Fetal Electrocardiogram Signal Extraction Based on Fast Independent Component Analysis and Singular Value Decomposition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. FECG Signal Extraction Based on the FastICA and SVD Algorithm
2.3. FECG Signal Detection Based on the Wavelet Mode Maximum Method
- R-R interval: When R-R interval is too long and fetal heart is too slow, we can consider late hypoxia; when the R-R interval is unequal, e.g., the change of fetal heart rate is more than 25–30 bpm, we can preliminarily diagnose premature beats, cardiac arrest, irregular rhythm and other abnormalities, and we should do the next examination immediately.
- QRS waves: The normal duration of fetal QRS ranges from 0.02 s to 0.05 s. If it exceeds 0.05 s, it is abnormal. According to the duration and amplitude of QRS waves, fetal weights can be estimated, such as macrosomia or fetal growth retardation. For the fetus with hemolytic anemia, the severity of anemia can be determined [25].
- ST segment: ST segment is the potential line from the end of QRS waves to the beginning of T-waves, and the normal ST segment is equipotent. Metabolic acidosis should be considered when the ST segment is significantly depressed or elevated. Changes in ST segments are also an important indicator to evaluate whether the fetus is healthy [26]. Abnormal ST segments and T-waves of FECG signals indicate fetal electrolyte disorder, myocardial hypoxia, and abnormal myocardial metabolism.
2.3.1. Wavelet Mode Maximum Method
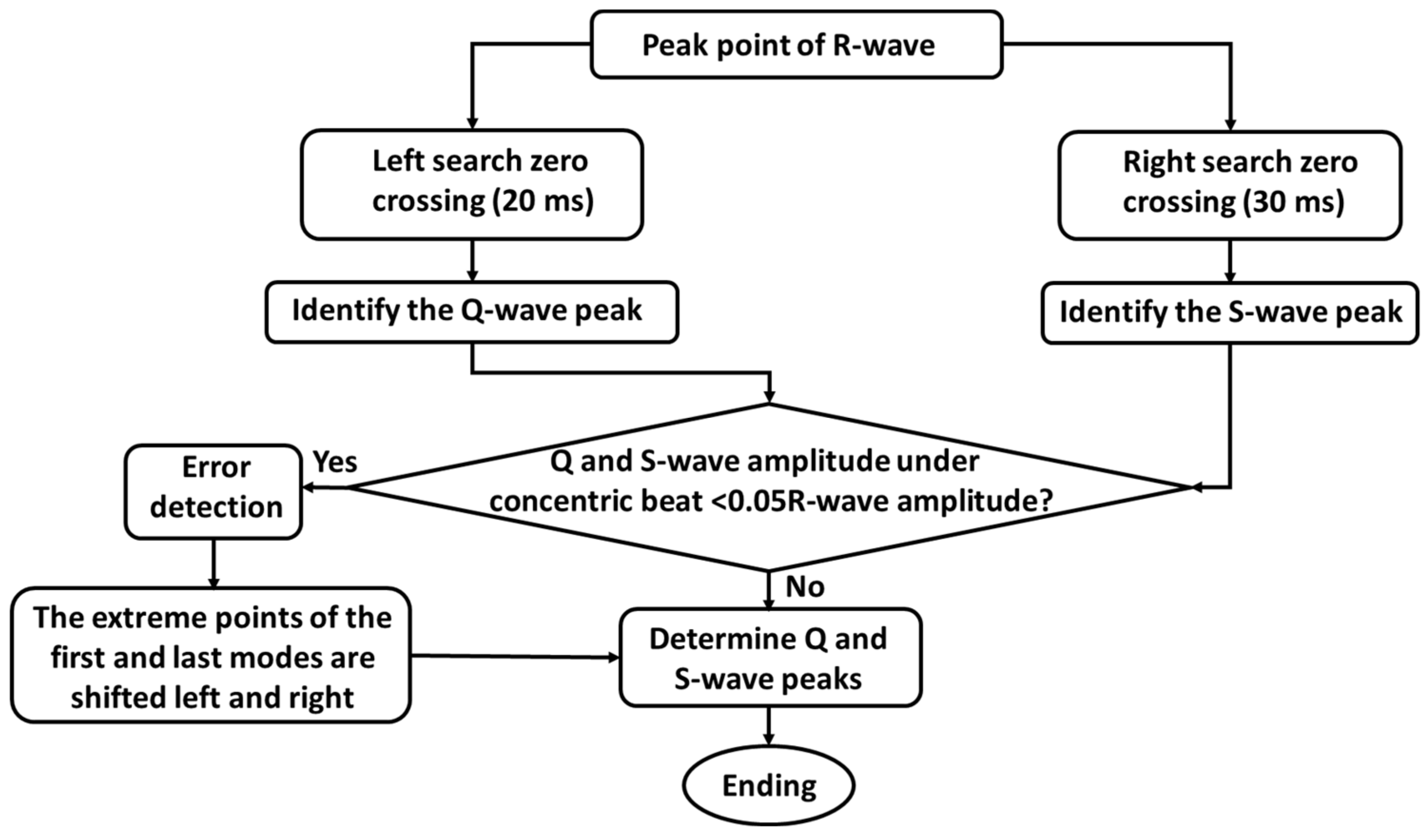
2.3.2. QRS Wave Detection

2.3.3. Detection of P-waves, T-waves, and ST Segments
2.4. Evaluation Methods
3. Results
3.1. Extraction of FECG Signals
3.2. Detection of FECG Signals
4. Discussion
4.1. Significance of This Study
4.2. Comparison with Related Work
| Authors | Methods | Year | Database | Se/% * | PPV/% | F1/% |
|---|---|---|---|---|---|---|
| Gurve et al. [32] | ICA | 2020 | ADFECGD and PhysioNet2013 | 93.30 | 94.00 | 93.60 |
| Gurve et al. [32] | NNMF + ICA | 2020 | ADFECGD and PhysioNet2013 | 95.30 | 94.60 | 94.80 |
| Taha et al. [5] | FastICA | 2020 | PhysioNet2013 | 97.30 | 93.30 | 95.70 |
| Barnova et al. [6] | EEMD | 2021 | ADFECGD and PhysioNet2013 | 81.79 | 87.16 | 84.08 |
| Barnova et al.[6] | EEMD + RLS + ICA | 2021 | ADFECGD and PhysioNet2013 | 95.09 | 96.36 | 95.69 |
| Sarafan et al. [12] | FastICA + TS + EKF | 2020 | PhysioNet2013 | - | - | 92.61 |
| Zhang et al.[33] | K-means + PCA | 2019 | PhysioNet2013 | 96.23 | 95.35 | 95.78 |
| Panigrahy et al.[28] | EKS + DE + ANFIS | 2017 | PhysioNet2013 | 91.47 | 92.18 | - |
| Jaba et al. [34] | PSF + ANC | 2021 | DaISy and PhysioNet2013 | 97.92 | 94.66 | 96.12 |
| Liu et al. [35] | SQA + FTM | 2014 | PhysioNet2013 | 94.13 | 93.74 | 93.90 |
| Mollakazemi et al. [36] | PCA + WT | 2021 | PhysioNet2013 | - | - | 98.77 |
| Jallouli et al. [27] | Clifford Wavelet Entropy | 2021 | PhysioNet2013 | 99.76 | 99.2 | 99.47 |
| Rasti-Meymandi et al. [37] | AECG-DecompNet | 2021 | PhysioNet2013 | 97.40 | 93.52 | 95.42 |
| Fotiadou et al. [3] | DNN+ LSTM | 2020 | PhysioNet2013 | 98.10 | - | - |
| Vo et al. [16] | 1-D Octave convolution | 2020 | PhysioNet2013 | - | - | 90.70 |
| Ting et al. [17] | 2-D-CNN | 2021 | PhysioNet2013 | 95.20 | - | - |
| Mohebian et al. [31] | Conv1D + CycleGAN | 2021 | ADFECGDand NI-FECG | - | - | 99.70 |
| Our method | FastICA + SVD + WT | 2022 | ADFECGD and PhysioNet2013 | 96.90 | 98.23 | 95.24 |
4.3. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monson, M.; Heuser, C.; Einerson, B.D.; Esplin, I.; Snow, G.; Varner, M.; Esplin, M.S. Evaluation of an external fetal electrocardiogram monitoring system: A randomized controlled trial. Am. J. Obstet. Gynecol. 2020, 223, e1–e244. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, F.; Jongbloed, M.R.M.; Van Geloven, N.; Ten Harkel, A.D.J.; Van Lith, J.M.M.; Haak, M.C. Assessment of human fetal cardiac autonomic nervous system development using color tissue Doppler imaging. Echocardiography 2021, 38, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Fotiadou, E.; Xu, M.; Van Erp, B.; Van Sloun, R.J.G.; Vullings, R. Deep Convolutional Long Short-Term Memory Network for Fetal Heart Rate Extraction. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 608–611. [Google Scholar]
- Sulas, E.; Urru, M.; Tumbarello, R.; Raffo, L.; Pani, D. Systematic analysis of single-and multi-reference adaptive filters for non-invasive fetal electrocardiography. Math. Biosci. Eng. 2019, 17, 286–308. [Google Scholar] [CrossRef] [PubMed]
- Taha, L.; Abdel-Raheem, E. A null space-based blind source separation for fetal electrocardiogram signals. Sensors 2020, 20, 3536. [Google Scholar] [CrossRef]
- Barnova, K.; Martinek, R.; Jaros, R.; Kahankova, R.; Matonia, A.; Jezewski, M.; Czabanski, R.; Horoba, K.; Jezewski, J. A novel algorithm based on ensemble empirical mode decomposition for non-invasive fetal ECG extraction. PLoS ONE 2021, 16, e0256154. [Google Scholar] [CrossRef]
- Wu, S.; Shen, Y.; Zhou, Z.; Lin, L.; Zeng, Y.; Gao, X. Research of fetal ECG extraction using wavelet analysis and adaptive filtering. Comput. Biol. Med. 2013, 43, 1622–1627. [Google Scholar] [CrossRef]
- Vasudeva, B.; Deora, P.; Pradhan, P.M.; Dasgupta, S. Efficient implementation of LMS adaptive filter-based FECG extraction on an FPGA. Healthc. Technol. Lett. 2020, 7, 125–131. [Google Scholar] [CrossRef]
- Ferranti, M.; Le, T.H.; Vandebril, R. A comparison between the complex symmetric based and classical computation of the singular value decomposition of normal matrices. Numer. Algorithms 2021, 67, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Tomar, H.; Mehla, V.K.; Komaragiri, R.; Kumar, M. Stationary wavelet transform based ECG signal denoising method. ISA Trans. 2021, 114, 251–262. [Google Scholar] [CrossRef]
- Martinek, R.; Kahankova, R.; Jezewski, J.; Jaros, R.; Mohylova, J.; Fajkus, M.; Nedoma, J.; Janku, P.; Nazeran, H. Comparative effectiveness of ICA and PCA in extraction of fetal ECG from abdominal signals: Toward non-invasive fetal monitoring. Front. Physiol. 2018, 9, 648. [Google Scholar] [CrossRef] [Green Version]
- Sarafan, S.; Le, T.; Naderi, A.M.; Nguyen, Q.D.; Kuo, B.T.Y.; Ghirmai, T.; Han, H.D.; Lau, M.P.H.; Cao, H. Investigation of methods to extract fetal electrocardiogram from the mother’s abdominal signal in practical scenarios. Technologies 2020, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999, 10, 626–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Zhou, Z.; Yuan, Y.; Wu, S. An improved FastICA method for fetal ECG extraction. Comput. Math. Methods Med. 2018, 2018, 7061456. [Google Scholar] [CrossRef] [PubMed]
- Kaleem, A.M.; Kokate, R.D. A survey on FECG extraction using neural network and adaptive filter. Soft Comput. 2021, 25, 4379–4392. [Google Scholar] [CrossRef]
- Vo, K.; Le, T.; Rahmani, A.M.; Dutt, N.; Cao, H. An efficient and robust deep learning method with 1-D octave convolution to extract fetal electrocardiogram. Sensors 2020, 20, 3757. [Google Scholar] [CrossRef]
- Ting, Y.C.; Lo, F.W.; Tsai, P.Y. Implementation for fetal ECG detection from multi-channel abdominal recordings with 2D convolutional neural network. J. Signal. Process. Syst. 2021, 93, 1101–1113. [Google Scholar] [CrossRef]
- Zhou, Z.Q.; Huang, K.J.; Qiu, Y.; Shen, H.B.; Ming, Z.Y. Morphology extraction of fetal electrocardiogram by slow-fast LSTM network. Biomed. Signal. Process. 2020, 68, 102664. [Google Scholar] [CrossRef]
- Malek, A.S.; Elnahrawy, A.; Anwar, H.; Naeem, M. Automated detection of premature ventricular contraction in ECG signals using enhanced template matching algorithm. Biomed. Phys. Eng. Express. 2020, 6, 015024. [Google Scholar] [CrossRef]
- Singla, M.; Azeemuddin, S.; Sistla, P. Learning-Based Model for Central Blood Pressure Estimation using Feature Extracted from ECG and PPG signals. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 855–858. [Google Scholar]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Kaleem, A.M.; Kokate, R.D. Performance evaluation of fetal ECG extraction algorithms. In Emerging Research in Electronics, Computer Science and Technology; Springer: Singapore, 2019; pp. 187–194. [Google Scholar]
- Dessì, A.; Pani, D.; Raffo, L. An advanced algorithm for fetal heart rate estimation from non-invasive low electrode density recordings. Physiol. Meas. 2014, 35, 1621–1636. [Google Scholar] [CrossRef]
- Peri, E.; Xu, L.; Ciccarelli, C.; Vandenbussche, N.L.; Xu, H.; Long, X.; Overeem, S.; Dijk, J.P.; Mischi, M. Singular Value Decomposition for removal of cardiac interference from trunk electromyogram. Sensors 2021, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Le, J. Clinical value of QRS, ST segment and T wave in fetal electrocardiogram. Obstet. Gynecol. Int. 1992, 5, 275–276. [Google Scholar]
- Olofsson, P.; Norén, H.; Carlsson, A.; Rosén, K.G. Identifying newborns with umbilical cord blood metabolic acidosis by intrapartum cardiotography combined with fetal ECG ST analysis (STAN): Comparison of the new and old FIGO systems to classify cardiotocograms. J. Matern. Fetal. Neonatal. Med. 2020, 33, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Jallouli, M.; Arfaoui, S.; Ben, M.A.; Cattani, C. Clifford Wavelet Entropy for fetal ECG Extraction. Entropy 2021, 23, 844. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, D.; Sahu, P.K. Extraction of fetal ECG signal by an improved method using extended Kalman smoother framework from single channel abdominal ECG signal. Australas. Phys. Eng. Sci. Med. 2017, 40, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, V.; Ummar, S.; Varghese, T.J.; Shibu, A.E. ECG noise classification using deep learning with feature extraction. Signal. Image Video P. 2022, 1–7. [Google Scholar] [CrossRef]
- Tseng, K.K.; Wang, C.; Xiao, T.J.; Chen, C.M.; Hassan, M.M.; Albuquerque, V.H.C. Sliding large kernel of deep learning algorithm for mobile electrocardiogram diagnosis. Comput. Electr. Eng. 2021, 96, 107521. [Google Scholar] [CrossRef]
- Mohebbian, M.R.; Vedaei, S.S.; Wahid, K.A.; Dinh, A.; Marateb, H.R.; Tavakolian, K. Fetal ECG extraction from maternal ECG using attention-based CycleGAN. IEEE. J. Biomed. Health 2022, 26, 515–526. [Google Scholar] [CrossRef]
- Gurve, D.; Krishnan, S. Separation of fetal-ECG from single-channel abdominal ECG using activation scaled non-negative matrix factorization. IEEE. J. Biomed. Health Inform. 2020, 24, 669–680. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, S. Single-lead noninvasive fetal ECG extraction by means of combining clustering and principal components analysis. Med. Biol. Eng. Comput. 2020, 58, 419–432. [Google Scholar] [CrossRef]
- Jaba, D.K.A.; Dhanalakshmi, S.R.K. An improved parallel sub-filter adaptive noise canceler for the extraction of fetal ECG. Biomed. Tech. 2021, 66, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, P.; Di, M.C.; Zhao, L.; Zhang, H.; Chen, Z. A multi-step method with signal quality assessment and fine-tuning procedure to locate maternal and fetal QRS complexes from abdominal ECG recordings. Physiol. Meas. 2014, 35, 1665–1683. [Google Scholar] [CrossRef] [PubMed]
- Mollakazemi, M.J.; Asadi, F.; Tajnesaei, M.; Ghaffari, A. Fetal QRS Detection in Noninvasive Abdominal Electrocardiograms Using Principal Component Analysis and Discrete Wavelet Transforms with Signal Quality Estimation. J. Biomed. Phys. Eng. 2021, 11, 197–204. [Google Scholar]
- Rasti-Meymandi, A.; Ghaffari, A. AECG-DecompNet: Abdominal ECG signal decomposition through deep-learning model. Phys. Meas. 2021, 42, 33706298. [Google Scholar] [CrossRef] [PubMed]







| Data | FastICA | SVD | FastICA + SVD | |||
|---|---|---|---|---|---|---|
| R01 | 1.158 | 0.369 | 2.982 | 6.956 | 3.194 | 7.759 |
| R04 | 1.549 | 0.184 | 2.494 | 5.265 | 2.749 | 6.637 |
| R07 | 2.305 | 5.125 | 1.872 | 3.391 | 5.349 | 26.632 |
| R08 | 1.072 | 0.142 | 1.186 | 9.682 | 3.125 | 45.028 |
| R10 | 2.927 | 7.797 | 2.873 | 5.118 | 3.258 | 10.362 |
| Data | FastICA | SVD | FastICA + SVD | |||
|---|---|---|---|---|---|---|
| a01 | 0.739 | 0.051 | 0.732 | 0.342 | 2.296 | 4.073 |
| a03 | 1.281 | 1.394 | 1.787 | 2.177 | 1.796 | 2.183 |
| a04 | 0.992 | 0.112 | 1.122 | 1.081 | 2.018 | 3.185 |
| a06 | 0.538 | −0.644 | 0.747 | 0.220 | 3.197 | 7.529 |
| a08 | 1.071 | 0.142 | 1.185 | 0.681 | 3.120 | 7.767 |
| a10 | 0.793 | 0.093 | 1.261 | 0.557 | 4.357 | 11.178 |
| a12 | 1.067 | 0.900 | 1.492 | 1.138 | 14.556 | 11.202 |
| a13 | 7.146 | 1.007 | 5.773 | 0.668 | 9.461 | 5.916 |
| a17 | 6.324 | 1.188 | 6.242 | 0.982 | 9.159 | 2.734 |
| a21 | 1.058 | 0.749 | 0.648 | 0.088 | 4.046 | 15.640 |
| Data | Correct Detection | Missing Detection | Incorrect Detection | Se/% * | PPV/% | F1/% |
|---|---|---|---|---|---|---|
| R01 | 627 | 17 | 2 | 97.36 | 99.68 | 97.06 |
| R04 | 620 | 12 | 19 | 98.10 | 97.02 | 95.24 |
| R07 | 613 | 14 | 21 | 97.77 | 96.69 | 94.60 |
| R08 | 623 | 19 | 5 | 97.04 | 99.20 | 96.29 |
| R10 | 620 | 37 | 9 | 95.65 | 98.57 | 93.09 |
| average | 3103 | 99 | 56 | 96.90 | 98.23 | 95.24 |
| Data | Maternal Characteristic Parameters | Fetal Characteristic Parameters | |||||
|---|---|---|---|---|---|---|---|
| MECG QRS Numbers | Mean R-R Intervals (s) | Heart Rates (bpm) | FECG QRS Numbers | Mean R-R Intervals (s) | Fetal Heart Rates (bpm) | Baseline Fetal Heart Rates (bpm) | |
| R01 | 413 | 0.728 | 82.40 | 644 | 0.490 | 122.45 | 122 |
| R04 | 433 | 0.690 | 86.96 | 632 | 0.490 | 122.45 | 122 |
| R07 | 401 | 0.747 | 80.32 | 627 | 0.478 | 125.52 | 126 |
| R08 | 411 | 0.729 | 82.30 | 642 | 0.460 | 130.43 | 130 |
| R10 | 480 | 0.642 | 93.46 | 657 | 0.456 | 131.58 | 132 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, J.; Yang, Y.; Zhou, Z.; Wu, S. Fetal Electrocardiogram Signal Extraction Based on Fast Independent Component Analysis and Singular Value Decomposition. Sensors 2022, 22, 3705. https://doi.org/10.3390/s22103705
Hao J, Yang Y, Zhou Z, Wu S. Fetal Electrocardiogram Signal Extraction Based on Fast Independent Component Analysis and Singular Value Decomposition. Sensors. 2022; 22(10):3705. https://doi.org/10.3390/s22103705
Chicago/Turabian StyleHao, Jingyu, Yuyao Yang, Zhuhuang Zhou, and Shuicai Wu. 2022. "Fetal Electrocardiogram Signal Extraction Based on Fast Independent Component Analysis and Singular Value Decomposition" Sensors 22, no. 10: 3705. https://doi.org/10.3390/s22103705
APA StyleHao, J., Yang, Y., Zhou, Z., & Wu, S. (2022). Fetal Electrocardiogram Signal Extraction Based on Fast Independent Component Analysis and Singular Value Decomposition. Sensors, 22(10), 3705. https://doi.org/10.3390/s22103705







