Early Diagnosis of Multiple Sclerosis Using Swept-Source Optical Coherence Tomography and Convolutional Neural Networks Trained with Data Augmentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Database
2.2. OCT Method
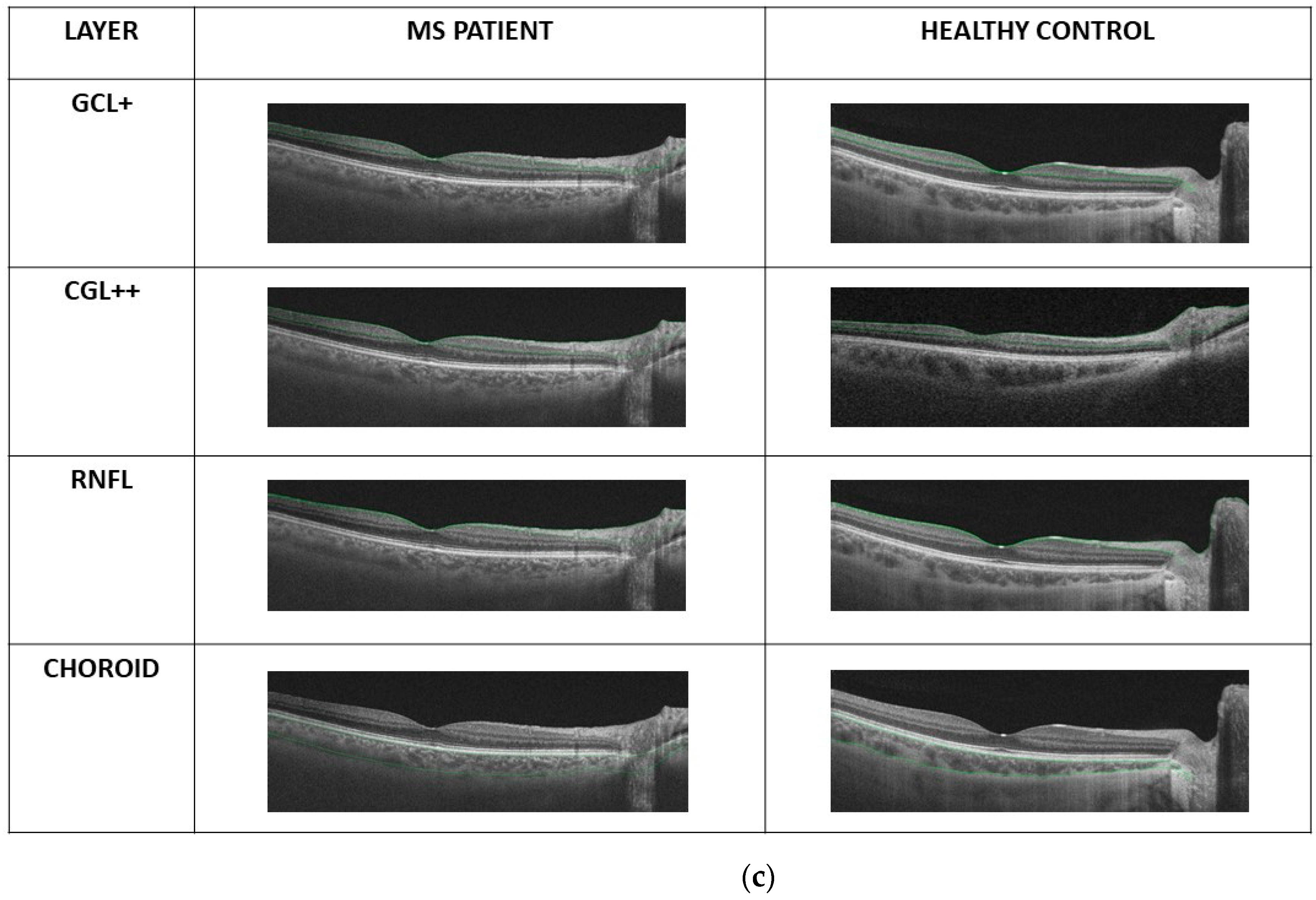
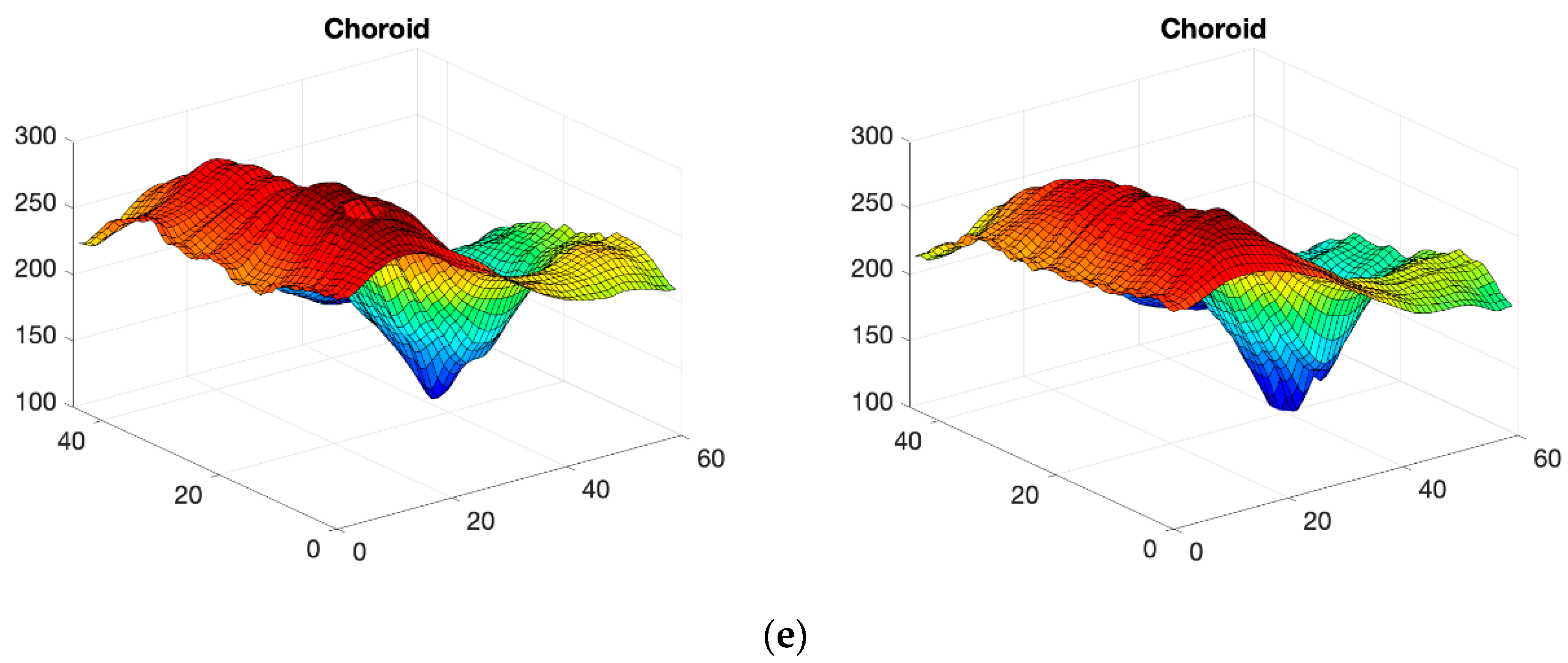
2.3. OCT Map Processing
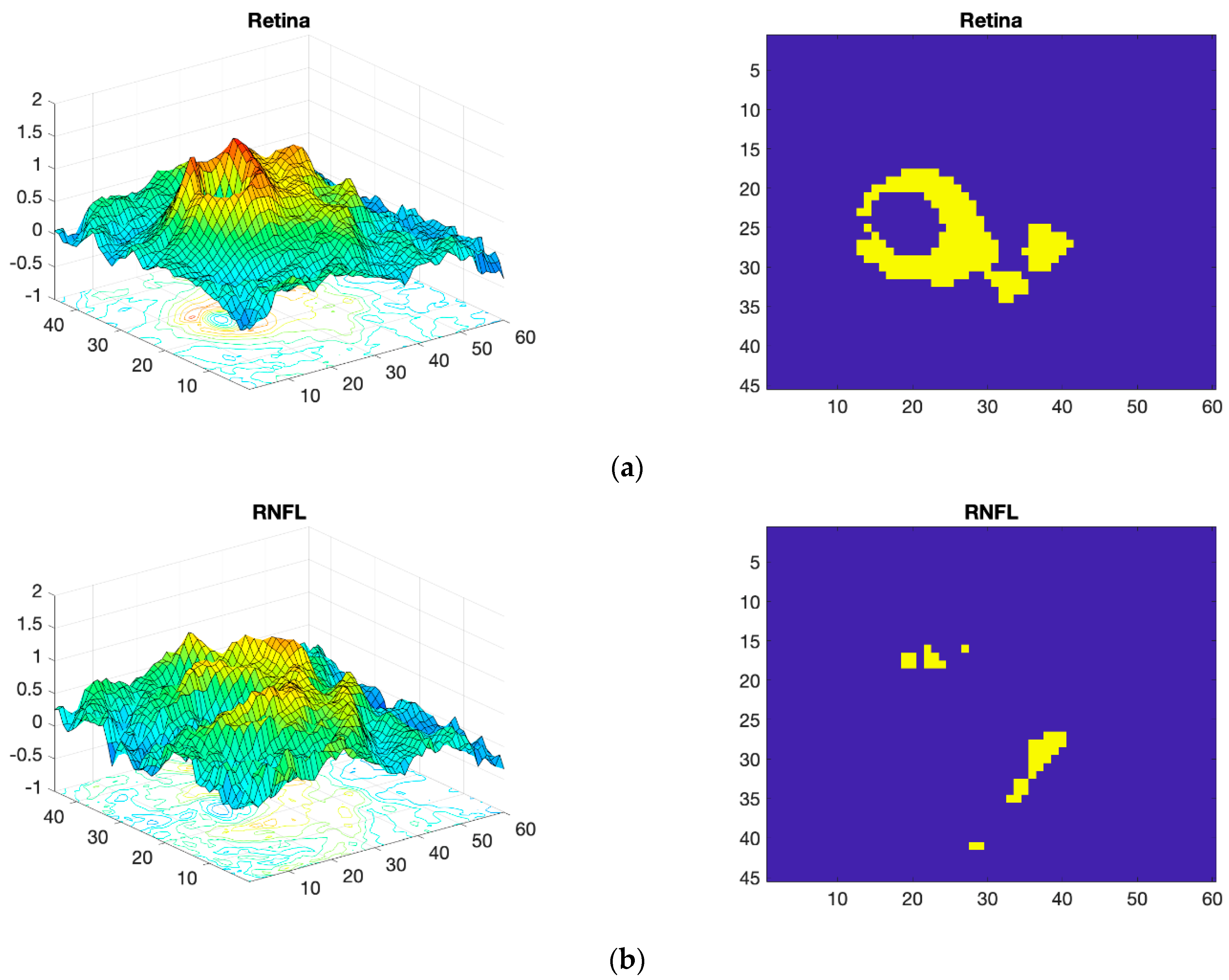
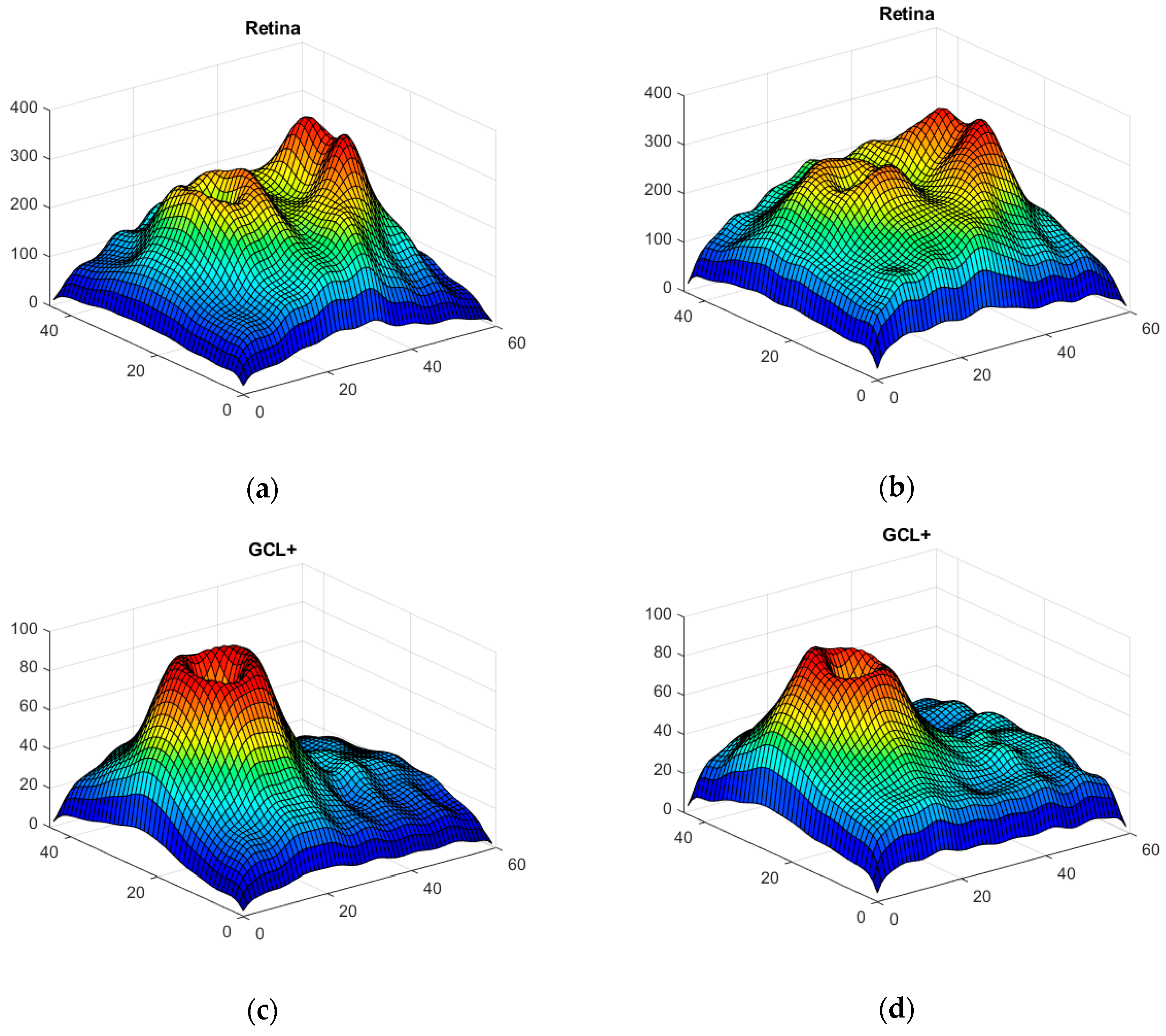
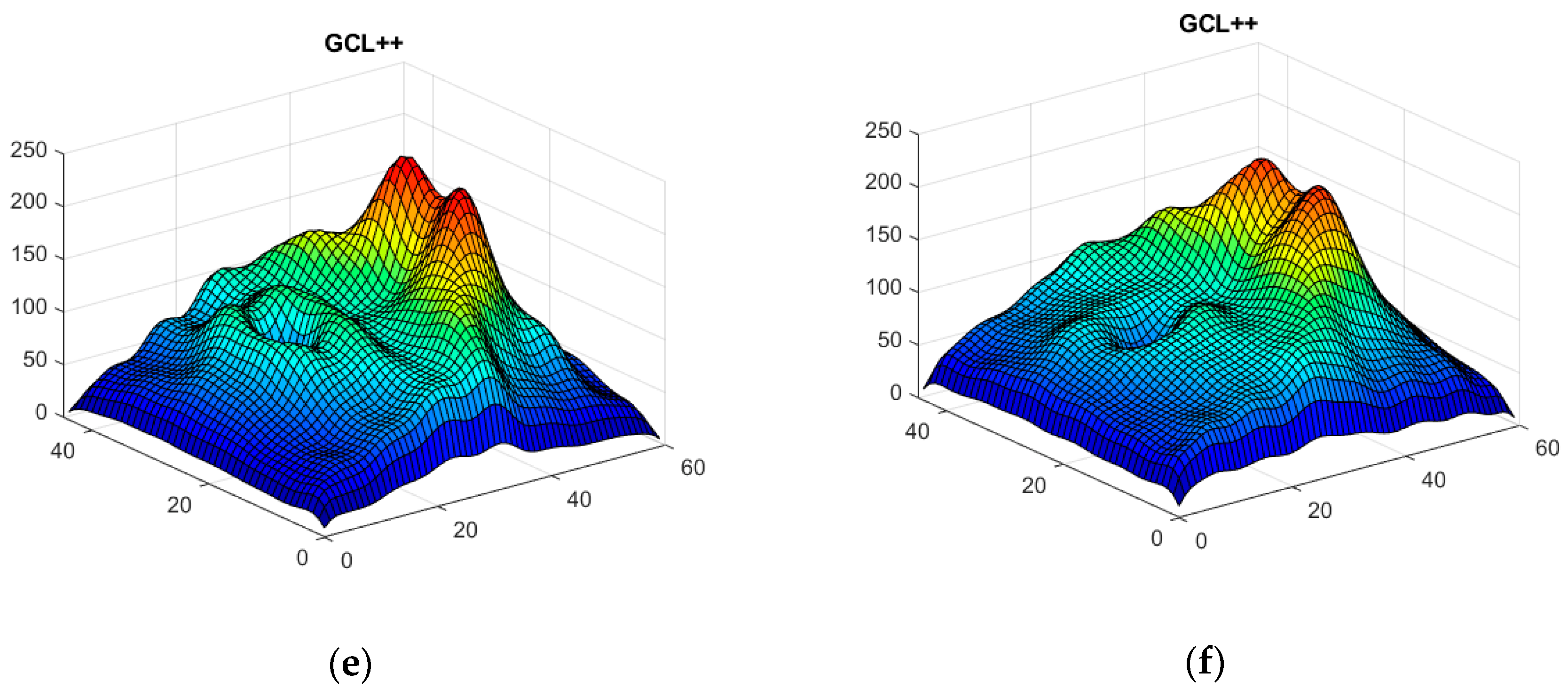
Thickness Image Pre-Processing
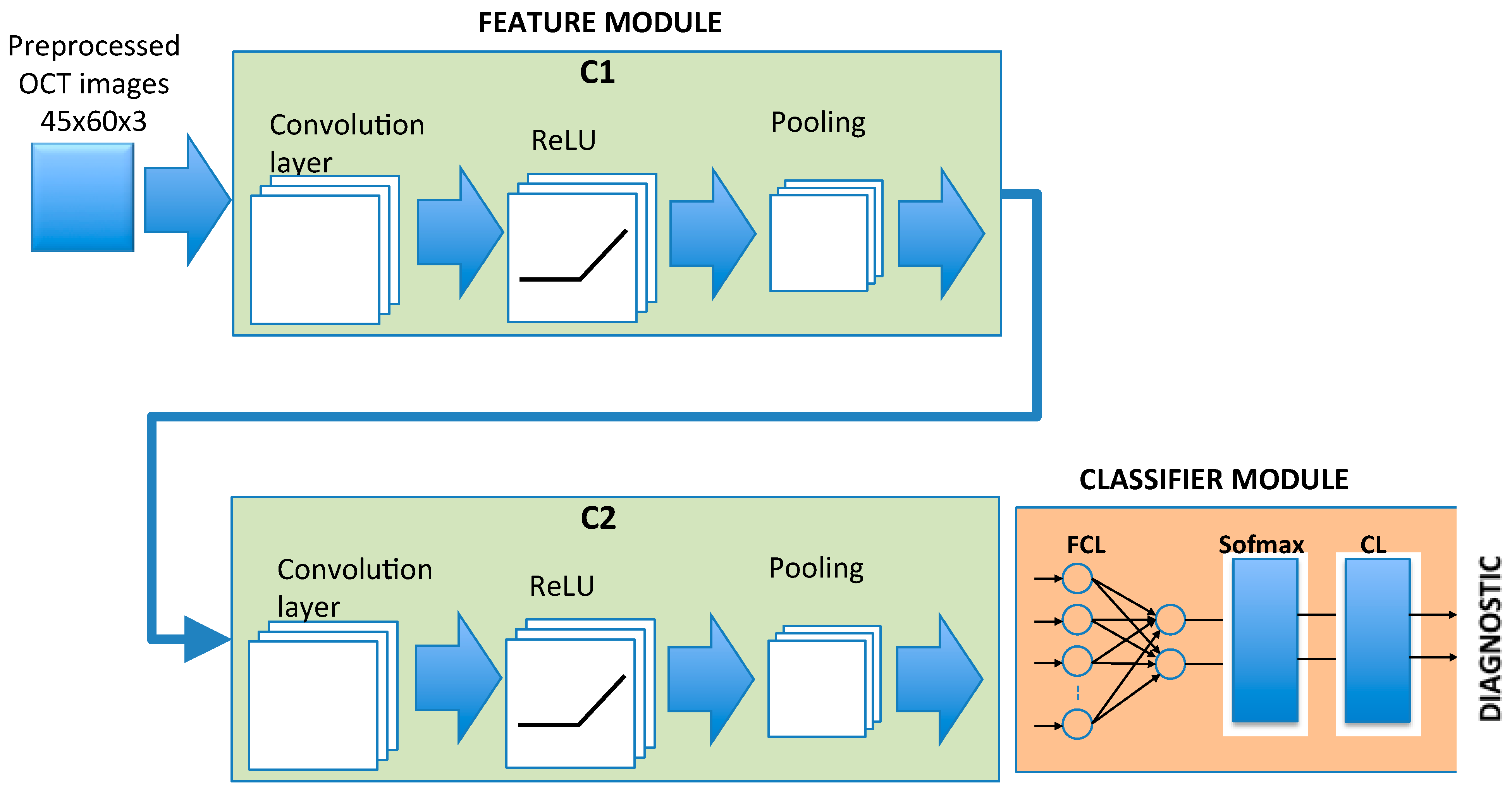
2.4. CNN Architecture
2.5. Training of the CNN
- An eye of a control subject that will not be used in training neither is selected. The remaining 47 control subject eyes are used to train a GAN (Section 2.6) to generate n = 100 synthetic control images, while the 48 MS patient eyes are used to train another GAN to generate n = 100 synthetic MS images. The process is performed on the complete retina, GCL+ and GCL++.
- The Cohen thresholding described above is applied to the total number of images available for each of the 3 layers (147 control eyes, 148 MS eyes), which are used to train the CNN.
- The trained CNN is tested on the images of the eye that was not used either to generate the synthetic images or to train the CNN. The result of the classification is taken into account with regard to the data in the confusion matrix.
- Points 1–3 are repeated until all the control eyes have been tested.
- Points 1–4 are repeated, but in this case leaving out, one by one, all the MS patient eyes.
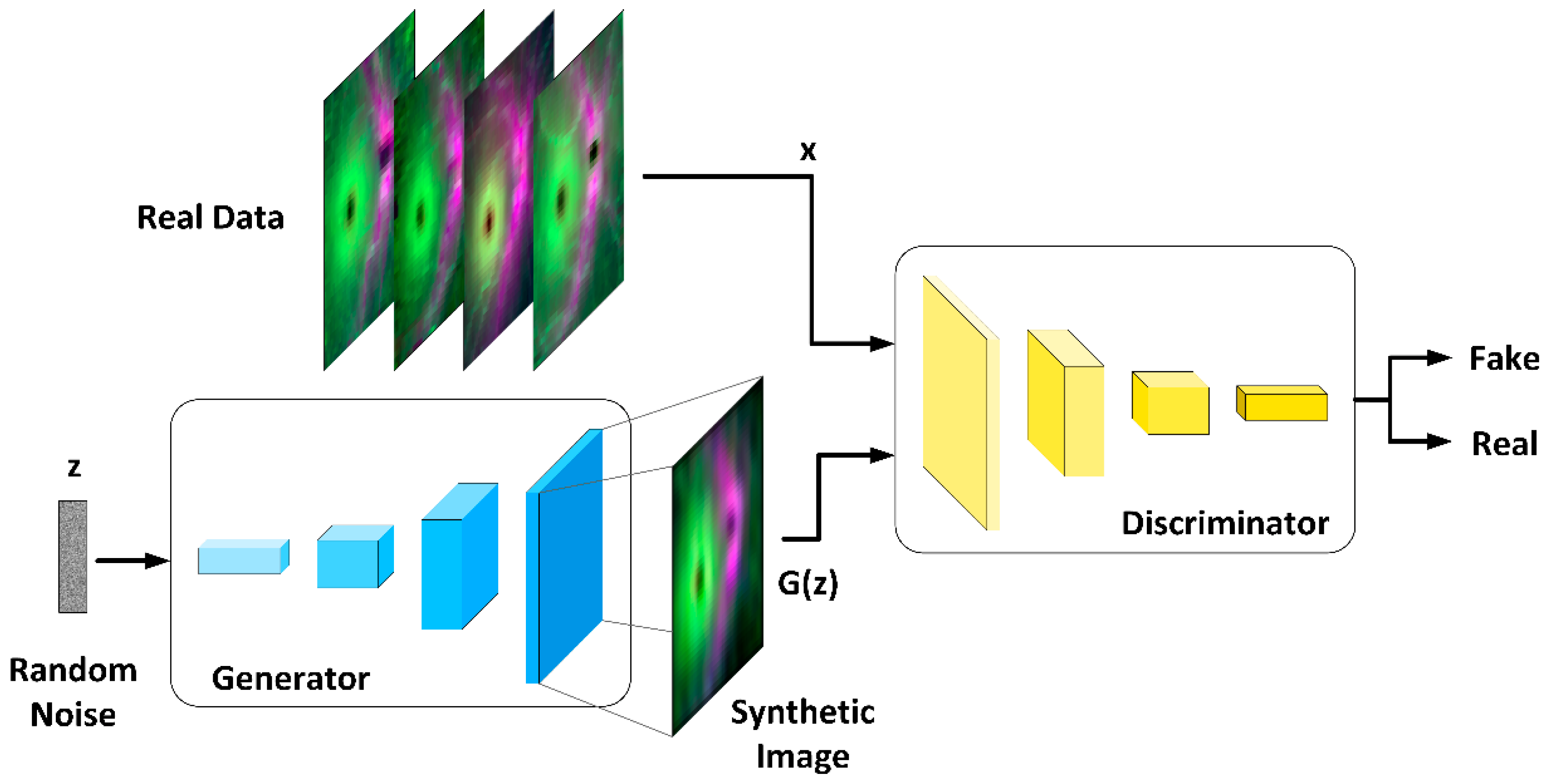
2.6. OCT Data Augmentation
2.6.1. Generator Architecture
2.6.2. Discriminator Architecture
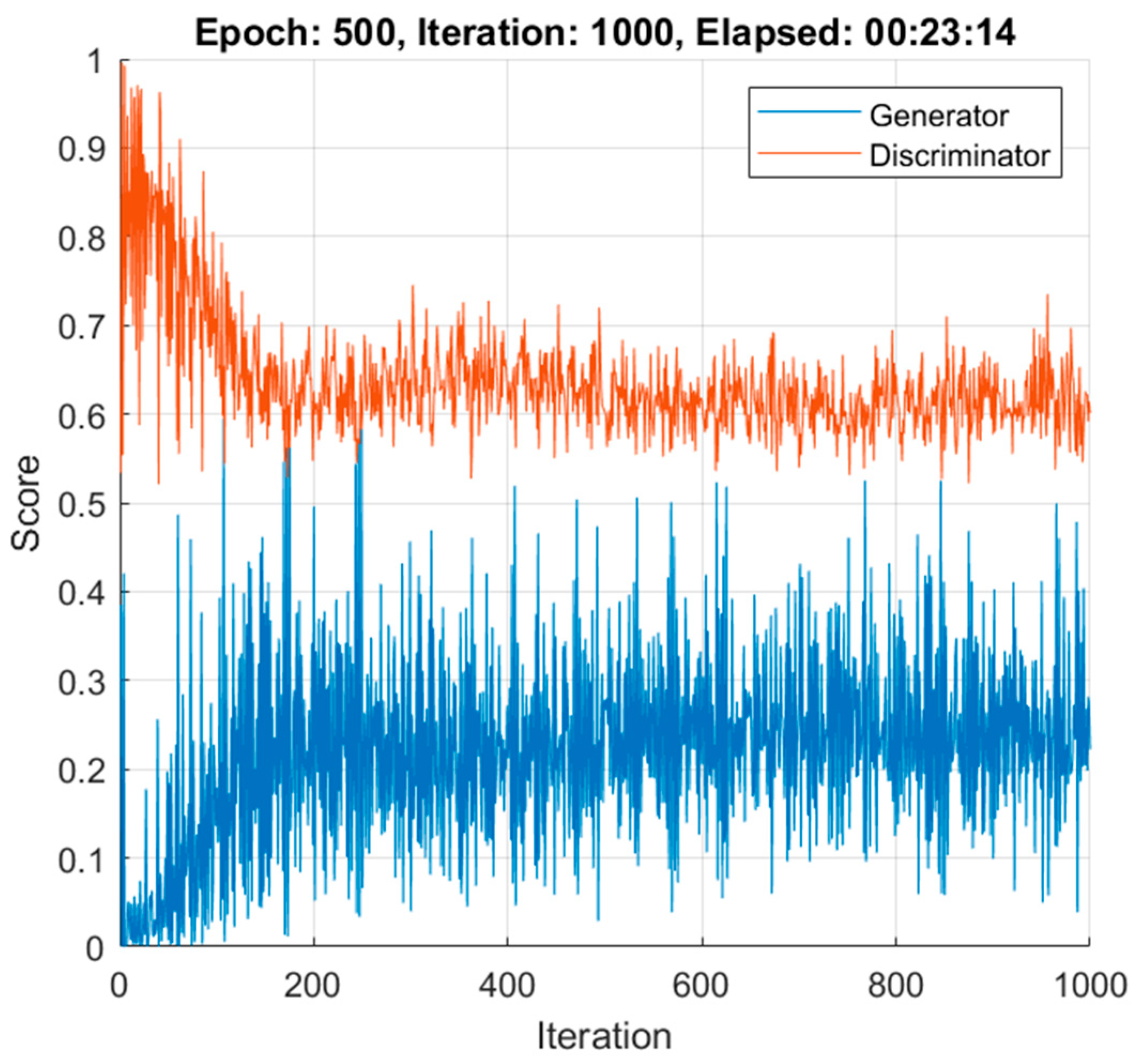
2.6.3. DCGAN Training
3. Results
3.1. Database
3.2. OCT Image Pre-Processing
3.3. Data Augmentation
3.4. Classification Results
3.5. Running Time Evaluation
- Generation of n = 100 synthetic patient images + 100 synthetic control images: 23 min.
- CNN training (147 control images, 148 patient images): 25 min.
- Testing of a single control subject’s images: 0.2 s.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Salem, M.; Valverde, S.; Cabezas, M.; Pareto, D.; Oliver, A.; Salvi, J.; Rovira, A.; Llado, X. Multiple Sclerosis Lesion Synthesis in MRI Using an Encoder-Decoder U-NET. IEEE Access 2019, 7, 25171–25184. [Google Scholar] [CrossRef]
- McKinley, R.; Wepfer, R.; Grunder, L.; Aschwanden, F.; Fischer, T.; Friedli, C.; Muri, R.; Rummel, C.; Verma, R.; Weisstanner, C.; et al. Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence. NeuroImage Clin. 2020, 25, 102104. [Google Scholar] [CrossRef] [PubMed]
- Marzullo, A.; Kocevar, G.; Stamile, C.; Durand-Dubief, F.; Terracina, G.; Calimeri, F.; Sappey-Marinier, D. Classification of Multiple Sclerosis Clinical Profiles via Graph Convolutional Neural Networks. Front. Neurosci. 2019, 13, 594. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; de Vos, B.D.; Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef] [PubMed]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J. Magn. Reson. Imaging 2019, 49, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 051001. [Google Scholar] [CrossRef]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef]
- Gautam, R.; Sharma, M. Prevalence and Diagnosis of Neurological Disorders Using Different Deep Learning Techniques: A Meta-Analysis. J. Med. Syst. 2020, 44, 49. [Google Scholar] [CrossRef]
- Khare, S.K.; Bajaj, V. Time-Frequency Representation and Convolutional Neural Network-Based Emotion Recognition. IEEE Trans. Neural Netw. Learn. Syst. 2020, 1–9. [Google Scholar] [CrossRef]
- Takahashi, H.; Emami, A.; Shinozaki, T.; Kunii, N.; Matsuo, T.; Kawai, K. Convolutional neural network with autoencoder-assisted multiclass labelling for seizure detection based on scalp electroencephalography. Comput. Biol. Med. 2020, 125, 104016. [Google Scholar] [CrossRef]
- Ganapathy, N.; Rao Veeranki, Y.; Swaminathan, R. Convolutional Neural Network based Emotion Classification using Electrodermal Activity Signals and Time-Frequency Features. Expert Syst. Appl. 2020, 159, 113571. [Google Scholar] [CrossRef]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG Arrhythmia Classification Using STFT-Based Spectrogram and Convolutional Neural Network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Panda, R.; Jain, S.; Tripathy, R.; Acharya, U.R. Detection of shockable ventricular cardiac arrhythmias from ECG signals using FFREWT filter-bank and deep convolutional neural network. Comput. Biol. Med. 2020, 124, 103939. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Peng, L.; Varadarajan, A.V.; Keane, P.A.; Burlina, P.M.; Chiang, M.F.; Schmetterer, L.; Pasquale, L.R.; Bressler, N.M.; Webster, D.R.; et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog. Retin. Eye Res. 2019, 72, 100759. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef]
- Wen, J.C.; Lee, C.S.; Keane, P.A.; Xiao, S.; Rokem, A.S.; Chen, P.P.; Wu, Y.; Lee, A.Y. Forecasting future Humphrey Visual Fields using deep learning. PLoS ONE 2019, 14, e0214875. [Google Scholar] [CrossRef]
- Raman, R.; Srinivasan, S.; Virmani, S.; Sivaprasad, S.; Rao, C.; Rajalakshmi, R. Fundus photograph-based deep learning algorithms in detecting diabetic retinopathy. Eye 2019, 33, 97–109. [Google Scholar] [CrossRef]
