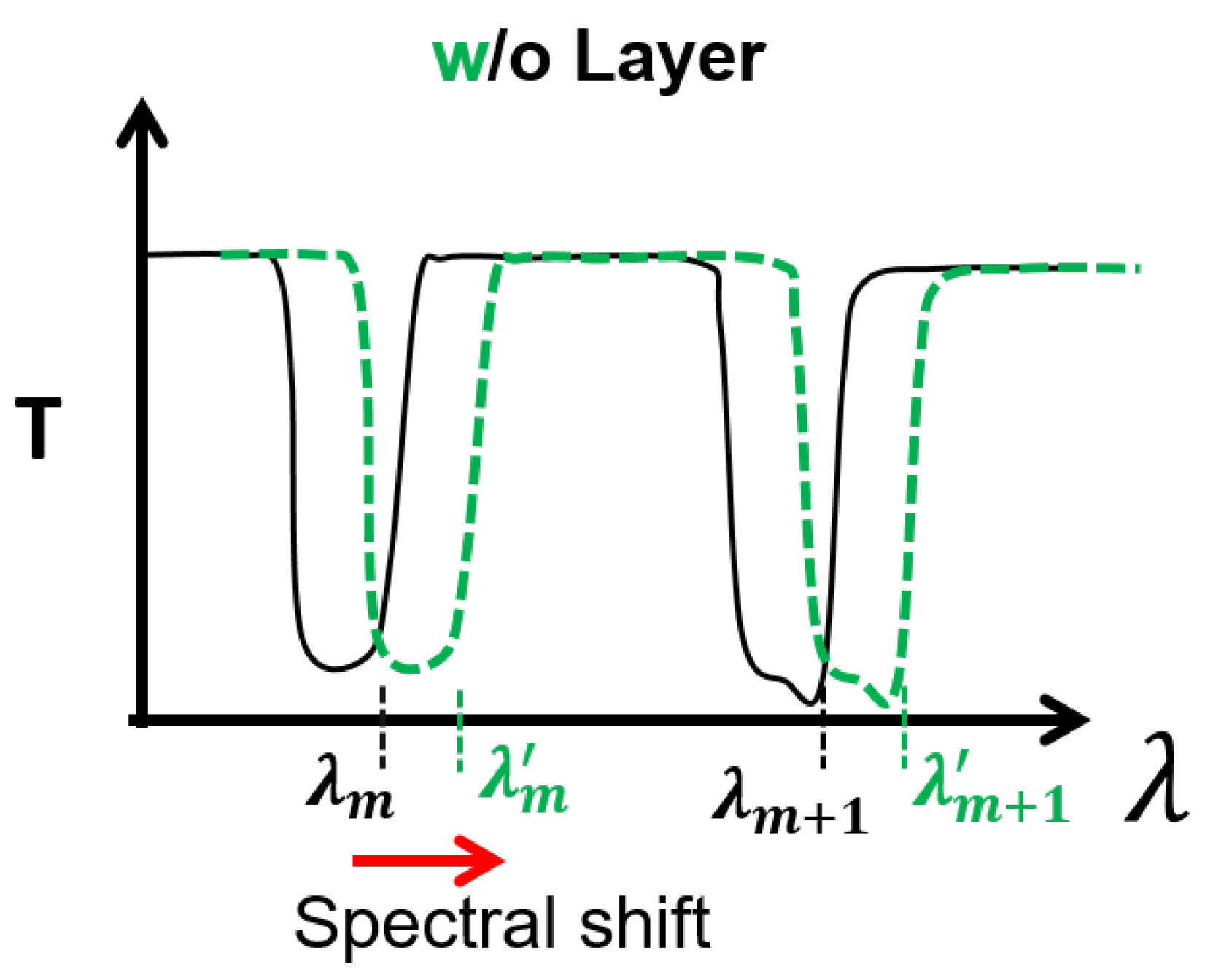
2. Hollow-Core Fiber-Based Biosensor
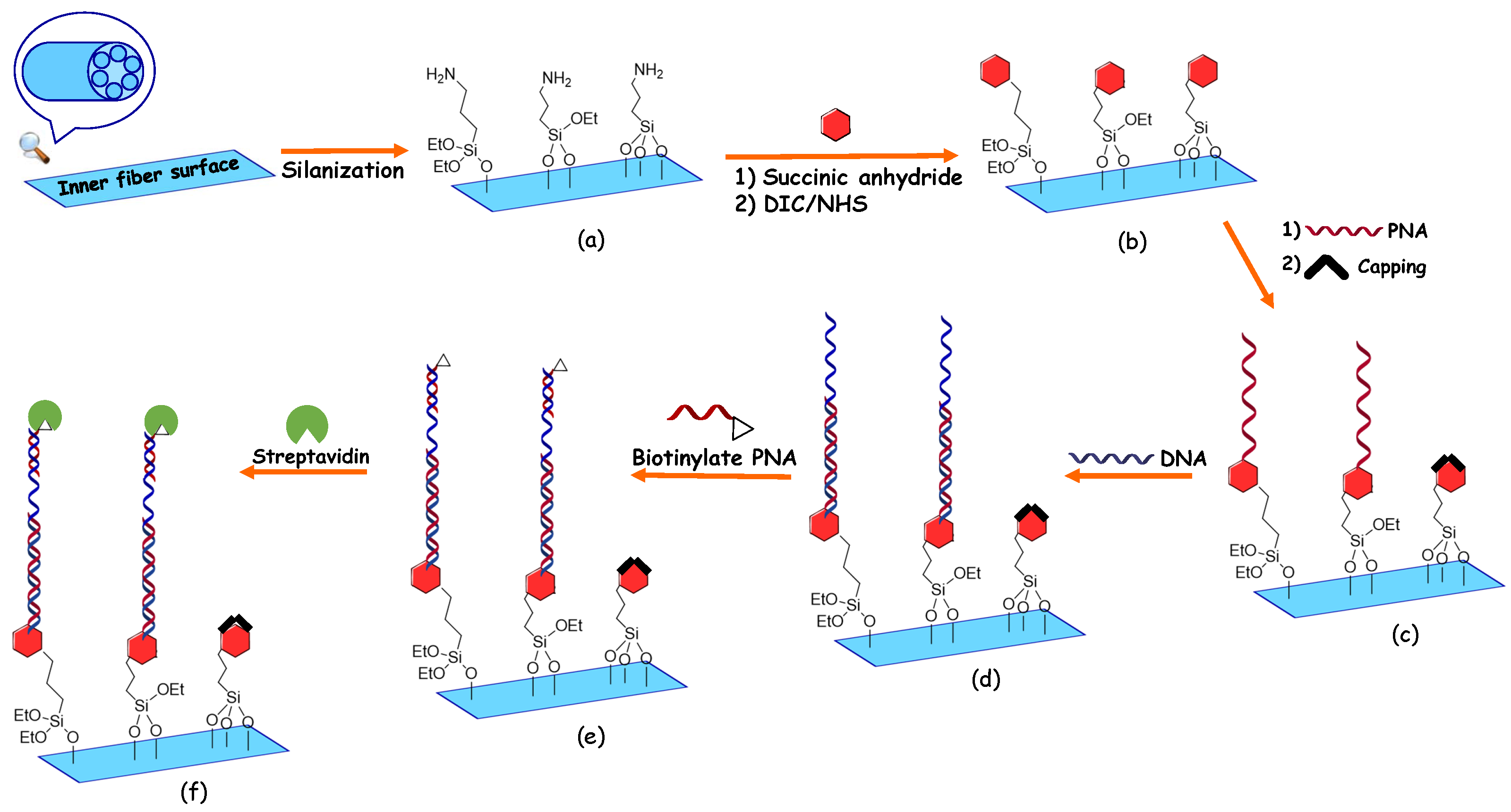
3. Chemical Functionalization Protocol
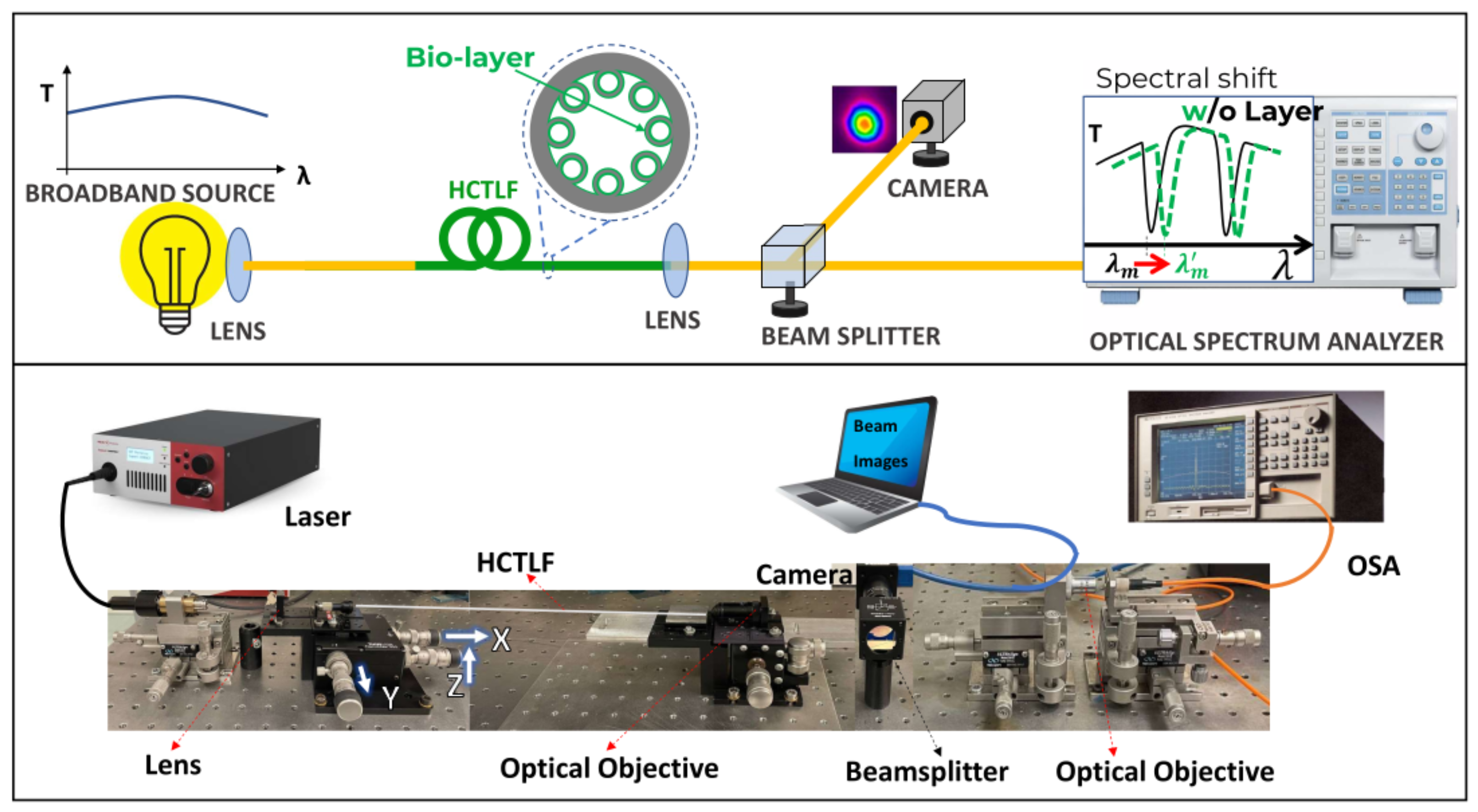
4. Optical Setup
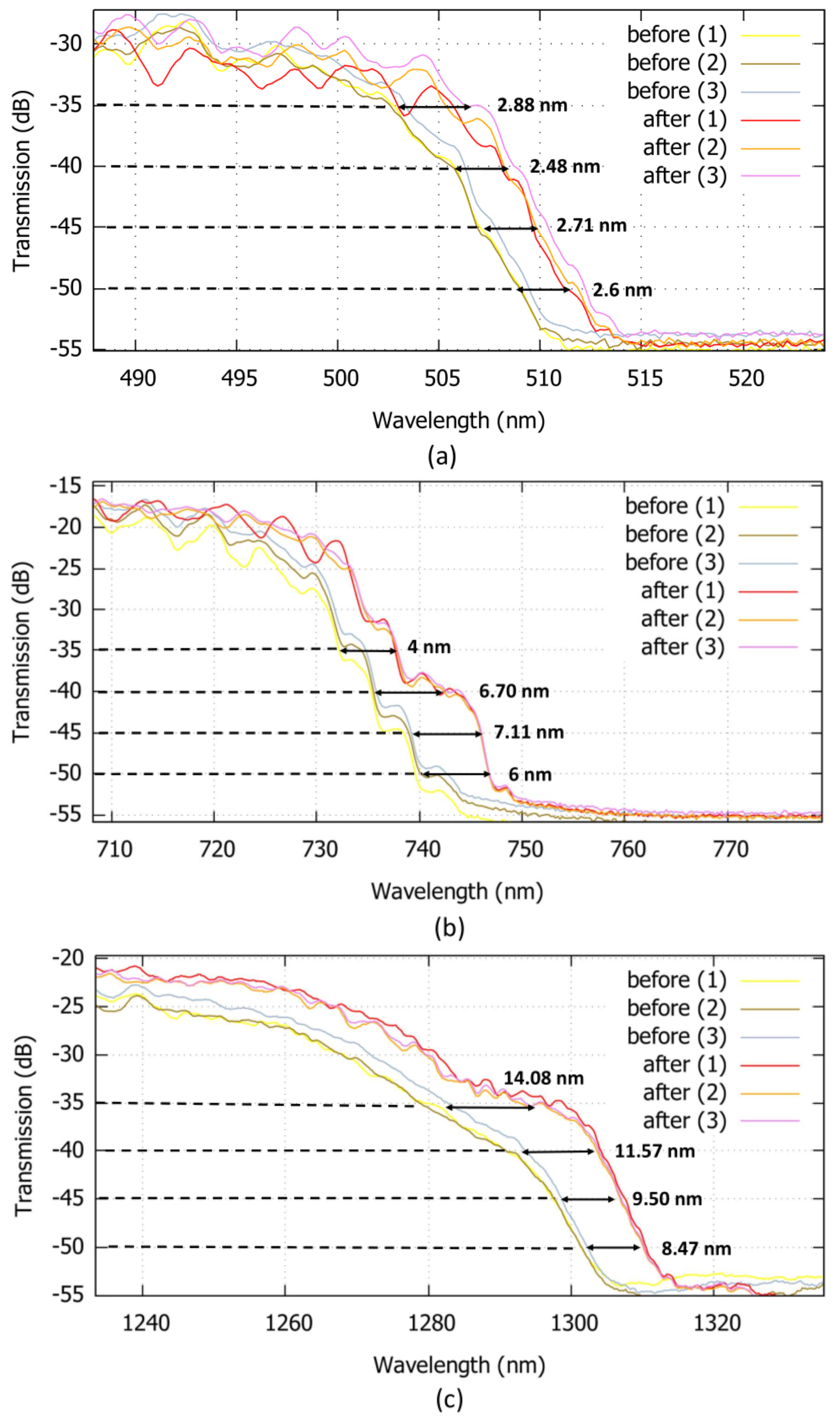
5. Results and Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haes, A.J.; Chang, L.; Klein, W.L.; Van Duyne, R.P. Detection of a biomarker for Alzheimer’s disease from synthetic and clinical samples using a nanoscale optical biosensor. J. Am. Chem. Soc. 2005, 127, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-free optical biosensors for food and biological sensor applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Kumar, H.; Rani, R. Development of biosensors for the detection of biological warfare agents: Its issues and challenges. Sci. Prog. 2013, 96, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Giardi, M.T.; Scognamiglio, V.; Rea, G.; Rodio, G.; Antonacci, A.; Lambreva, M.; Pezzotti, G.; Johanningmeier, U. Optical biosensors for environmental monitoring based on computational and biotechnological tools for engineering the photosynthetic D1 protein of Chlamydomonas reinhardtii. Biosens. Bioelectron. 2009, 25, 294–300. [Google Scholar] [CrossRef]
- Andrews, D.L. Photonics: Biomedical Photonics, Spectroscopy, and Microscopy; John Wiley & Sons: Hoboken, NJ, USA, 2015; Volume 4. [Google Scholar]
- Newman, J.D.; Turner, A.P. Biosensors: Principles and practice. Essays Biochem. 1992, 27, 147–159. [Google Scholar]
- Pohanka, M.; Skládal, P. Electrochemical biosensors—Principles and applications. J. Appl. Biomed. 2008, 6, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Zafar, M.S.; Dastgeer, G.; Kalam, A.; Al-Sehemi, A.G.; Imran, M.; Kim, Y.H.; Chae, H. Precise and Prompt Analyte Detection via Ordered Orientation of Receptor in WSe2-Based Field Effect Transistor. Nanomaterials 2022, 12, 1305. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Morales-Narváez, E.; Merkoçi, A. Graphene oxide as an optical biosensing platform: A progress report. Adv. Mater. 2019, 31, 1805043. [Google Scholar] [CrossRef] [Green Version]
- Chamorro-Posada, P. Asymmetric concentric microring resonator label-free biosensors. In Proceedings of the 10th International Conference on Photonics, Optics and Laser Technology, Online, 10–11 February 2022; Volume 9, p. 27. [Google Scholar]
- Khozeymeh, F.; Razaghi, M. Cylindrical optical resonators: Fundamental properties and bio-sensing characteristics. J. Opt. 2018, 20, 045301. [Google Scholar] [CrossRef]
- Khozeymeh, F.; Razaghi, M. Parallel-Coupled Dual SiOxNy Racetrack Resonators as Biosensors with High Improved Intrinsic Limit of Detection. Phys. Rev. Appl. 2019, 12, 054045. [Google Scholar] [CrossRef]
