Reverse Scan Conversion and Efficient Deep Learning Network Architecture for Ultrasound Imaging on a Mobile Device
Abstract
1. Introduction
2. Methods
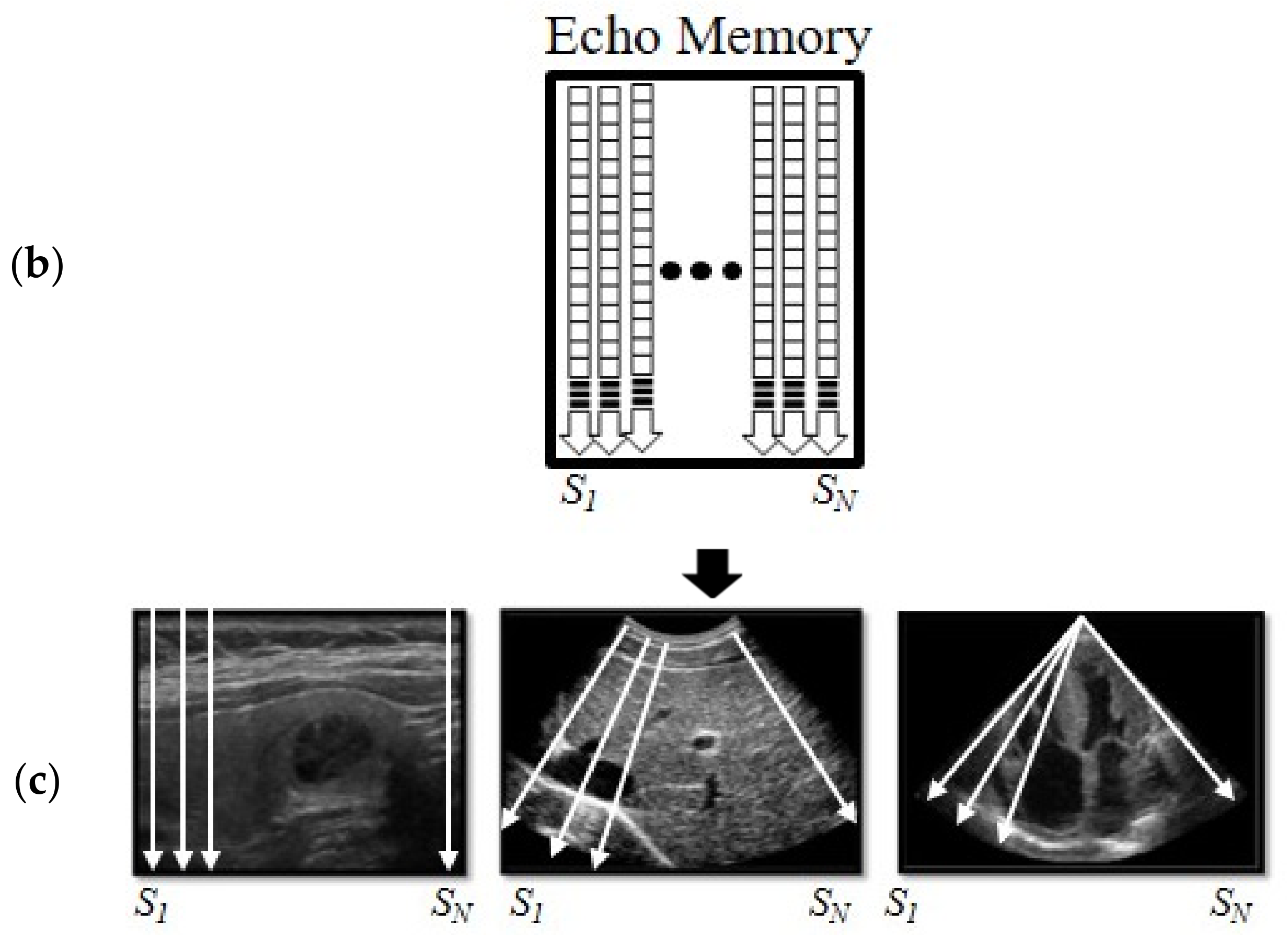
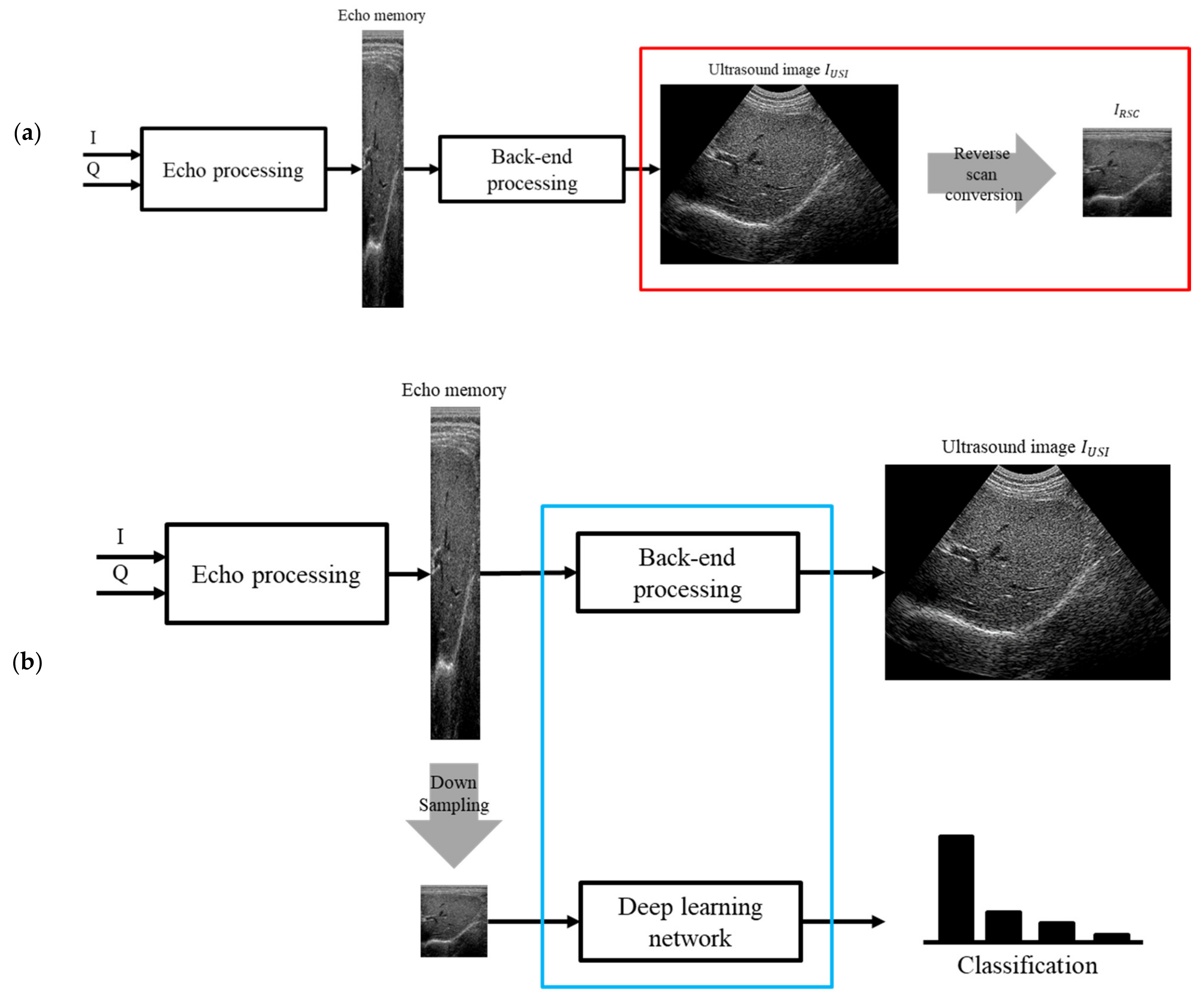
2.1. Reverse Scan Conversion
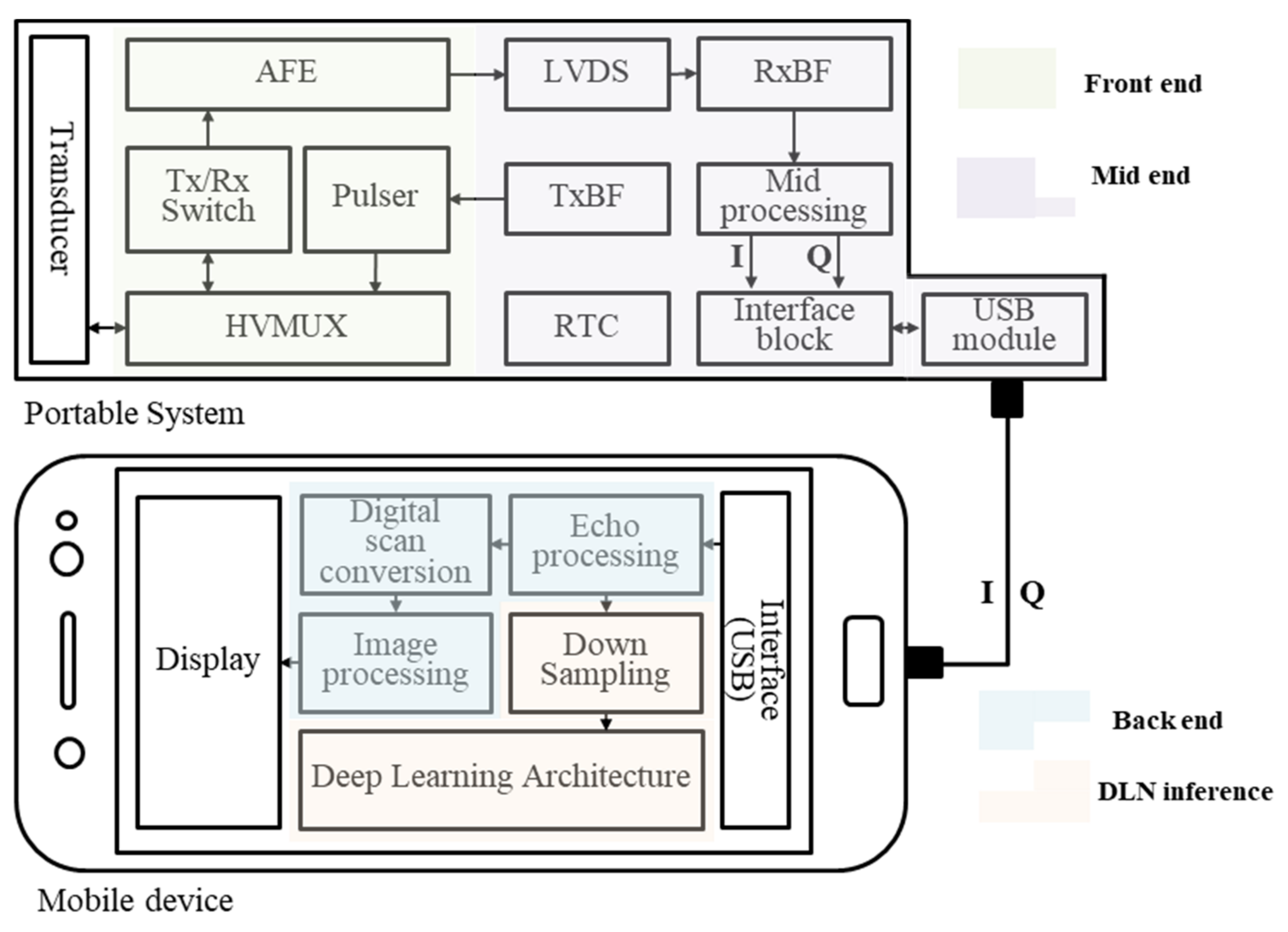
2.2. Structure on a Mobile Device–Frame Asynchronous Classification (FAC)
3. Experiments
3.1. Data
3.2. Embedded System
3.3. Network
4. Results
4.1. RSC without FOV Dependency
4.2. Structure for Real-Time Processing
5. Discussions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, C.L.; Copel, J.A. Point-of-care ultrasonography. N. Eng. J. Med. 2011, 364, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Ailon, J.; Mourad, O.; Nadjafi, M.; Cavalcanti, R. Point-of-care ultrasound as a competency for general internists: A survey of internal medicine training programs in Canada. Can. Med. Educ. J. 2016, 7, e51. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.C.; Rao, V.M.; Parker, L.; Frangos, A.J. Noncardiac point-of-care ultrasound by nonradiologist physicians: How widespread is it? J. Am. Coll. Radiol. 2011, 8, 772–775. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhang, F.; Li, X. Machine learning in ultrasound computer-aided diagnostic systems: A survey. BioMed Res. Int. 2018, 2018, 5137904. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Fletcher, T.P.; Smith, S.; Koch, L.M.; Kainz, B.; Rueckert, D. SonoNet: Real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans. Med. Imaging 2017, 36, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Sermanet, P.; Eigen, D.; Zhang, X.; Mathieu, M.; Fergus, R.; LeCun, Y. Overfeat: Integrated recognition, localization and detection using convolutional networks. In Proceedings of the 2nd International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing properties of neural networks. In Proceedings of the 2nd International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- Cheng, P.M.; Malhi, H.S. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. J. Digit. Imaging 2017, 30, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Huo, Y.; Park, J.; Landman, B.; Milkowski, A.; Grbic, S.; Zhou, S. Less is more: Simultaneous view classification and landmark detection for abdominal ultrasound images. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 711–719. [Google Scholar]
- Guo, T. Cloud-based or on-device: An empirical study of mobile deep inference. In Proceedings of the 2018 IEEE International Conference on Cloud Engineering (IC2E), San Francisco, CA, USA, 2–7 July 2018; pp. 184–190. [Google Scholar]
- Song, I.; Kim, H.-J.; Jeon, P.B. Deep learning for real-time robust facial expression recognition on a smartphone. In Proceedings of the 2014 IEEE International Conference on Consumer Electronics (ICCE), Taipei, Taiwan, 26–28 May 2014; pp. 564–567. [Google Scholar]
- Hosseini, M.-P.; Soltanian-Zadeh, H.; Elisevich, K.; Pompili, D. Cloud-based deep learning of big EEG data for epileptic seizure prediction. In Proceedings of the 2016 IEEE Global Conference on Signal and Information Processing, Greater Washington, DC, USA, 7–9 December 2016; pp. 1151–1155. [Google Scholar]
