Integrated Relaxation Pressure Classification and Probe Positioning Failure Detection in High-Resolution Esophageal Manometry Using Machine Learning
Abstract
1. Introduction
- To filter out the images for which the input probe was not correctly positioned.
- To determine the IRP parameter for the correct images.
2. Material and Methods
2.1. Raw Data Analysis
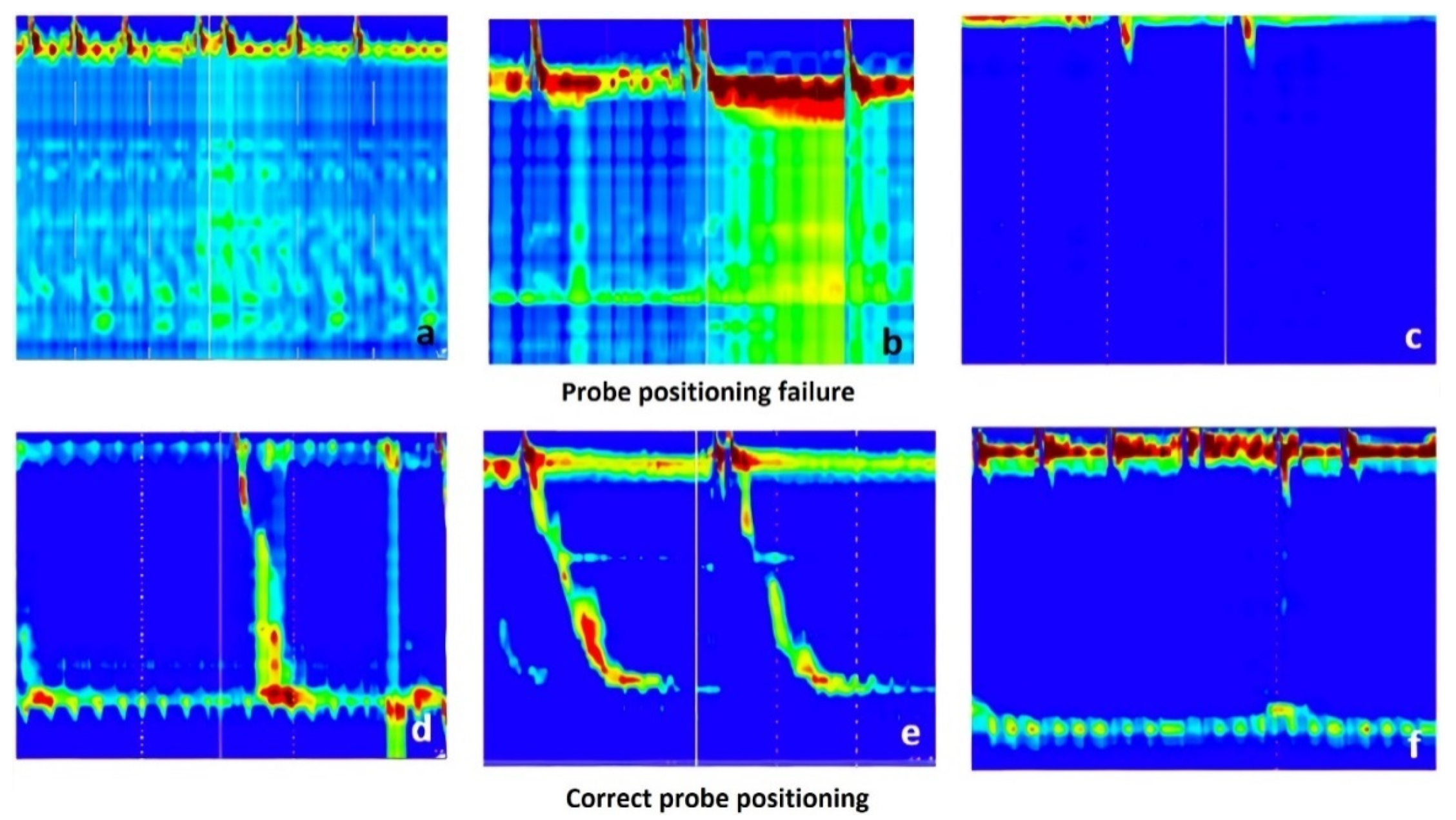
2.2. Preprocessing of the Esophageal Pressure Topography Maps
2.3. Transfer Learning
3. Results
3.1. Solution Pipeline
- Delete the margins of the image; every image has a 15 pixels top margin, 120 pixels left margin, and a 30 pixels bottom margin which should be removed.
- Binarize the image.
- Calculate the histogram of white pixels for each x-axis position based on the binarized image created previously.
- Find the maximum value of the previously calculated histogram. The x-axis position of the founded maximum value will be the x-axis position of the white vertical line.
- Crop the original image (the colored image without the margins) starting from the previously found x-axis position until the end of the image.
3.2. Metrics
- Accuracy: The number of correct classifications compared to the total number of examples.
- Precision: The ratio of the correctly classified positives to the total number of positive classifications.
- Recall: The ratio of the correctly classified positives to the total number of positives from the dataset.
- F1-Score: The harmonic mean of Precision and Recall.
- Confusion Matrix: A confusion matrix is a summary of prediction results on a classification problem. The number of correct and incorrect predictions are summarized with count values and broken down by each class. The confusion matrix shows how the classification model is confused when it makes predictions.
3.3. IRP Classification Results
3.4. Probe Positioning Failure Classification Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yadlapati, R. High-resolution esophageal manometry: Interpretation in clinical practice. Curr. Opin. Gastroenterol. 2017, 33, 301–309. [Google Scholar] [CrossRef]
- Laing, P.; Bress, A.P.; Fang, J.; Peterson, K.; Adler, D.G.; Gawron, A.J. Trends in diagnoses after implementation of the Chicago classification for esophageal motility disorders (V3.0) for high-resolution manometry studies. Dis. Esophagus 2017, 30, 1–6. [Google Scholar] [CrossRef]
- Kahrilas, P.J.; Bredenoord, A.J.; Fox, M.; Gyawali, C.P.; Roman, S.; Smout, A.J.; Pandolfino, J.E.; International High Resolution Manometry Working Group. The Chicago Classification of esophageal motility disorders, v3.0. Neurogastroenterol. Motil. 2015, 27, 160–174. [Google Scholar] [CrossRef]
- Monrroy, H.; Cisternas, D.; Bilder, C.; Ditaranto, A.; Remes-Troche, J.; Meixueiro, A.; Zavala, M.A.; Serra, J.; Marín, I.; de León, R.A.; et al. The Chicago Classification 3.0 Results in More Normal Findings and Fewer Hypotensive Findings with No Difference in Other Diagnoses. Am. J. Gastroenterol. 2017, 112, 606–612. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.E.; Cho, Y.K.; Lim, C.H.; Park, M.I.; Hwang, J.W.; Jang, J.S.; Oh, M.; Motility Study Club of Korean Society of Neurogastroenterology and Motility. Factors Determining the Inter-observer Variability and Diagnostic Accuracy of High-resolution Manometry for Esophageal Motility Disorders. J. Neurogastroenterol. Motil. 2018, 24, 506. [Google Scholar] [CrossRef]
- Fox, M.R.; Pandolfino, J.E.; Sweis, R.; Sauter, M.; Abreu, Y.; Abreu, A.T.; Anggiansah, A.; Bogte, A.; Bredenoord, A.J.; Dengler, W.; et al. Inter-observer agreement for diagnostic classification of esophageal motility disorders defined in high-resolution manometry. Dis. Esophagus 2015, 28, 711–719. [Google Scholar] [CrossRef]
- Lu, J.; Behbood, V.; Hao, P.; Zuo, H.; Xue, S.; Zhang, G. Transfer learning using computational intelligence: A survey. Knowl.-Based Syst. 2015, 80, 14–23. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), San Juan, PR, USA, 17–19 June 1997. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Lin, M.; Chen, Q.; Yan, S. Network in Network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Diederik, P.K.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Yang, Y.J.; Bang, C.S. Application of artificial intelligence in gastroenterology. World J. Gastroenterol. 2019, 25, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.D.; Tsoulfas, G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World J. Gastroenterol. 2021, 27, 6191–6223. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K.; et al. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef]
- Hujoel, I.A.; Murphree, D.H., Jr.; Van Dyke, C.T.; Choung, R.S.; Sharma, A.; Murray, J.A.; Rubio-Tapia, A. Machine Learning in Detection of Undiagnosed Celiac Disease. Clin. Gastroenterol. Hepatol. 2018, 16, 1354–1355.e1. [Google Scholar] [CrossRef]
- Augustin, S.; Muntaner, L.; Altamirano, J.T.; González, A.; Saperas, E.; Dot, J.; Abu–Suboh, M.; Armengol, J.R.; Malagelada, J.R.; Esteban, R.; et al. Predicting early mortality after acute varicealhemorrhage based on classification and regression tree analysis. Clin. Gastroenterol. Hepatol. 2009, 7, 1347–1354. [Google Scholar] [CrossRef]
- Piscaglia, F.; Cucchetti, A.; Benlloch, S.; Vivarelli, M.; Berenguer, J.; Bolondi, L.; Pinna, A.D.; Berenguer, M. Prediction of significant fibrosis in hepatitis C virus infected liver transplant recipients by artificial neural network analysis of clinical factors. Eur. J. Gastroenterol. Hepatol. 2006, 18, 1255–1261. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Nakamura, H.; Kataoka, S.; Maeda, Y.; Kudo, T.; Hayashi, T.; Wakamura, K.; Miyachi, H.; et al. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology 2016, 150, 1531–1532.e3. [Google Scholar] [CrossRef]
- Rogers, B.; Samanta, S.; Ghobadi, K.; Patel, A.; Savarino, E.; Roman, S.; Sifrim, D.; Gyawali, C.P. Artificial intelligence automates and augments baseline impedance measurements from pH-impedance studies in gastroesophageal reflux disease. J. Gastroenterol. 2021, 56, 34–41. [Google Scholar] [CrossRef]
- Hoffman, M.R.; Mielens, J.D.; Omari, T.I.; Rommel, N.; Jiang, J.J.; McCulloch, T.M. Artificial neural network classification of pharyngeal high-resolution manometry with impedance data. Laryngoscope 2013, 123, 713–720. [Google Scholar] [CrossRef]
- Mielens, J.D.; Hoffman, M.R.; Ciucci, M.R.; McCulloch, T.M.; Jiang, J.J. Application of classification models to pharyngeal high-resolution manometry. J. Speech Lang. Hear. Res. 2012, 55, 892–902. [Google Scholar] [CrossRef][Green Version]
- Lee, T.H.; Lee, J.S.; Hong, S.J.; Lee, J.S.; Jeon, S.R.; Kim, W.J.; Kim, H.G.; Cho, J.Y.; Kim, J.O.; Cho, J.H.; et al. High-resolution manometry: Reliability of automated analysis of upper esophageal sphincter relaxation parameters. Turk. J. Gastroenterol. 2014, 25, 473–480. [Google Scholar] [CrossRef]
- Jungheim, M.; Busche, A.; Miller, S.; Schilling, N.; Schmidt-Thieme, L.; Ptok, M. Calculation of upper esophageal sphincter restitution time from high resolution manometry data using machine learning. Physiol. Behav. 2016, 165, 413–424. [Google Scholar] [CrossRef]
- Geng, Z.; Hoffman, M.R.; Jones, C.A.; McCulloch, T.M.; Jiang, J.J. Three-dimensional analysis of pharyngeal high-resolution manometry data. Laryngoscope 2013, 123, 1746–1753. [Google Scholar] [CrossRef]
- Frigo, A.; Costantini, M.; Fontanella, C.G.; Salvador, R.; Merigliano, S.; Carniel, E.L. A Procedure for the Automatic Analysis of High-Resolution Manometry Data to Support the Clinical Diagnosis of Esophageal Motility Disorders. IEEE Trans. Biomed. Eng. 2018, 65, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Kou, W.; Carlson, D.A.; Baumann, A.J.; Donnan, E.; Luo, Y.; Pandolfino, J.E.; Etemadi, M. A deep-learning-based unsupervised model on esophageal manometry using variational autoencoder. Artif. Intell. Med. 2021, 112, 102006. [Google Scholar] [CrossRef] [PubMed]
- Jell, A.; Kuttler, C.; Ostler, D.; Hüser, N. How to Cope with Big Data in Functional Analysis of the Esophagus. Visc. Med. 2020, 36, 439–442. [Google Scholar] [CrossRef]









| Class | Precision | Recall | F1-Score |
|---|---|---|---|
| IRP higher than cut-off | 95% | 100% | 98% |
| Normal IRP | 100% | 92% | 96% |
| Overall Accuracy | 97% | ||
| Class | Precision | Recall | F1-Score |
|---|---|---|---|
| Wrong | 91% | 95% | 93% |
| Normal | 89% | 80% | 84% |
| Overall Accuracy | 91% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czako, Z.; Surdea-Blaga, T.; Sebestyen, G.; Hangan, A.; Dumitrascu, D.L.; David, L.; Chiarioni, G.; Savarino, E.; Popa, S.L. Integrated Relaxation Pressure Classification and Probe Positioning Failure Detection in High-Resolution Esophageal Manometry Using Machine Learning. Sensors 2022, 22, 253. https://doi.org/10.3390/s22010253
Czako Z, Surdea-Blaga T, Sebestyen G, Hangan A, Dumitrascu DL, David L, Chiarioni G, Savarino E, Popa SL. Integrated Relaxation Pressure Classification and Probe Positioning Failure Detection in High-Resolution Esophageal Manometry Using Machine Learning. Sensors. 2022; 22(1):253. https://doi.org/10.3390/s22010253
Chicago/Turabian StyleCzako, Zoltan, Teodora Surdea-Blaga, Gheorghe Sebestyen, Anca Hangan, Dan Lucian Dumitrascu, Liliana David, Giuseppe Chiarioni, Edoardo Savarino, and Stefan Lucian Popa. 2022. "Integrated Relaxation Pressure Classification and Probe Positioning Failure Detection in High-Resolution Esophageal Manometry Using Machine Learning" Sensors 22, no. 1: 253. https://doi.org/10.3390/s22010253
APA StyleCzako, Z., Surdea-Blaga, T., Sebestyen, G., Hangan, A., Dumitrascu, D. L., David, L., Chiarioni, G., Savarino, E., & Popa, S. L. (2022). Integrated Relaxation Pressure Classification and Probe Positioning Failure Detection in High-Resolution Esophageal Manometry Using Machine Learning. Sensors, 22(1), 253. https://doi.org/10.3390/s22010253







