Live-Cell Systems in Real-Time Biomonitoring of Water Pollution: Practical Considerations and Future Perspectives
Abstract
:1. Introduction
2. Established Online Biomonitors Using Bacteria and Algae
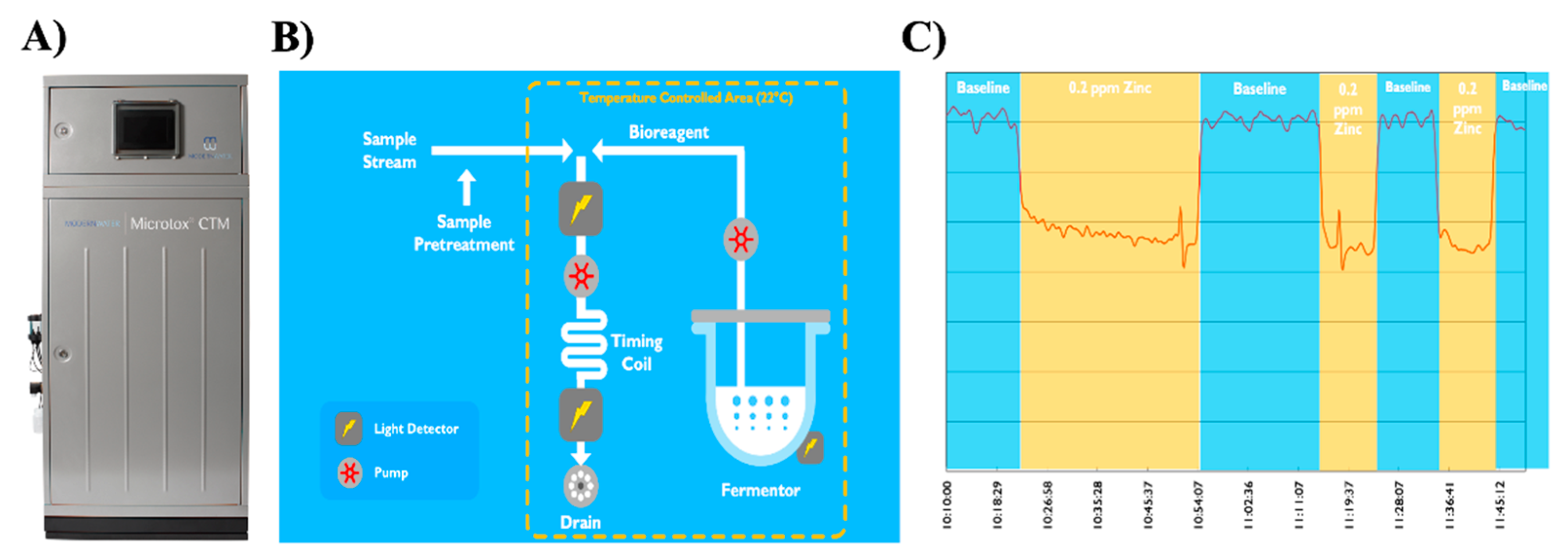
2.1. Online Microtox System
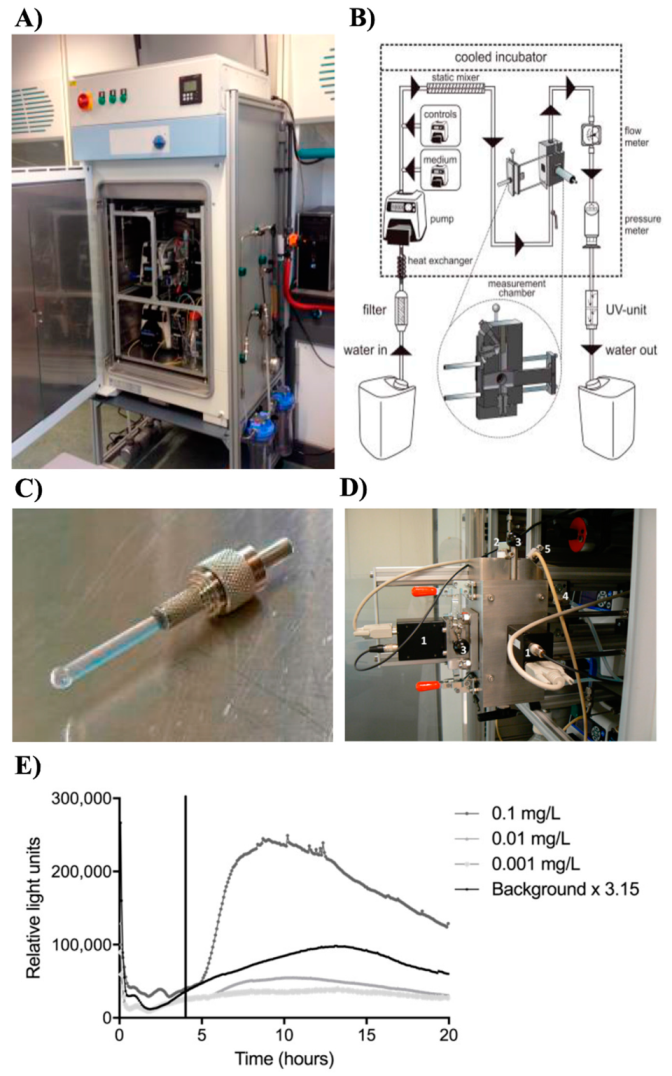
2.2. ToxAlarm Toximeter
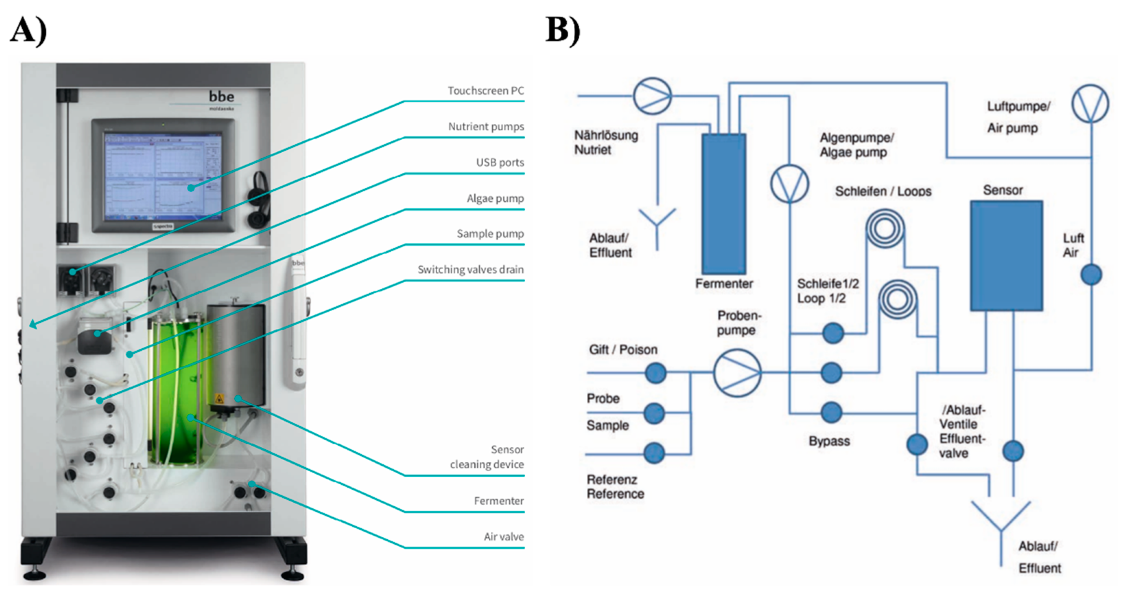
2.3. Algae Toximeter II
3. Automated Flow Cytometry and Online Fluorimetry
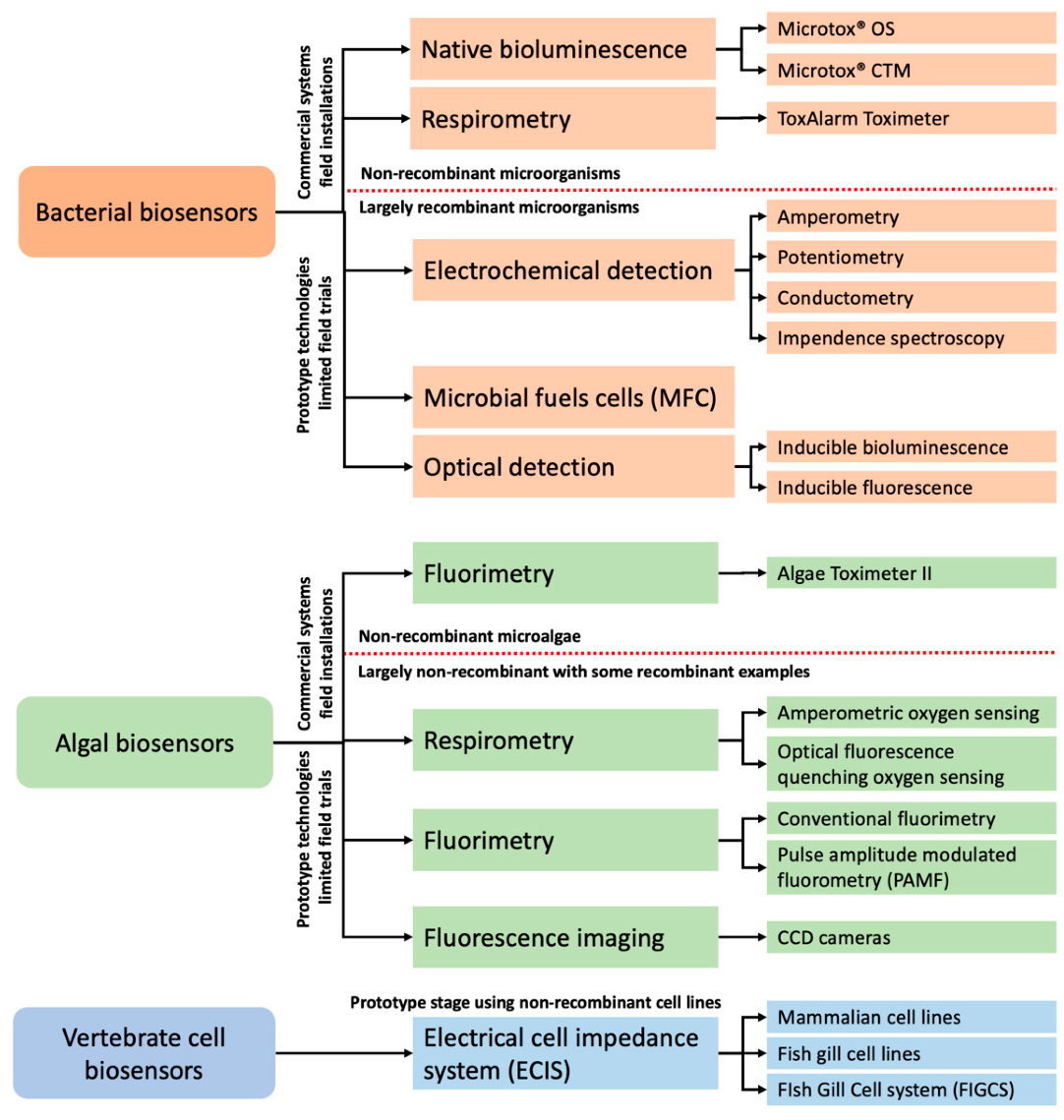
4. Bacterial Biosensors
4.1. Electrochemical Sensing
4.2. Microbial Fuels Cell (MFC)-Based Biosensors
4.3. Optical Sensing
4.3.1. Bioluminescence Methods
4.3.2. Fluorescence Methods
4.4. Practical Aspects of Bacterial Sensing Technologies in Real-Time Water Biomonitoring
5. Cyanobacteria Biosensing Technologies
6. Algal Biosensing Technologies
6.1. Respirometry
6.2. Fluorimetry
6.3. Chlorophyl Fluorescence Imaging
6.4. Practical Aspects of Algal Technologies in Real-Time Water Biomonitoring
7. Biosensing with Vertebrate Cells
8. Limitations of Live-Cell Online Biomonitoring in Practical Deployment Scenarios
8.1. Non-Quantitative Nature
8.2. Analysis Time
8.3. Maintenance of Cell Cultures
8.4. Sterilisation Protocols
8.5. Pre-Processing of Water Samples
8.6. Waste Disposal
8.7. Thresholds of Sensitivity
8.8. Reliability of Alarm Events
9. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bae, M.J.; Park, Y.S. Biological early warning system based on the responses of aquatic organisms to disturbances: A review. Sci. Total Environ. 