- Russakoff, D.B.; Lamin, A.; Oakley, J.D.; Dubis, A.M.; Sivaprasad, S. Deep Learning for Prediction of AMD Progression: A Pilot Study. Investig. Opthalmol. Vis. Sci. 2019, 60, 712. [Google Scholar] [CrossRef]
- Brown, J.M.; Kalpathy-Cramer, J.; Campbell, J.P.; Beers, A.; Chang, K.; Ostmo, S.; Chan, R.V.P.; Erdogmus, D.; Ioannidis, S.; Chiang, M.F.; et al. Fully automated disease severity assessment and treatment monitoring in retinopathy of prematurity using deep learning. In Proceedings of the Medical Imaging 2018: Imaging Informatics for Healthcare, Research, and Applications, SPIE Medical Imaging, Houston, TX, USA, 13–15 February 2018; Volume 10579, p. 22. [Google Scholar]
- Kihara, Y.; Heeren, T.F.C.; Lee, C.S.; Wu, Y.; Xiao, S.; Tzaridis, S.; Holz, F.G.; Charbel Issa, P.; Egan, C.A.; Lee, A.Y. Estimating Retinal Sensitivity Using Optical Coherence Tomography With Deep-Learning Algorithms in Macular Telangiectasia Type 2. JAMA Netw. Open 2019, 2, e188029. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106–154. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Alonso, R.; Gonzalez-Moron, D.; Garcea, O. Optical coherence tomography as a biomarker of neurodegeneration in multiple sclerosis: A review. Mult. Scler. Relat. Disord. 2018, 22, 77–82. [Google Scholar] [CrossRef]
- Saxena, S.; Caprnda, M.; Ruia, S.; Prasad, S.; Ankita; Fedotova, J.; Kruzliak, P.; Krasnik, V. Spectral domain optical coherence tomography based imaging biomarkers for diabetic retinopathy. Endocrine 2019, 66, 509–516. [Google Scholar] [CrossRef]
- Rolle, T.; Dallorto, L.; Bonetti, B. Retinal and macular ganglion cell count estimated with optical coherence tomography RTVUE-100 as a candidate biomarker for glaucoma. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5772–5779. [Google Scholar] [CrossRef]
- Chrysou, A.; Jansonius, N.M.; van Laar, T. Retinal layers in Parkinson’s disease: A meta-analysis of spectral-domain optical coherence tomography studies. Park. Relat. Disord. 2019, 64, 40–49. [Google Scholar] [CrossRef]
- Cavaliere, C.; Vilades, E.; Alonso-Rodríguez, M.C.; Rodrigo, M.J.; Pablo, L.E.; Miguel, J.M.; López-Guillén, E.; Morla, E.M.S.; Boquete, L.; Garcia-Martin, E. Computer-Aided Diagnosis of Multiple Sclerosis Using a Support Vector Machine and Optical Coherence Tomography Features. Sensors 2019, 19, 5323. [Google Scholar] [CrossRef]
- Garcia-Martin, E.; Ortiz, M.; Boquete, L.; Sánchez-Morla, E.M.; Barea, R.; Cavaliere, C.; Vilades, E.; Orduna, E.; Rodrigo, M.J. Early diagnosis of multiple sclerosis by OCT analysis using Cohen’s d method and a neural network as classifier. Comput. Biol. Med. 2021, 129, 104165. [Google Scholar] [CrossRef]