- Center, P.R. Internet seen as positive influence on education but negative on morality in emerging and developing nations. Pew Res. Cent. 2015, 13–20. [Google Scholar]
- Hon, W.K.; Millard, C.; Walden, I. The problem of ‘personal data’ in cloud computing: What information is regulated?—The cloud of unknowing. Int. Data Priv. Law 2011, 1, 211–228. [Google Scholar] [CrossRef]
- Liu, L.; Li, H.; Gruteser, M. Edge assisted real-time object detection for mobile augmented reality. In Proceedings of the 25th Annual International Conference on Mobile Computing and Networking, Los Cabos, Mexico, 21–25 October 2019; pp. 1–16. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Liu, J.Y.; Xu, J.; Forsberg, F.; Liu, J.-B. CMUT/CMOS-based butterfly IQ-A portable personal sonoscope. Adv. Ultrasound Diagn. Ther. 2019, 3, 115–118. [Google Scholar]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit. Med. 2018, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Szabo, T.L. Diagnostic Ultrasound Imaging: Inside Out; Academic Press: Cambridge, MA, USA, 2004. [Google Scholar]
- McGahan, J.P.; Goldberg, B.B. Diagnostic Ultrasound: A Logical Approach; LWW: Philadelphia, PA, USA, 1998. [Google Scholar]
- Kim, J.H.; Yeo, S.; Kim, M.; Kye, S.; Lee, Y.; Song, T. A smart-phone based portable ultrasound imaging system for point-of-care applications. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; pp. 1–5. [Google Scholar]
- Angelsen, B.A.; Torp, H.; Holm, S.; Kristoffersen, K.; Whittingham, T.A. Which transducer array is best? Eur. J. Ultrasound 1995, 2, 151–164. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E. Use of the hough transformation to detect lines and curves in pictures. Commun. ACM 1972, 15, 11–15. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Bonet-Carne, E.; Eixarch, E.; Crispi, F.; Gratacós, E. Evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes. Sci. Rep. 2020, 10, 10200. [Google Scholar] [CrossRef] [PubMed]
- Palmer, P.E. Manual of Diagnostic Ultrasound; World Health Organization: Geneva, Switzerland, 1995. [Google Scholar]
- Shreiner, D.; Sellers, G.; Kessenich, J.; Licea-Kane, B. OpenGL Programming Guide: The Official Guide to Learning OpenGL, Version 4.3; Addison-Wesley: Boston, MA, USA, 2013. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Chollet, F. Keras. Available online: https://github.com/keras-team/keras (accessed on 8 April 2021).
- Wan, L.; Zeiler, M.; Zhang, S.; Le Cun, Y.; Fergus, R. Regularization of neural networks using dropconnect. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. 1058–1066. [Google Scholar]
- Stocksley, M. Abdominal Ultrasound; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]







| Parameters | Calculations | |
|---|---|---|
| AlexNet | 7.1 M | 305.3 M |
| ShallowNet | 2.1 M | 22.2 M |
| MobileNet | 3.2 M | 44.3 M |
| Xception | 16.0 M | 25.7 M |
| USI | RSC | |||||
|---|---|---|---|---|---|---|
| AC | SE | SP | AC | SE | SP | |
| AlexNet | 93.90 | 94.29 | 96.99 | 98.55 | 95.94 | 97.90 |
| MobileNet | 62.72 | 51.99 | 76.54 | 68.73 | 55.56 | 78.72 |
| ShallowNet | 84.00 | 50.65 | 76.42 | 94.17 | 73.62 | 87.47 |
| Xception | 88.96 | 67.42 | 84.34 | 93.69 | 74.33 | 84.32 |
| Average | 85.53 | 66.09 | 83.57 | 88.79 | 74.86 | 87.10 |
| Processing Time (ms) | #Dividing | ||
|---|---|---|---|
| CP | AlexNet | 447 | 22 |
| MobileNet | 43 | 2 | |
| ShallowNet | 49 | 2 | |
| Xception | 61 | 3 | |
| IRP | 4 | (not divided) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Kim, M.; Lim, C.; Song, T.-K. Reverse Scan Conversion and Efficient Deep Learning Network Architecture for Ultrasound Imaging on a Mobile Device. Sensors 2021, 21, 2629. https://doi.org/10.3390/s21082629
Lee K, Kim M, Lim C, Song T-K. Reverse Scan Conversion and Efficient Deep Learning Network Architecture for Ultrasound Imaging on a Mobile Device. Sensors. 2021; 21(8):2629. https://doi.org/10.3390/s21082629
Chicago/Turabian StyleLee, Kunkyu, Min Kim, Changhyun Lim, and Tai-Kyong Song. 2021. "Reverse Scan Conversion and Efficient Deep Learning Network Architecture for Ultrasound Imaging on a Mobile Device" Sensors 21, no. 8: 2629. https://doi.org/10.3390/s21082629
APA StyleLee, K., Kim, M., Lim, C., & Song, T.-K. (2021). Reverse Scan Conversion and Efficient Deep Learning Network Architecture for Ultrasound Imaging on a Mobile Device. Sensors, 21(8), 2629. https://doi.org/10.3390/s21082629