2014, 466–467, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Bownik, A.; Wlodkowic, D. Advances in real-time monitoring of water quality using automated analysis of animal behaviour. Sci. Total Environ. 2021, 789, 147796. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, A.; Ingram, M.K.; Kang, I.J.; Ulitzur, S. In situ on-line toxicity biomonitoring in water: Recent developments. Environ. Toxicol. Chem. 2006, 25, 2263–2271. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, A. Online Biomonitoring for integrated smart real-time water management. Water Solut. 2020, 3, 20–23. [Google Scholar]
- Fitch, J.P.; Raber, E.; Imbro, D.R. Technology challenges in responding to biological or chemical attacks in the civilian sector. Science 2003, 302, 1350–1354. [Google Scholar] [CrossRef]
- Eubanks, L.M.; Dickerson, T.J.; Janda, K.D. Technological advancements for the detection of and protection against biological and chemical warfare agents. Chem. Soc. Rev. 2007, 36, 458–470. [Google Scholar] [CrossRef]
- Green, U.; Kremer, J.H.; Zillmer, M.; Moldaenke, C. Detection of chemical threat agents in drinking water by an early warning real-time biomonitor. Environ. Toxicol. 2003, 18, 368–374. [Google Scholar] [CrossRef]
- Storey, M.V.; van der Gaag, B.; Burns, B.P. Advances in on-line drinking water quality monitoring and early warning systems. Water Res. 2011, 45, 741–747. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, L.; Liu, Y.; Chon, T.; Ren, Z.; Wang, Z.; Zhao, J.; Zhao, Y. A new online monitoring and management system for accidental pollution events developed for the regional water basin in Ningbo, China. Water Sci. Technol. 2011, 64, 1828–1834. [Google Scholar] [CrossRef]
- Kramer, K.J.M.; Foekema, E.M. The “Musselmonitor®” as Biological Early Warning System. In Biomonitors and Biomarkers as Indicators of Environmental Change 2; Butterworth, F.M., Gunatilaka, A., Gonsebatt, M.E., Eds.; Springer: New York, NY, USA, 2000; pp. 59–87. [Google Scholar]
- Diehl, P.; Gerke, T.; Jeuken, A.; Lowis, L.; Steen, R.; van Steenwijk, J.; Stoks, P.; Willemsen, H.G. Early Warning Strategies and Practices Along the River Rhine. In The Rhine; Knepper, T.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 99–124. [Google Scholar]
- Eltzov, E.; Marks, R.S. Whole-cell aquatic biosensors. Anal. Bioanal. Chem. 2011, 400, 895–913. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef]
- Gu, M.B.; Mitchell, R.J.; Kim, B.C. Whole-cell-based biosensors for environmental biomonitoring and application. Adv. Biochem. Eng. Biotechnol. 2004, 87, 269–305. [Google Scholar] [PubMed]
- Matejczyk, M. The potential of application of microbial biosensors. Postep. Mikrobiol. 2010, 49, 297–304. [Google Scholar]
- Woutersen, M.; Belkin, S.; Brouwer, B.; van Wezel, A.P.; Heringa, M.B. Are luminescent bacteria suitable for online detection and monitoring of toxic compounds in drinking water and its sources? Anal. Bioanal. Chem. 2011, 400, 915–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawazumi, H.; Gobi, V.; Ogino, K.; Maeda, H.; Miura, N. Compact surface plasmon resonance (SPR) immunosensor using multichannel for simultaneous detection of small molecule compounds. Sens. Actuators B Chem. 2005, 108, 791–796. [Google Scholar] [CrossRef]
- Mauriz, E.; Calle, A.; Abad, A.; Montoya, A.; Hildebrandt, A.; Barcelo, D.; Lechuga, L.M. Determination of carbaryl in natural water samples by a surface plasmon resonance flow-through immunosensor. Biosens. Bioelectron. 2006, 21, 2129–2136. [Google Scholar] [CrossRef]
- Nabok, A.V.; Tsargorodskaya, A.; Hassan, A.K.; Starodub, N.F. Total internal reflection ellipsometry and SPR detection of low molecular weight environmental toxins. Appl. Surf. Sci. 2005, 246, 381–386. [Google Scholar] [CrossRef]
- van der Schalie, W.H.; Shedd, T.R.; Knechtges, P.L.; Widder, M.W. Using higher organisms in biological early warning systems for real-time toxicity detection. Biosens. Bioelectron. 2001, 16, 457–465. [Google Scholar] [CrossRef]
- Brayner, R.; Coute, A.; Livage, J.; Perrette, C.; Sicard, C. Micro-algal biosensors. Anal. Bioanal. Chem. 2011, 401, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Taylor, P.B.; Leach, F.R. Use of the Microtox assay system for environmental samples. Bull. Environ. Contam. Toxicol. 1981, 26, 150–156. [Google Scholar] [CrossRef]
- Dutka, B.J.; Kwan, K.K. Comparison of three microbial toxicity screening tests with the Microtox test. Bull. Environ. Contam. Toxicol. 1981, 27, 753–757. [Google Scholar] [CrossRef]
- Coleman, R.N.; Qureshi, A.A. Microtox and Spirillum volutans tests for assessing toxicity of environmental samples. Bull. Environ. Contam. Toxicol. 1985, 35, 443–451. [Google Scholar] [CrossRef]
- LAWA. Recommendations on the Deployment of Continuous Biomonitors for the Monitoring of Surface Waters; Working Group of the Federal States on Water Problems (LAWA): Stuttgart, Germany, 1998.