- Sánchez-Morla, E.M.; Fuentes, J.L.; Miguel-Jiménez, J.M.; Boquete, L.; Ortiz, M.; Orduna, E.; Satue, M.; Garcia-Martin, E. Automatic Diagnosis of Bipolar Disorder Using Optical Coherence Tomography Data and Artificial Intelligence. J. Pers. Med. 2021, 11, 803. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S. Impact of swept source optical coherence tomography on ophthalmology. Taiwan J. Ophthalmol. 2016, 6, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Pekala, M.; Joshi, N.; Liu, T.Y.A.; Bressler, N.M.; DeBuc, D.C.; Burlina, P. Deep learning based retinal OCT segmentation. Comput. Biol. Med. 2019, 114, 103445. [Google Scholar] [CrossRef] [PubMed]
- Masood, S.; Fang, R.; Li, P.; Li, H.; Sheng, B.; Mathavan, A.; Wang, X.; Yang, P.; Wu, Q.; Qin, J.; et al. Automatic Choroid Layer Segmentation from Optical Coherence Tomography Images Using Deep Learning. Sci. Rep. 2019, 9, 3058. [Google Scholar] [CrossRef]
- He, Y.; Carass, A.; Liu, Y.; Jedynak, B.M.; Solomon, S.D.; Saidha, S.; Calabresi, P.A.; Prince, J.L. Deep learning based topology guaranteed surface and MME segmentation of multiple sclerosis subjects from retinal OCT. Biomed. Opt. Express 2019, 10, 5042. [Google Scholar] [CrossRef]
- Photocoagulation for Diabetic Macular Edema. Arch. Ophthalmol. 1985, 103, 1796. [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Puntmann, V.O. How-to guide on biomarkers: Biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad. Med. J. 2009, 85, 538–545. [Google Scholar] [CrossRef]
- Solomon, A.J.; Pettigrew, R.; Naismith, R.T.; Chahin, S.; Krieger, S.; Weinshenker, B. Challenges in multiple sclerosis diagnosis: Misunderstanding and misapplication of the McDonald criteria. Mult. Scler. J. 2021, 27, 250–258. [Google Scholar] [CrossRef]
- Kaisey, M.; Solomon, A.J.; Luu, M.; Giesser, B.S.; Sicotte, N.L. Incidence of multiple sclerosis misdiagnosis in referrals to two academic centers. Mult. Scler. Relat. Disord. 2019, 30, 51–56. [Google Scholar] [CrossRef]
- Midaglia, L.; Sastre-Garriga, J.; Pappolla, A.; Quibus, L.; Carvajal, R.; Vidal-Jordana, A.; Arrambide, G.; Río, J.; Comabella, M.; Nos, C.; et al. The frequency and characteristics of MS misdiagnosis in patients referred to the multiple sclerosis centre of Catalonia. Mult. Scler. J. 2021, 27, 913–921. [Google Scholar] [CrossRef]
- Mobasheri, F.; Jaberi, A.R.; Hasanzadeh, J.; Fararouei, M. Multiple sclerosis diagnosis delay and its associated factors among Iranian patients. Clin. Neurol. Neurosurg. 2020, 199, 106278. [Google Scholar] [CrossRef]
- Patti, F.; Chisari, C.G.; Arena, S.; Toscano, S.; Finocchiaro, C.; Fermo, S.L.; Judica, M.L.; Maimone, D. Factors driving delayed time to multiple sclerosis diagnosis: Results from a population-based study. Mult. Scler. Relat. Disord. 2021, 103361. [Google Scholar] [CrossRef]
- Thompson, A.J.; Baranzini, S.E.; Geurts, J.; Hemmer, B.; Ciccarelli, O. Multiple sclerosis. Lancet 2018, 391, 1622–1636. [Google Scholar] [CrossRef]
- Miguel, J.M.; Roldán, M.; Pérez-Rico, C.; Ortiz, M.; Boquete, L.; Blanco, R. Using advanced analysis of multifocal visual-evoked potentials to evaluate the risk of clinical progression in patients with radiologically isolated syndrome. Sci. Rep. 2021, 11, 2036. [Google Scholar] [CrossRef]
- Pinto, M.F.; Oliveira, H.; Batista, S.; Cruz, L.; Pinto, M.; Correia, I.; Martins, P.; Teixeira, C. Prediction of disease progression and outcomes in multiple sclerosis with machine learning. Sci. Rep. 2020, 10, 21038. [Google Scholar] [CrossRef]
- De Brouwer, E.; Becker, T.; Moreau, Y.; Havrdova, E.K.; Trojano, M.; Eichau, S.; Ozakbas, S.; Onofrj, M.; Grammond, P.; Kuhle, J.; et al. Longitudinal machine learning modeling of MS patient trajectories improves predictions of disability progression. Comput. Methods Programs Biomed. 2021, 208, 106180. [Google Scholar] [CrossRef]
- Montolío, A.; Martín-Gallego, A.; Cegoñino, J.; Orduna, E.; Vilades, E.; Garcia-Martin, E.; Palomar, A.P. del Machine learning in diagnosis and disability prediction of multiple sclerosis using optical coherence tomography. Comput. Biol. Med. 2021, 133, 104416. [Google Scholar] [CrossRef]
- Parisi, V.; Manni, G.; Spadaro, M.; Colacino, G.; Restuccia, R.; Marchi, S.; Bucci, M.G.; Pierelli, F. Correlation between morphological and functional retinal impairment in multiple sclerosis patients. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2520–2527. [Google Scholar]
- Petzold, A.; Balcer, L.J.; Calabresi, P.A.; Costello, F.; Frohman, T.C.; Frohman, E.M.; Martinez-Lapiscina, E.H.; Green, A.J.; Kardon, R.; Outteryck, O.; et al. Retinal layer segmentation in multiple sclerosis: A systematic review and meta-analysis. Lancet Neurol. 2017, 16, 797–812. [Google Scholar] [CrossRef]
- Britze, J.; Pihl-Jensen, G.; Frederiksen, J.L. Retinal ganglion cell analysis in multiple sclerosis and optic neuritis: A systematic review and meta-analysis. J. Neurol. 2017, 264, 1837–1853. [Google Scholar] [CrossRef]
- Garcia-Martin, E.; Ara, J.R.; Martin, J.; Almarcegui, C.; Dolz, I.; Vilades, E.; Gil-Arribas, L.; Fernandez, F.J.; Polo, V.; Larrosa, J.M.; et al. Retinal and Optic Nerve Degeneration in Patients with Multiple Sclerosis Followed up for 5 Years. Ophthalmology 2017, 124, 688–696. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems—Volume 2, Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; pp. 2672–2680. [Google Scholar]
- Alqahtani, H.; Kavakli-Thorne, M.; Kumar, G. Applications of Generative Adversarial Networks (GANs): An Updated Review. Arch. Comput. Methods Eng. 2021, 28, 525–552. [Google Scholar] [CrossRef]
- Aggarwal, A.; Mittal, M.; Battineni, G. Generative adversarial network: An overview of theory and applications. Int. J. Inf. Manag. Data Insights 2021, 1, 100004. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative Adversarial Networks: An Overview. IEEE Signal. Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Yang, W.; Bi, F.; Yu, F.R. A State-of-the-Art Review on Image Synthesis With Generative Adversarial Networks. IEEE Access 2020, 8, 63514–63537. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Sorin, V.; Barash, Y.; Konen, E.; Klang, E. Creating Artificial Images for Radiology Applications Using Generative Adversarial Networks (GANs)—A Systematic Review. Acad. Radiol. 2020, 27, 1175–1185. [Google Scholar] [CrossRef]
- Lashgari, E.; Liang, D.; Maoz, U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods 2020, 346, 108885. [Google Scholar] [CrossRef]
- Mousavi, Z.; Yousefi Rezaii, T.; Sheykhivand, S.; Farzamnia, A.; Razavi, S.N. Deep convolutional neural network for classification of sleep stages from single-channel EEG signals. J. Neurosci. Methods 2019, 324, 108312. [Google Scholar] [CrossRef]
- Wulan, N.; Wang, W.; Sun, P.; Wang, K.; Xia, Y.; Zhang, H. Generating electrocardiogram signals by deep learning. Neurocomputing 2020, 404, 122–136. [Google Scholar] [CrossRef]
- Barile, B.; Marzullo, A.; Stamile, C.; Durand-Dubief, F.; Sappey-Marinier, D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Comput. Methods Programs Biomed. 2021, 206, 106113. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, F.; Yu, T.; Barquero, G.; Thiran, J.-P.; Granziera, C.; Bach Cuadra, M. MPRAGE to MP2RAGE UNI translation via generative adversarial network improves the automatic tissue and lesion segmentation in multiple sclerosis patients. Comput. Biol. Med. 2021, 132, 104297. [Google Scholar] [CrossRef] [PubMed]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic criteria for multiple sclerosis: 2010 Revisions to the McDonald criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef]
- Petzold, A.; Albrecht, P.; Balcer, L.; Bekkers, E.; Brandt, A.U.; Calabresi, P.A.; Deborah, O.G.; Graves, J.S.; Green, A.; Keane, P.A.; et al. Artificial intelligence extension of the OSCAR-IB criteria. Ann. Clin. Transl. Neurol. 2021, acn3.51320. [Google Scholar] [CrossRef]
- Cruz-Herranz, A.; Balk, L.J.; Oberwahrenbrock, T.; Saidha, S.; Martinez-Lapiscina, E.H.; Lagreze, W.A.; Schuman, J.S.; Villoslada, P.; Calabresi, P.; Balcer, L.; et al. The APOSTEL recommendations for reporting quantitative optical coherence tomography studies. Neurology 2016, 86, 2303–2309. [Google Scholar] [CrossRef]
- Cohen, J. The statistical power of abnormal-social psychological research: A review. J. Abnorm. Soc. Psychol. 1962, 65, 145–153. [Google Scholar] [CrossRef]
- Shore, J.; Johnson, R. Properties of cross-entropy minimization. IEEE Trans. Inf. Theory 1981, 27, 472–482. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2014. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Dumoulin, V.; Visin, F. A guide to convolution arithmetic for deep learning. arXiv 2018, arXiv:1603.07285v2. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning ICML, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Noordzij, M.; Dekker, F.W.; Zoccali, C.; Jager, K.J. Sample Size Calculations. Nephron Clin. Pract. 2011, 118, c319–c323. [Google Scholar] [CrossRef]
- López-Dorado, A.; Pérez, J.; Rodrigo, M.J.; Miguel-Jiménez, J.M.; Ortiz, M.; de Santiago, L.; López-Guillén, E.; Blanco, R.; Cavalliere, C.; Morla, E.M.S.; et al. Diagnosis of multiple sclerosis using multifocal ERG data feature fusion. Inf. Fusion 2021, 76, 157–167. [Google Scholar] [CrossRef]
- de Santiago, L.; del Castillo, M.O.; Garcia-Martin, E.; Rodrigo, M.J.; Morla, E.M.S.; Cavaliere, C.; Cordón, B.; Miguel, J.M.; López, A.; Boquete, L. Empirical mode decomposition-based filter applied to multifocal electroretinograms in multiple sclerosis diagnosis. Sensors 2020, 20, 7. [Google Scholar] [CrossRef]
- Ford, H. Clinical presentation and diagnosis of multiple sclerosis. Clin. Med. 2020, 20, 380–383. [Google Scholar] [CrossRef]
- Fjeldstad, A.S.; Carlson, N.G.; Rose, J.W. Optical coherence tomography as a biomarker in multiple sclerosis. Expert Opin. Med. Diagn. 2012, 6, 593–604. [Google Scholar] [CrossRef]
- Vermersch, P.; Outteryck, O.; Petzold, A. Optical Coherence Tomography—A New Monitoring Tool for Multiple Sclerosis? Eur. Neurol. Rev. 2010, 5, 73. [Google Scholar] [CrossRef]
- Petzold, A.; de Boer, J.F.; Schippling, S.; Vermersch, P.; Kardon, R.; Green, A.; Calabresi, P.A.; Polman, C. Optical coherence tomography in multiple sclerosis: A systematic review and meta-analysis. Lancet Neurol. 2010, 9, 921–932. [Google Scholar] [CrossRef]
- Hu, Y.-Q.; Yu, Y. A technical view on neural architecture search. Int. J. Mach. Learn. Cybern. 2020, 11, 795–811. [Google Scholar] [CrossRef]




| Actual MS | Actual Control | |
|---|---|---|
| Predict MS | TP = 48 | FP = 0 |
| Predict control | FN = 0 | TN = 48 |
| Method | Confusion Matrix Results | ||||
|---|---|---|---|---|---|
| TN | FP | FN | TP | Accuracy | |
| Average thicknesses. Gaussian SVM [30] | 44 | 5 | 43 | 4 | 0.90 |
| Wide protocol. Cohen’s d. Linear SVM Classifier [31] | 41 | 7 | 7 | 41 | 0.85 |
| Wide protocol. Cohen’s d. Quadratic SVM Classifier [31] | 40 | 8 | 6 | 42 | 0.83 |
| Wide protocol. Cohen’s d. Cubic SVM Classifier [31] | 38 | 10 | 5 | 43 | 0.79 |
| Wide protocol. Cohen’s d. Fine Gaussian SVM Classifier [31] | 43 | 5 | 29 | 19 | 0.89 |
| Wide protocol. Cohen’s d. Medium Gaussian SVM Classifier [31] | 41 | 7 | 6 | 42 | 0.85 |
| Wide protocol. Cohen’s d. Coarse Gaussian SVM Classifier [31] | 36 | 12 | 6 | 42 | 0.75 |
| Wide protocol. Cohen’s d. FFNN 5 neurons hidden layer [31] | 46 | 2 | 5 | 43 | 0.95 |
| Wide protocol. Cohen’s d. FFNN 10 neurons hidden layer [31] | 47 | 1 | 1 | 47 | 0.98 |
| Wide protocol. Cohen’s d. FFNN 15 neurons hidden layer [31] | 47 | 1 | 2 | 46 | 0.97 |
| Wide protocol. Cohen’s d. Convolutional Neural Network | 48 | 0 | 0 | 48 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Dorado, A.; Ortiz, M.; Satue, M.; Rodrigo, M.J.; Barea, R.; Sánchez-Morla, E.M.; Cavaliere, C.; Rodríguez-Ascariz, J.M.; Orduna-Hospital, E.; Boquete, L.; et al. Early Diagnosis of Multiple Sclerosis Using Swept-Source Optical Coherence Tomography and Convolutional Neural Networks Trained with Data Augmentation. Sensors 2022, 22, 167. https://doi.org/10.3390/s22010167
López-Dorado A, Ortiz M, Satue M, Rodrigo MJ, Barea R, Sánchez-Morla EM, Cavaliere C, Rodríguez-Ascariz JM, Orduna-Hospital E, Boquete L, et al. Early Diagnosis of Multiple Sclerosis Using Swept-Source Optical Coherence Tomography and Convolutional Neural Networks Trained with Data Augmentation. Sensors. 2022; 22(1):167. https://doi.org/10.3390/s22010167
Chicago/Turabian StyleLópez-Dorado, Almudena, Miguel Ortiz, María Satue, María J. Rodrigo, Rafael Barea, Eva M. Sánchez-Morla, Carlo Cavaliere, José M. Rodríguez-Ascariz, Elvira Orduna-Hospital, Luciano Boquete, and et al. 2022. "Early Diagnosis of Multiple Sclerosis Using Swept-Source Optical Coherence Tomography and Convolutional Neural Networks Trained with Data Augmentation" Sensors 22, no. 1: 167. https://doi.org/10.3390/s22010167
APA StyleLópez-Dorado, A., Ortiz, M., Satue, M., Rodrigo, M. J., Barea, R., Sánchez-Morla, E. M., Cavaliere, C., Rodríguez-Ascariz, J. M., Orduna-Hospital, E., Boquete, L., & Garcia-Martin, E. (2022). Early Diagnosis of Multiple Sclerosis Using Swept-Source Optical Coherence Tomography and Convolutional Neural Networks Trained with Data Augmentation. Sensors, 22(1), 167. https://doi.org/10.3390/s22010167