- Khozeymeh, F.; Razaghi, M. Crystalline MgF2 Whispering Gallery Mode Resonators as Optical Refractometric Sensors with Ultra-High Improved Sensitivity. IEEE Sens. J. 2019, 20, 2416–2423. [Google Scholar] [CrossRef]
- Khozeymeh, F.; Razaghi, M.; Chalyan, T.; Pavesi, L. Fast analytical modelling of a SOI micro-ring resonator for bio-sensing application. J. Phys. D Appl. Phys. 2018, 28, 285401. [Google Scholar] [CrossRef]
- Khozeymeh, F.; Razaghi, M. Sensitivity optimization in whispering gallery mode optical cylindrical biosensors. J. Phys. Conf. Ser. 2018, 956, 012008. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Hill, K.; Warren-Smith, S.; Monro, T. Interferometric-type optical biosensor based on exposed core microstructured optical fiber. Sens. Actuators B Chem. 2015, 221, 320–327. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Chen, Y.; Yu, F.; Tang, L.; Li, M.; He, J.J. High-sensitivity optical biosensor based on cascaded Mach–Zehnder interferometer and ring resonator using Vernier effect. Opt. Lett. 2014, 39, 6363–6366. [Google Scholar] [CrossRef]
- Cunningham, B.; Qiu, J.; Li, P.; Lin, B. Enhancing the surface sensitivity of colorimetric resonant optical biosensors. Sens. Actuators B Chem. 2002, 87, 365–370. [Google Scholar] [CrossRef]
- Khozeymeh, F.; Razaghi, M. Characteristics optimization in single and dual coupled silicon-on-insulator ring (disk) photonic biosensors. Sens. Actuators B Chem. 2019, 281, 998–1008. [Google Scholar] [CrossRef]
- TalebiFard, S.; Schmidt, S.; Shi, W.; Wu, W.; Jaeger, N.A.; Kwok, E.; Ratner, D.M.; Chrostowski, L. Optimized sensitivity of silicon-on-insulator (SOI) strip waveguide resonator sensor. Biomed. Opt. Express 2017, 8, 500–511. [Google Scholar] [CrossRef]
- González-Guerrero, A.B.; Maldonado, J.; Herranz, S.; Lechuga, L.M. Trends in photonic lab-on-chip interferometric biosensors for point-of-care diagnostics. Anal. Methods 2016, 8, 8380–8394. [Google Scholar] [CrossRef] [Green Version]
- Ricciardi, A.; Crescitelli, A.; Vaiano, P.; Quero, G.; Consales, M.; Pisco, M.; Esposito, E.; Cusano, A. Lab-on-fiber technology: A new vision for chemical and biological sensing. Analyst 2015, 140, 8068–8079. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, C.; Newhouse, R.; Zhang, J.Z.; Gu, C. Hollow-core photonic crystal fibers for surface-enhanced Raman scattering probes. Int. J. Opt. 2011, 2011, 754610. [Google Scholar] [CrossRef] [Green Version]
- Nasirifar, R.; Danaie, M.; Dideban, A. Hollow-core graded index optical fiber refractive index sensor based on surface plasmon resonance. Opt. Quantum Electron. 2020, 52, 341. [Google Scholar] [CrossRef]
- Coscelli, E.; Sozzi, M.; Poli, F.; Passaro, D.; Cucinotta, A.; Selleri, S.; Corradini, R.; Marchelli, R. Toward a highly specific DNA biosensor: PNA-modified suspended-core photonic crystal fibers. IEEE J. Sel. Top. Quantum Electron. 2009, 16, 967–972. [Google Scholar] [CrossRef]
- Vincetti, L.; Setti, V. Waveguiding mechanism in tube lattice fibers. Opt. Express 2010, 18, 23133–23146. [Google Scholar] [CrossRef] [PubMed]
- Giovanardi, F.; Cucinotta, A.; Vincetti, L. Inhibited coupling guiding hollow fibers for label-free DNA detection. Opt. Express 2017, 25, 26215–26220. [Google Scholar] [CrossRef]
- Giovanardi, F.; Cucinotta, A.; Rozzi, A.; Corradini, R.; Benabid, F.; Rosa, L.; Vincetti, L. Hollow core inhibited coupling fibers for biological optical sensing. J. Light. Technol. 2019, 37, 2598–2604. [Google Scholar] [CrossRef]
- Mudraboyina, A.K.; Sabarinathan, J. Protein binding detection using on-chip silicon gratings. Sensors 2011, 11, 11295–11304. [Google Scholar] [CrossRef]
- Jabin, M.A.; Luo, Y.; Peng, G.D.; Rana, M.J.; Ahmed, K.; Nguyen, T.K.; Paul, B.K.; Dhasarathan, V. Design and fabrication of amoeba faced photonic crystal fiber for biosensing application. Sens. Actuators A Phys. 2020, 313, 112204. [Google Scholar] [CrossRef]
- Bertucci, A.; Manicardi, A.; Candiani, A.; Giannetti, S.; Cucinotta, A.; Spoto, G.; Konstantaki, M.; Pissadakis, S.; Selleri, S.; Corradini, R. Detection of unamplified genomic DNA by a PNA-based microstructured optical fiber (MOF) Bragg-grating optofluidic system. Biosens. Bioelectron. 2015, 63, 248–254. [Google Scholar] [CrossRef] [PubMed]



| Oligo | Name | Sequence |
|---|---|---|
| PNA | PNA Soy RR | N H-O-TGC TAG AGT CAG CTT-NH C |
| DNA | 50-mer SOY | 5′- ACC CTA ATC ATT TCA TTT GGA GAG |
| RR | GAC ACG CTG ACA AGC TGA CTC TAG CA -3′ | |
| Biotinylated PNA | PNA 8 mer-Biotin | N Biotin-O-TGG GAT TA-Gly-NH C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khozeymeh, F.; Melli, F.; Capodaglio, S.; Corradini, R.; Benabid, F.; Vincetti, L.; Cucinotta, A. Hollow-Core Fiber-Based Biosensor: A Platform for Lab-in-Fiber Optical Biosensors for DNA Detection. Sensors 2022, 22, 5144. https://doi.org/10.3390/s22145144
Khozeymeh F, Melli F, Capodaglio S, Corradini R, Benabid F, Vincetti L, Cucinotta A. Hollow-Core Fiber-Based Biosensor: A Platform for Lab-in-Fiber Optical Biosensors for DNA Detection. Sensors. 2022; 22(14):5144. https://doi.org/10.3390/s22145144
Chicago/Turabian StyleKhozeymeh, Foroogh, Federico Melli, Sabrina Capodaglio, Roberto Corradini, Fetah Benabid, Luca Vincetti, and Annamaria Cucinotta. 2022. "Hollow-Core Fiber-Based Biosensor: A Platform for Lab-in-Fiber Optical Biosensors for DNA Detection" Sensors 22, no. 14: 5144. https://doi.org/10.3390/s22145144
APA StyleKhozeymeh, F., Melli, F., Capodaglio, S., Corradini, R., Benabid, F., Vincetti, L., & Cucinotta, A. (2022). Hollow-Core Fiber-Based Biosensor: A Platform for Lab-in-Fiber Optical Biosensors for DNA Detection. Sensors, 22(14), 5144. https://doi.org/10.3390/s22145144