- Noack, U.; Walter, J. The algae toximeter for continuous water monitoring. Schr. Ver. Wasser. Boden. Lufthyg. 1992, 89, 305–309. [Google Scholar]
- Gerhardt, V.; Putzger, J. A biotest for water monitoring based on delayed fluorescence of algae. Schr. Ver. Wasser. Boden. Lufthyg. 1992, 89, 277–284. [Google Scholar]
- Lechelt, M.; Blohm, W.; Kirschneit, B.; Pfeiffer, M.; Gresens, E.; Liley, J.; Holz, R.; Lüring, C.; Moldaenke, C. Monitoring of surface water by ultrasensitive Daphnia toximeter. Wit. Tr. Biomed. Health 2000, 15, 390–400. [Google Scholar]
- Machdar, E.; van der Steen, N.P.; Raschid-Sally, L.; Lens, P.N. Application of Quantitative Microbial Risk Assessment to analyze the public health risk from poor drinking water quality in a low income area in Accra, Ghana. Sci. Total Environ. 2013, 449, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Efstratiou, A.; Ongerth, J.E.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017, 114, 14–22. [Google Scholar] [CrossRef]
- Besmer, M.D.; Hammes, F. Short-term microbial dynamics in a drinking water plant treating groundwater with occasional high microbial loads. Water Res. 2016, 107, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Buysschaert, B.; Byloos, B.; Leys, N.; Van Houdt, R.; Boon, N. Reevaluating multicolor flow cytometry to assess microbial viability. Appl. Microbiol. Biot. 2016, 100, 9037–9051. [Google Scholar] [CrossRef]
- Stauber, J.L.; Franklin, N.M.; Adams, M.S. Applications of flow cytometry to ecotoxicity testing using microalgae. Trends Biotechnol. 2002, 20, 141–143. [Google Scholar] [CrossRef]
- Prest, E.I.; Hammes, F.; Kotzsch, S.; van Loosdrecht, M.C.; Vrouwenvelder, J.S. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 2013, 47, 7131–7142. [Google Scholar] [CrossRef]
- Besmer, M.D.; Epting, J.; Page, R.M.; Sigrist, J.A.; Huggenberger, P.; Hammes, F. Online flow cytometry reveals microbial dynamics influenced by concurrent natural and operational events in groundwater used for drinking water treatment. Sci. Rep. 2016, 6, 38462. [Google Scholar] [CrossRef]
- Sorensen, J.P.R.; Vivanco, A.; Ascott, M.J.; Gooddy, D.C.; Lapworth, D.J.; Read, D.S.; Rushworth, C.M.; Bucknall, J.; Herbert, K.; Karapanos, I.; et al. Online fluorescence spectroscopy for the real-time evaluation of the microbial quality of drinking water. Water Res. 2018, 137, 301–309. [Google Scholar] [CrossRef]
- Bridgeman, J.; Baker, A.; Brown, D.; Boxall, J.B. Portable LED fluorescence instrumentation for the rapid assessment of potable water quality. Sci. Total Environ. 2015, 524–525, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, J.P.; Lapworth, D.J.; Marchant, B.P.; Nkhuwa, D.C.; Pedley, S.; Stuart, M.E.; Bell, R.A.; Chirwa, M.; Kabika, J.; Liemisa, M.; et al. In-situ tryptophan-like fluorescence: A real-time indicator of faecal contamination in drinking water supplies. Water Res. 2015, 81, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Cohen, E.; Levy, G.J.; Borisover, M. Fluorescent components of organic matter in wastewater: Efficacy and selectivity of the water treatment. Water Res. 2014, 55, 323–334. [Google Scholar] [CrossRef]
- Bechor, O.; Smulski, D.R.; Van Dyk, T.K.; LaRossa, R.A.; Belkin, S. Recombinant microorganisms as environmental biosensors: Pollutants detection by Escherichia coli bearing fabA’::lux fusions. J. Biotechnol. 2002, 94, 125–132. [Google Scholar] [CrossRef]
- Zhou, T.; Han, H.; Liu, P.; Xiong, J.; Tian, F.; Li, X. Microbial Fuels Cell-Based Biosensor for Toxicity Detection: A Review. Sensors 2017, 17, 2230. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Skladal, P.; Morozova, N.O.; Reshetilov, A.N. Amperometric biosensors for detection of phenol using chemically modified electrodes containing immobilized bacteria. Biosens. Bioelectron. 2002, 17, 867–873. [Google Scholar] [CrossRef]
- Biran, I.; Babai, R.; Levcov, K.; Rishpon, J.; Ron, E.Z. Online and in situ monitoring of environmental pollutants: Electrochemical biosensing of cadmium. Environ. Microbiol. 2000, 2, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Borman, S. Biosensors: Potentiometric and amperometric. Anal. Chem. 1987, 59, 1091A–1098A. [Google Scholar] [PubMed]
- Lei, Y.; Mulchandani, P.; Chen, W.; Mulchandani, A. Biosensor for direct determination of fenitrothion and EPN using recombinant Pseudomonas putida JS444 with surface-expressed organophosphorous hydrolase. 2. Modified carbon paste electrode. Appl. Biochem. Biotechnol. 2007, 136, 243–250. [Google Scholar] [CrossRef]
- Farre, M.; Barcelo, D. Characterization of wastewater toxicity by means of a whole-cell bacterial biosensor, using Pseudomonas putida, in conjunction with chemical analysis. Fresenius J. Anal. Chem. 2001, 371, 467–473. [Google Scholar] [CrossRef]
- Mehala, N.; Rajendran, L. Analysis of mathematical modelling on potentiometric biosensors. ISRN Biochem. 2014, 2014, 582675. [Google Scholar] [CrossRef]
- Koncki, R. Recent developments in potentiometric biosensors for biomedical analysis. Anal. Chim. Acta 2007, 599, 7–15. [Google Scholar] [CrossRef]
- Gaberlein, S.; Spener, F.; Zaborosch, C. Microbial and cytoplasmic membrane-based potentiometric biosensors for direct determination of organophosphorus insecticides. Appl. Microbiol. Biotechnol. 2000, 54, 652–658. [Google Scholar] [CrossRef]
- Kumar, S.; Kundu, S.; Pakshirajan, K.; Dasu, V.V. Cephalosporins determination with a novel microbial biosensor based on permeabilized Pseudomonas aeruginosa whole cells. Appl. Biochem. Biotechnol. 2008, 151, 653–664. [Google Scholar] [CrossRef]
- Hnaien, M.; Lagarde, F.; Bausells, J.; Errachid, A.; Jaffrezic-Renault, N. A new bacterial biosensor for trichloroethylene detection based on a three-dimensional carbon nanotubes bioarchitecture. Anal. Bioanal. Chem. 2011, 400, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Gaberlein, S.; Knoll, M.; Spener, F.; Zaborosch, C. Disposable potentiometric enzyme sensor for direct determination of organophosphorus insecticides. Analyst 2000, 125, 2274–2279. [Google Scholar] [CrossRef] [PubMed]
- Okafor, C.; Grooms, D.; Alocilja, E.; Bolin, S. Fabrication of a Novel Conductometric Biosensor for Detecting Mycobacterium avium subsp. paratuberculosis Antibodies. Sensors 2008, 8, 6015–6025. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Lamarche, P.; