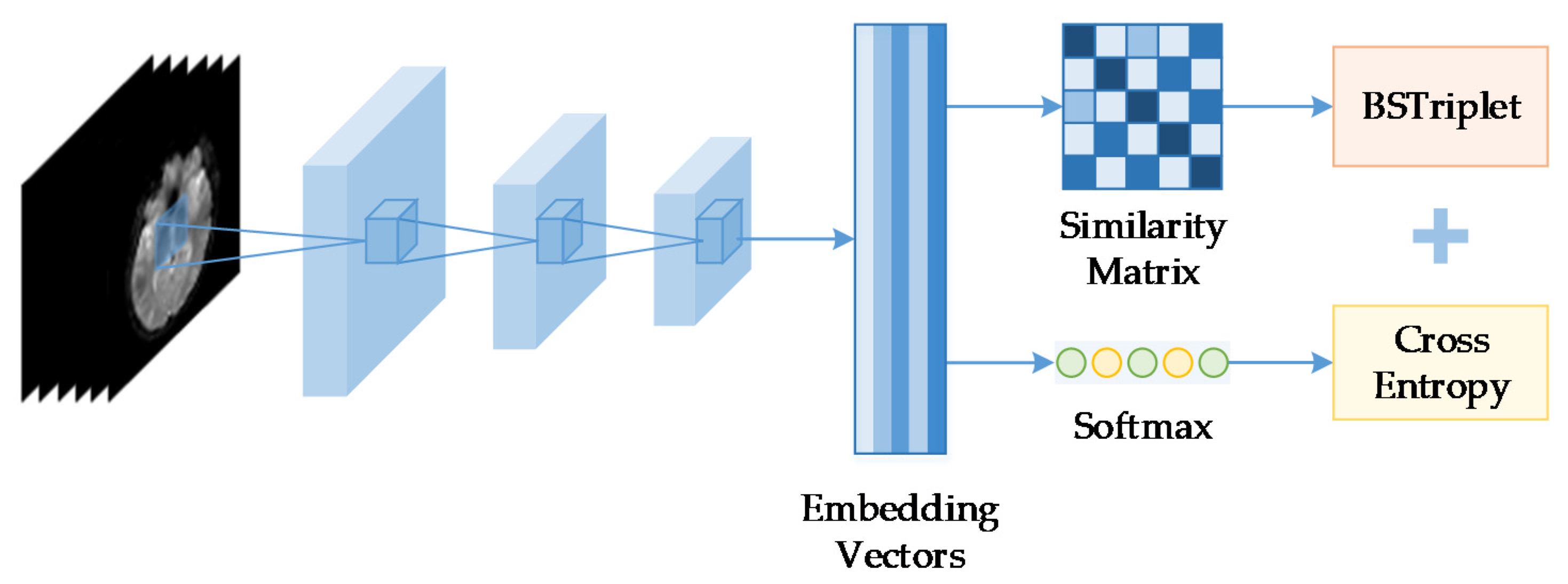
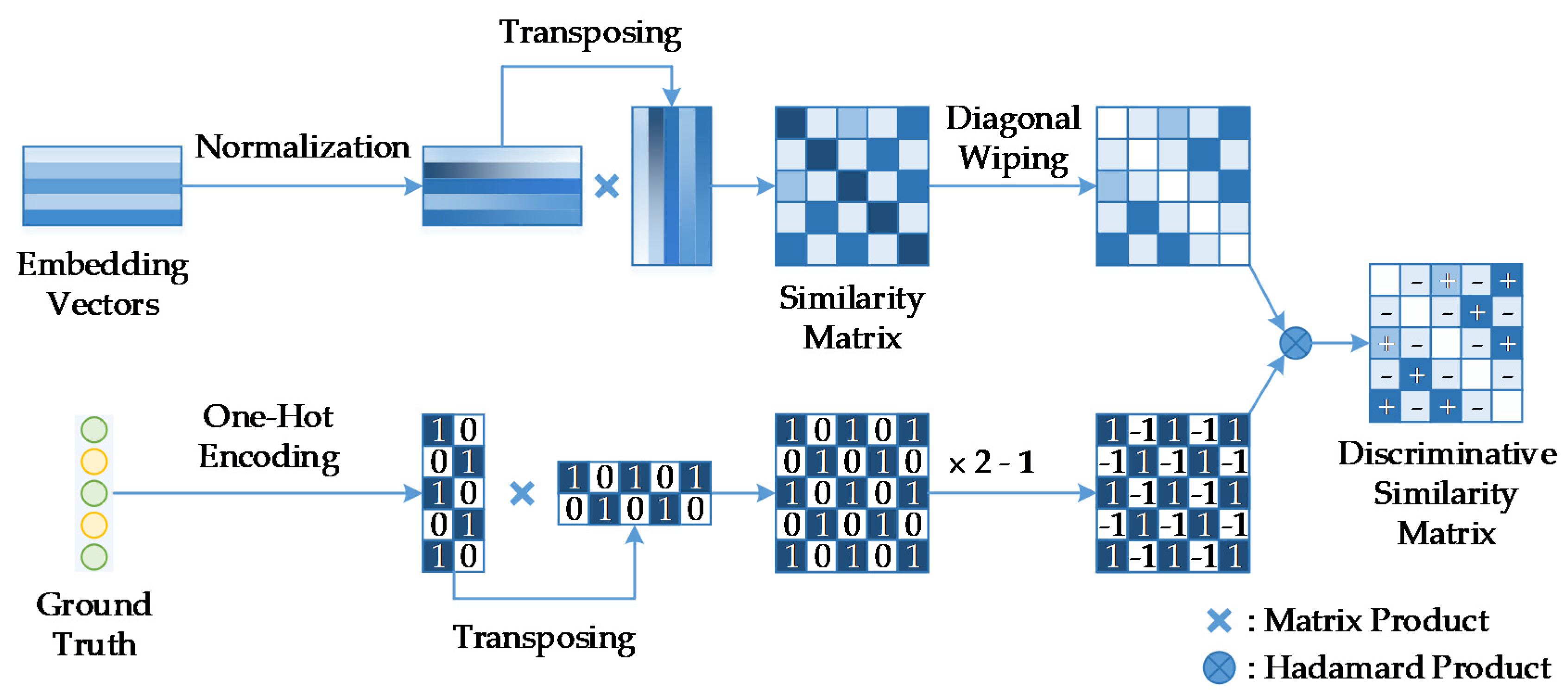
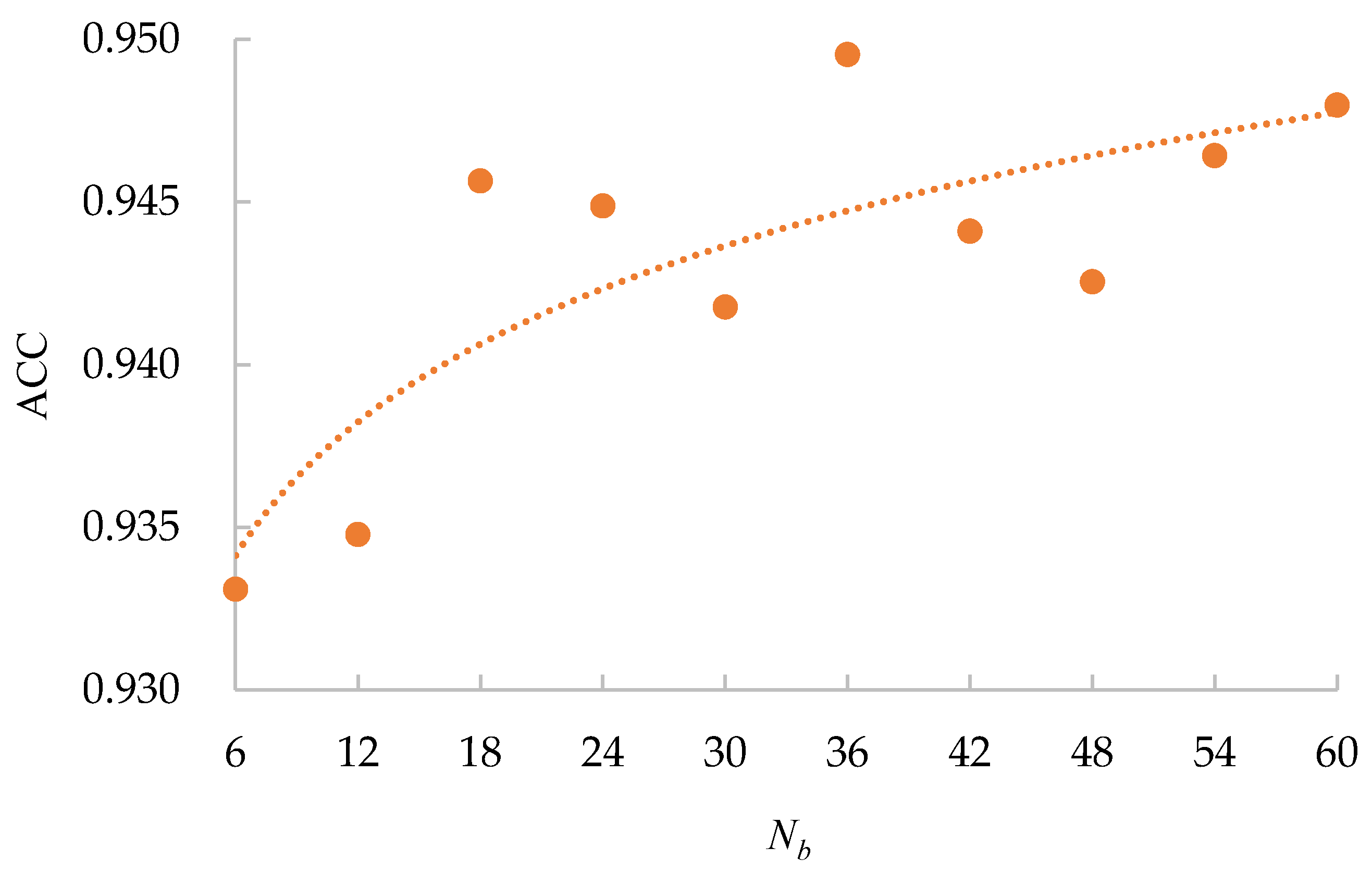3.2. The Influence of
The number of images in one batch
is an important parameter in the calculation of the BSTriplet loss, because it can influence the similarity matrix
and further affect the estimation for training data distribution. Besides this, it has an impact on the stability of the training process. To explore the influence of
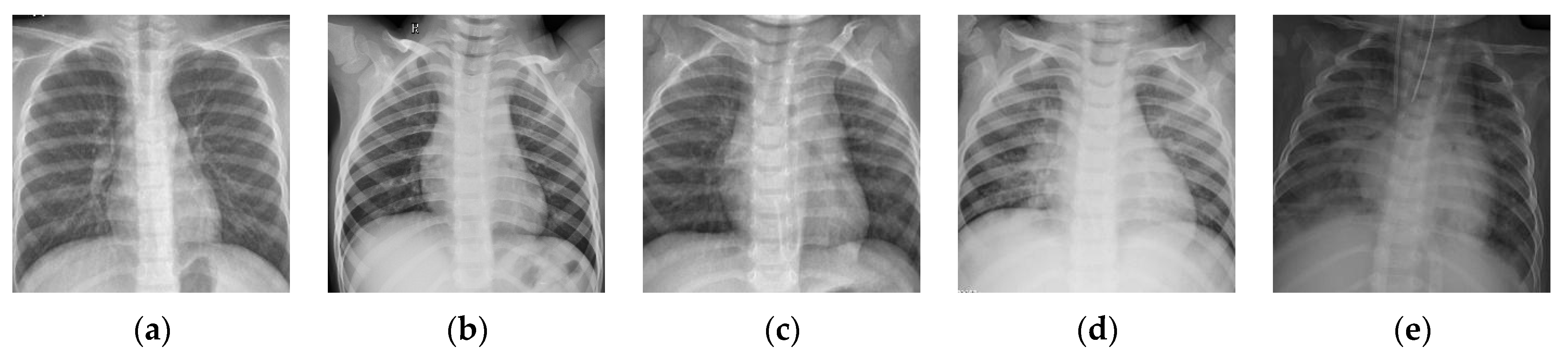
, we have downloaded a dataset of chest X-ray images [
41,
42] from the Kaggle [
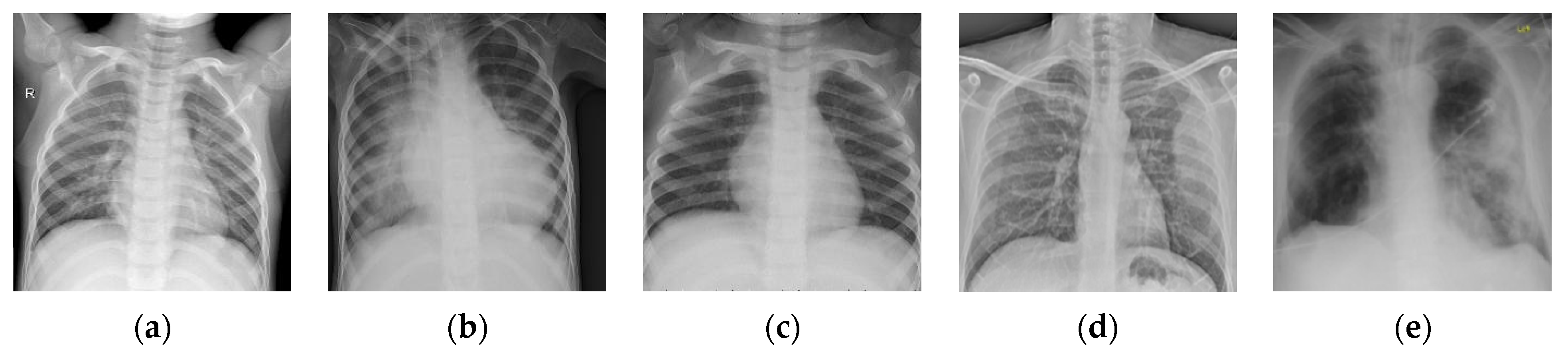
43] website. This dataset is denoted as “Chest-1” for brevity in the following. For this dataset, the X-ray images will be classified into three classes, including normal, (regular) pneumonia, and COVID-19. Several examples are shown in
Figure 4. Here, these images are cropped to squares for better clarity. All the images are resized as
to be input into the network, and the construction of the Chest-1 dataset is shown in
Table 2. According to the data mining strategy described above, we cluster the images in the Chest-1 dataset into six groups by K-means [
44], wherein there are two groups for each class. Based on the ratios of groups, we have sampled each group to build different sizes of input batches
. ModbileNet-V3-Small is used as the base framework and is trained by CE loss combined with the proposed BSTriplet loss. Some experiments have been performed with various
, while other hyper-parameters are fixed according to the method of controlling variables. The accuracy (
) is adopted as a metric to evaluate the performance, which is defined as:
where
and
are the numbers of true positive and true negative cases for the
-th class, respectively, and
is the number of test images.
All the obtained
values for different
s are shown in
Figure 5. From
Figure 5, we can see that the accuracy (94.95%) achieved using
is the highest, and the accuracy (93.31%) achieved using
is the lowest. Overall, the accuracy has a positive correlation with
, which is in line with the supposition that a bigger
will ensure that each input batch can represent the data distribution of the training dataset more precisely. Moreover, the curve converges quickly. The reason for this is that the CE loss provides a base accuracy, and it increases the stability of the curve. Furthermore, our data mining technique ensures the diversity of images in the batch. Therefore, a batch with a small
has a similar data distribution to that with a big
. Considering that a bigger
will lead to insufficient iteration times for network training in one epoch, we will use
as the default setting for the rest of the experiments.
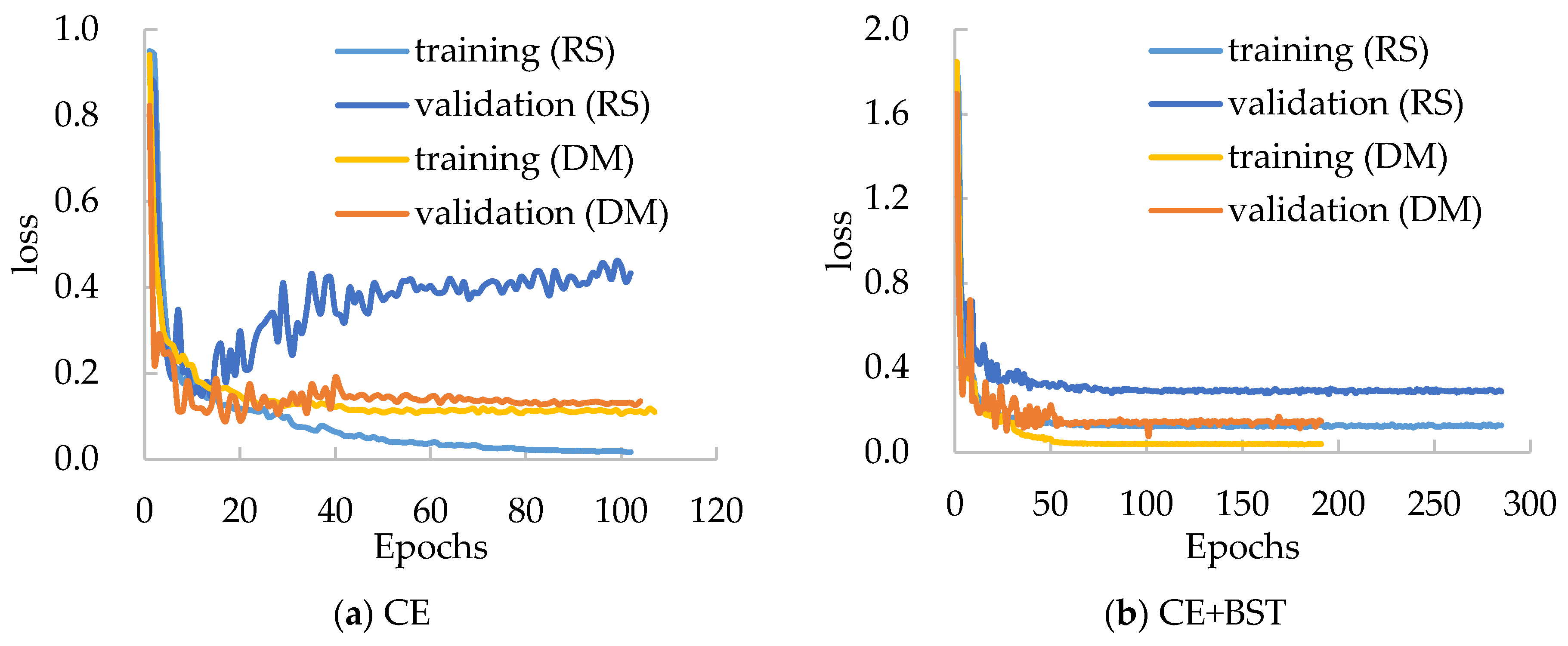
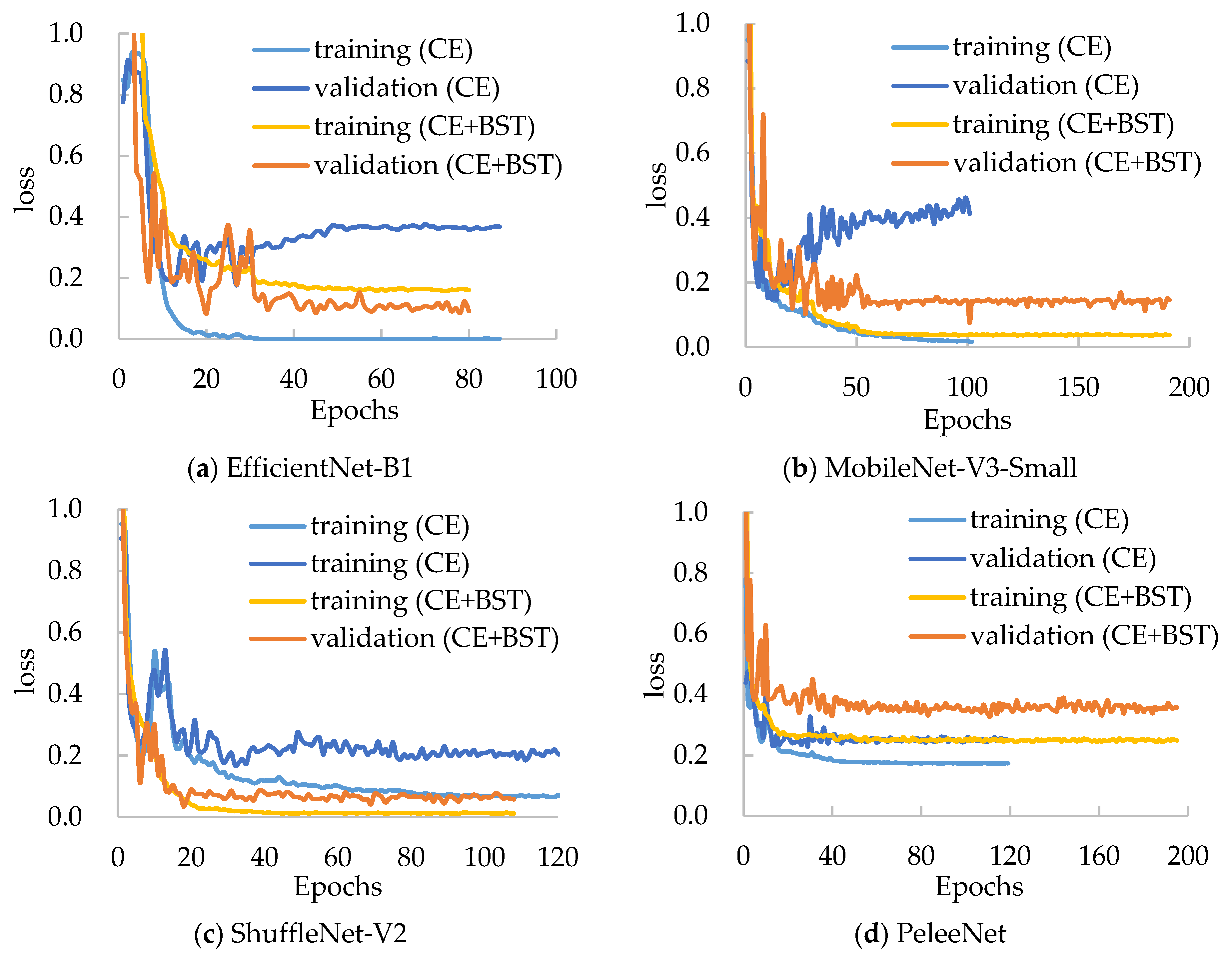
3.5. Applicability of the BSTriplet Loss
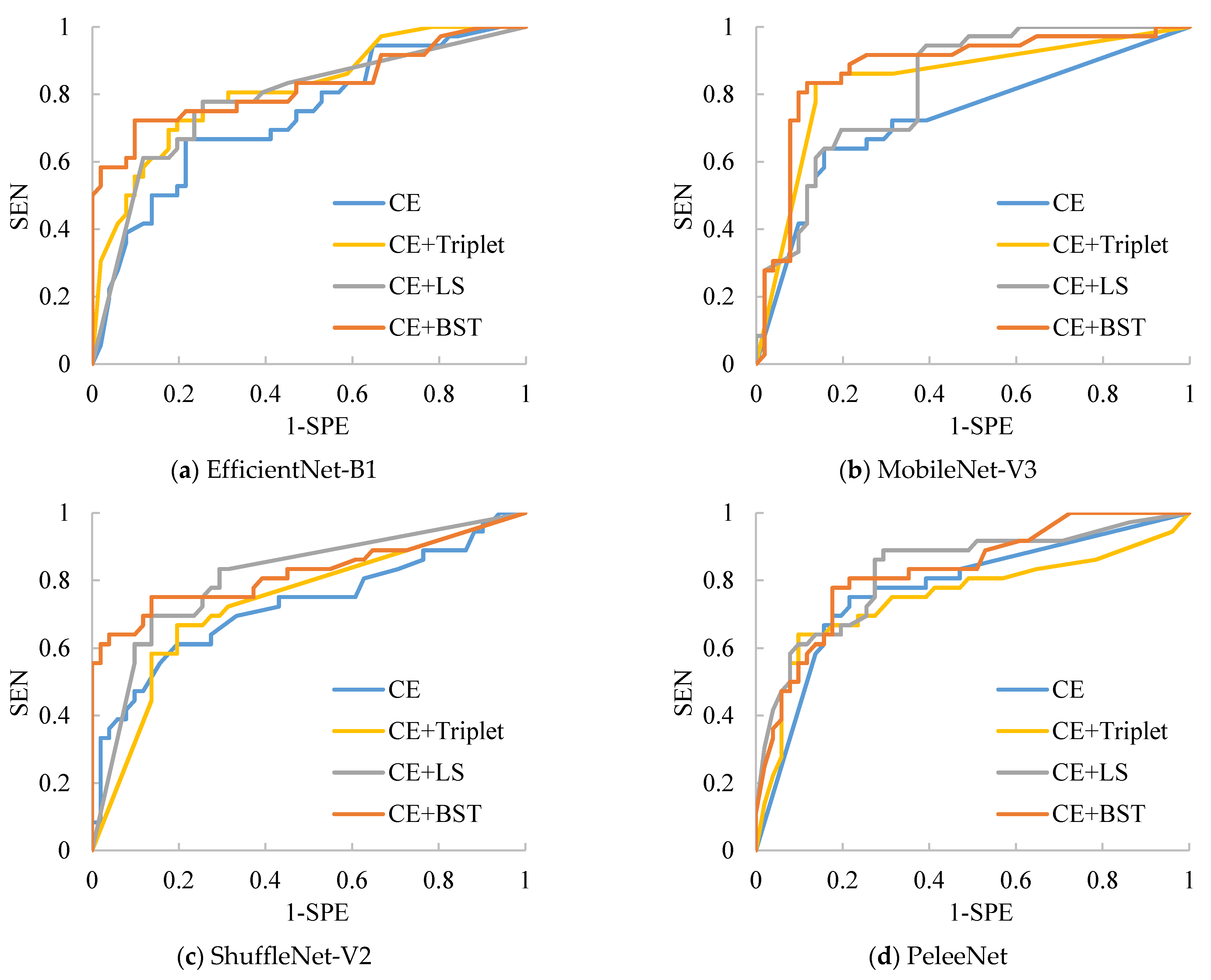
Moreover, we have compared the performances of several mentioned models with the BSTriplet loss to discover its applicability and effect. For each compared network, we have trained it with the CE loss and CE+BST loss. The training loss and validation loss are shown in
Figure 8. By comparing the four networks trained with CE loss, we can see that the validation loss of EfficientNet-B1 and MobileNet-V3-Small goes up when the time of iteration gets bigger. This observation means that there is a more severe over-fitting problem in their training process compared with the others. When these models are trained by CE+BST loss, the over-fitting problem is suppressed. Moreover, the gap between training loss and validation loss demonstrates that there is a smaller gap for CE+BST loss than for CE loss in such models as EfficientNet-B1, MobileNet-V3-Small and ShuffleNet-V2. As for PeleeNet, it has a relatively small over-fitting problem, which benefits from the fact that it has the lowest number of weights. Overall, the BSTriplet loss can suppress the over-fitting problem, which demonstrates that the BSTriplet loss can play the role of a regularization term for the CE loss.
To verify the effectiveness of the proposed combined loss, we have compared it with CE combined with triple loss [
45] and CE combined with the improved lifted structure loss [
46]. We test all these loss functions on four compared networks on the Chest-1 dataset. Since Chest-1 provides a multi-class classification task, the average sensitivity
and specificity
are used as metrics for evaluating the performance of the networks.
and
are calculated as
where
and
respectively represent the sensitivity and specificity of the
-th class, which shows the ability of the classifier to correctly find real positive cases and negative ones for the target disease;
and
denote the number of false positive and false negative cases for the
-th class, respectively. Besides this, the
and the area under the curve (AUC) of the receiver operating characteristic (ROC) are also employed to assess the classification ability of the trained models. The results are listed in
Table 4, where “Triplet”, “LS” and “BST” represent the regular triplet loss, the improved lifted structure loss and the proposed BSTriplet loss, respectively. For each evaluated model with different losses, the best value for every metric is indicated with bold in
Table 4. Clearly, ShuffleNet-V2 provides the highest accuracy among all the compared networks for each of the four different losses, which demonstrates the superiority of its structure. For each network alone, both the LS loss and the BSTriplet loss can improve the accuracy compared with the CE loss. By comparison, the regular triplet loss causes
to decrease from 92.70% to 92.24% for MobileNet-V3-Small, and
to decrease from 92.93% to 90.06% for PeleeNet. The reason is that the regular triplet loss identifies each input sample only according to one positive pair and one negative pair, which easily leads to an unstable training process and could make it difficult to search for the optimal solution. Furthermore, among all the compared losses, the CE+BST loss guides such models as EfficientNet-B1, MobileNet-V3-Small and ShuffleNet-V2 to gain the highest accuracy, sensitivity, specificity and AUC values. As for PeleeNet, the proposed CE+BST loss achieves the second highest AUC value (0.9841) and specificity (96.21%). Overall, the BSTriplet loss is helpful for training light-weighted CNN models when employed and combined with CE loss, and it is more effective than the regular triplet loss and lifted structure loss.
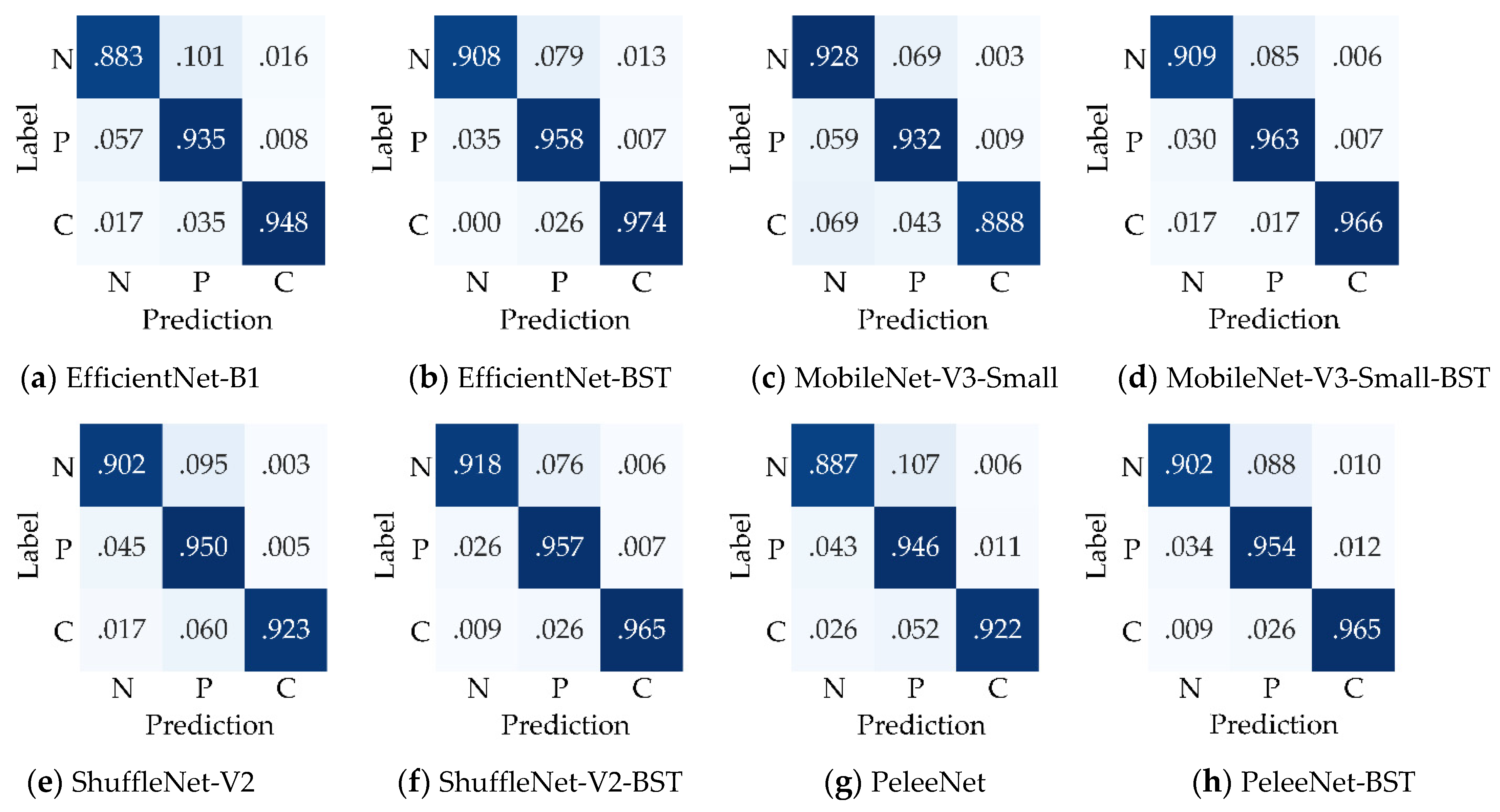
To intuitively show the superiority of the proposed BSTriplet loss, the confusion matrixes of the four networks trained with CE loss and CE+BST loss are given in
Figure 9. It can be seen that the normal images are relatively more easily misclassified as pneumonia images or COVID-19 images for all the compared models. Therefore, the proposed loss has the lowest average recognition rate (0.905) over all trained models. However, the average recognition rates for pneumonia images and COVID-19 images are 0.949 and 0.944, respectively. In addition, EfficientNet-B1, ShuffleNet-V2 and PeleeNet achieve higher recognition rates for every class when trained with CE+BST loss compared to when trained with CE loss. As for the MobileNet-V3-Small model, the proposed CE+BST loss causes the recognition rate for normal images to decrease from 0.928 to 0.909, but it has a much better ability to distinguish the images of the rest of the categories. Moreover, the recognition rate for COVID images is improved the most among the three classes when the BSTriplet loss is used, which indicates that it can resolve the problem of data imbalance.
To further verify the consistency of the proposed BSTriplet loss, we have carried out comparative experiments on another dataset of chest X-ray images denoted as “Chest-2”. This dataset is also downloaded from the Kaggle [
43] website. Here, Chest-2 is used for the classification of lung images into normal lung images and pneumonia images. Some examples are shown in
Figure 10. The construction of Chest-2 is listed in
Table 5. Each of the mentioned four light-weighted networks is trained with four kinds of losses on Chest-2. Similar to the experiments on Chest-1, all these images in Chest-2 are resized into
. We have performed the K-means algorithm to partition the training images into six groups, and randomly selected six samples from each group to build the input batch for the proposed CE+BST loss.
The results for the classification of Chest-2 are shown in
Table 6. From
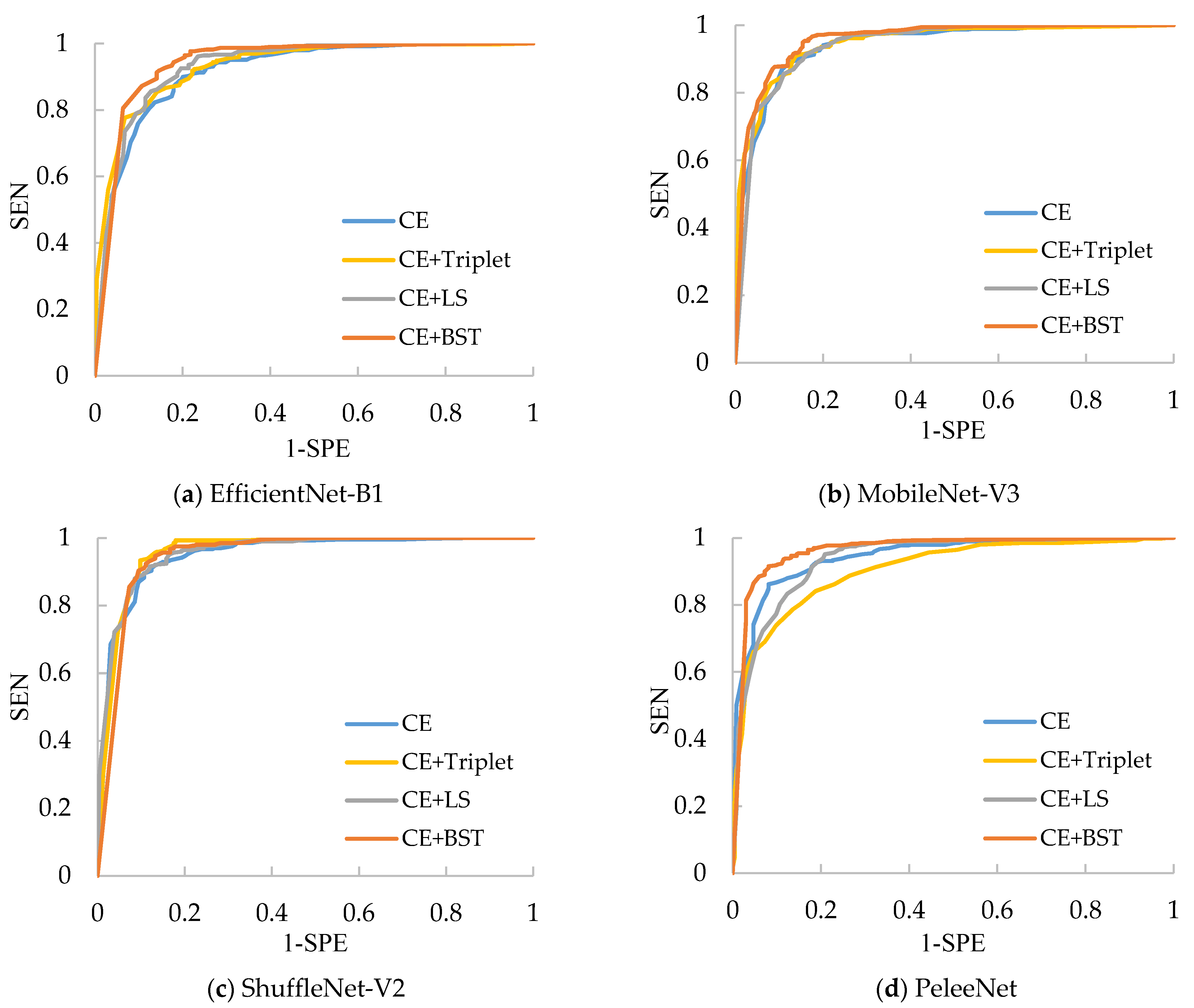
Table 6, we can see that ShuffleNet-V2 still has the best performance because it can achieve the highest average SEN (97.14%), average SPE (80.56%), average ACC (90.91%) and average AUC (0.9569) over four kinds of losses. Besides this, the CE+Triplet loss helps ShuffleNet-V2 to provide the highest ACC (92.47%), which is 3.21% higher than that of ShuffleNet-V2 trained with CE loss. Nevertheless, CE+Triplet loss achieves lower ACC and AUC compared with CE loss when they are applied to PeleeNet, which reveals that the consistency of the effectiveness of CE+Triplet loss cannot be guaranteed for various networks. In comparison, both CE+LS and our CE+BST loss have better consistency, which benefits from the analysis for all the pairs formed by the samples in the batch. Furthermore, the proposed CE+BST loss surpasses the CE+LS loss in terms of average values of SEN, SPE, ACC and AUC over four evaluated networks by 0.26%, 7.27%, 2.89% and 0.012%, respectively. The reason why BSTriplet outperforms LS is that the former adopts similarity instead of Euclidean distance, so that it has a clear upper bound, and the value of BSTriplet loss is close to CE loss, so that it can achieve better coordination than LS loss. We have also provided the ROC curves of all compared methods in
Figure 11. When the ROC curve is closer to the upper left corner, it means the corresponding classifier has a better classification ability. For ShuffleNet-V2, all the three combined losses show similar performances, and they all surpass the performance of CE loss. As for the rest of the networks, our CE+BST loss can provide the most significant improvement in classification performance for each network, especially when it is used in PeleeNet. In general, the proposed BSTriplet loss is more suitable for assisting CE loss in CNN training, and it outperforms the other compared DML losses.
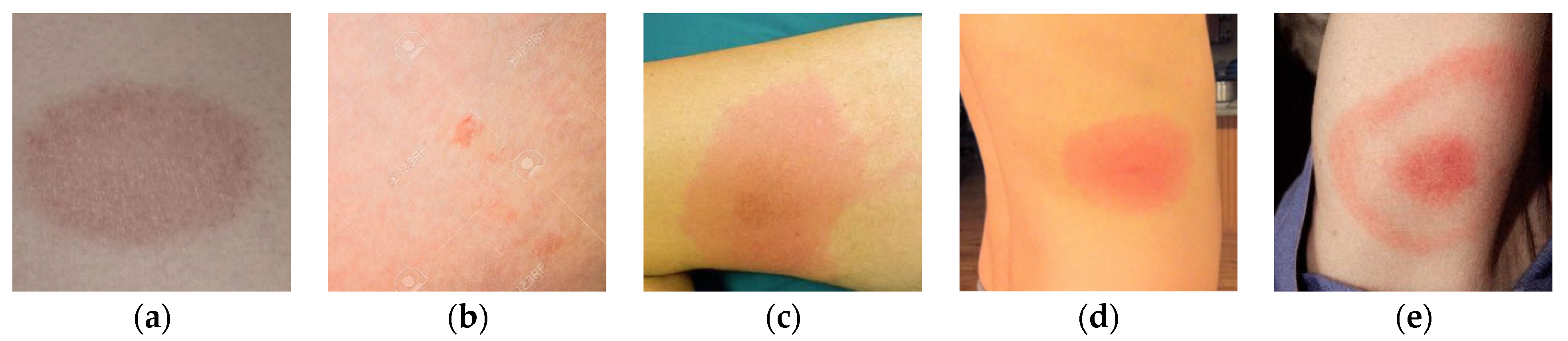
To further validate the effectiveness of the proposed BSTriplet loss, some experiments have been conducted on a skin rash image dataset, which is used to distinguish the images containing Lyme disease from those with other disease [
47]. The composition of the rash image dataset is listed in
Table 7.
Figure 12 shows some images in the rash image dataset. It can be seen that the images in this dataset are colorful optical images, which are much different from the above X-ray images. Besides this, the number of images in the rash image dataset is evidently less than that in the above datasets. To alleviate the over-fitting problem, we have augmented the training images by such methods as rotation by
,
and
, horizontal/vertical flipping, and horizontal/vertical translation for five pixels so that the number of training images is enlarged by seven-fold. As for the test images, the augmentation has not been implemented. We have clustered the images into four categories, and built the input batches for the proposed CE+BST loss according to the steps in
Section 2.3.
The results for each evaluated model are given in
Table 8. It can be seen that almost all the metrics are lower than those in the above experiments. The reason for this is that the rash images are full of varied backgrounds and the training images are insufficient. Nevertheless, when the loss involves the DML methods, the CNNs can gain higher accuracy compared with themselves trained with CE loss, except that CE+LS loss provides the same
(75.86%) for MobileNet-V3-Small. MobileNet-V3-Small provides the highest
(83.91%) and AUC (0.8755) when trained with the proposed CE+BST loss, and it can produce the second best
(82.76%) and AUC (0.8423) when it is trained with CE+Triplet loss. Both ShuffleNet-V2 and PeleeNet gain the second highest
(80.46%) when they are trained with our CE+BST loss. A comparison among the four results obtained by PeleeNet shows that CE+LS loss gives the highest
but the lowest
, while CE+Triplet loss gives the opposite results. In comparison, our CE+BST loss can provide both the second highest
(77.78%) and
(82.35%), as well as the highest
(80.46%), for PeleeNet. Besides this, if the results for the CE loss are used as the baseline for each model, the average improvement for each combined loss in four models can be calculated. The proposed CE+BST loss gains an advantage over CE+Triplet loss and CE+LS loss by providing the highest average improvements in
,
,
and AUC, by 9.03%, 5.39%, 6.90% and 0.1063, respectively.
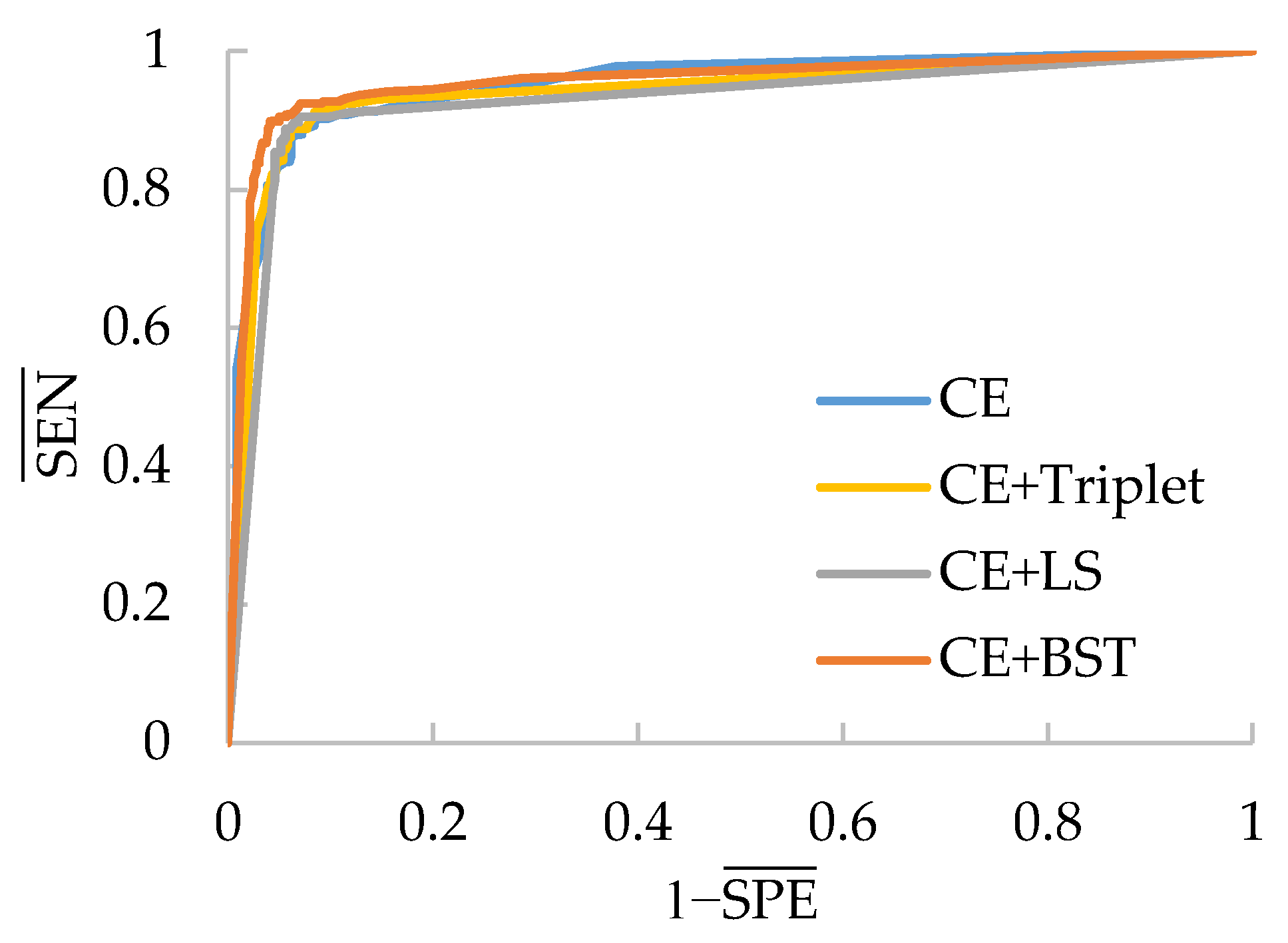
The ROC curves for every test model on the rash image dataset are shown in
Figure 13. It is clear that these curves are not as smooth as those in
Figure 11 due to the small number of test images. Almost all the combined losses surpass CE loss for each compared CNN, especially for MobileNet-V3-Small. Only CE+Triplet is inferior to CE loss when applied to PeleeNet, which reveals its instability again. In addition, the curves of the proposed CE+BST loss are much closer to the top left corner in each subfigure than the rest, which demonstrates its effectiveness for a small dataset and adaptability to different networks.
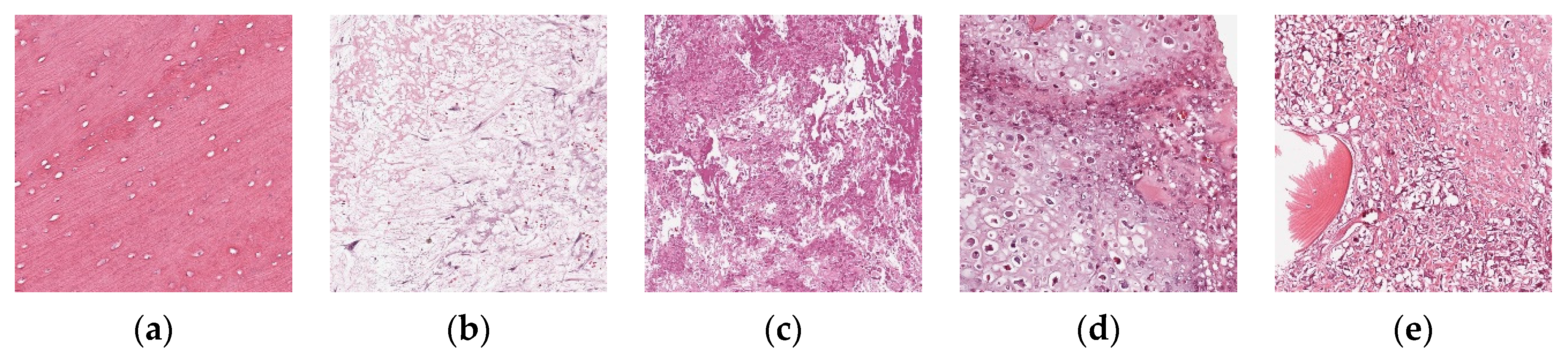
To further verify the applicability of the BSTriplet loss to other medical image modalities, additional experiments have been done on an osteosarcoma histology image dataset [
48,
49,
50], which can be accessed from the cancer imaging archive (TCIA) [
51]. There are three kinds of images in the osteosarcoma histology image dataset, as shown in
Figure 14. Compared with the above involved images, the histology images are colored images, and their backgrounds are simpler than those of the skin rash images. This dataset is a small sample dataset and its construction is listed in
Table 9. The augmentation for this dataset is the same as that for the rash images. We have clustered each kind of images into two groups for CE+BST loss according to the proposed data mining strategy. Using the accuracy
, average sensitivity
, average specificity
and AUC as metrics, we have tested MobileNet-V3-Small trained with several different loss functions. The results are provided in
Table 10 and
Figure 15.
Table 10 shows that CE+BST loss-based MobileNet-V3-Small provides the best performance in terms of all the metrics. In particular, the accuracy provided by CE+BST loss is 2.18% higher than the second best
, which is produced by CE+LS loss. Besides this, the regular triplet loss and the LS loss help the CE to obtain better
,
and
values, although their AUC values are smaller than those of CE loss. On the other hand, BSTriplet loss obtains the best AUC value in
Table 10 and ROC curve in
Figure 15. The results demonstrate that, compared with the LS loss and the regular triplet loss, our proposed BSTriplet loss has a better ability to assist CE loss in improving the performance of the CNNs on small sample datasets.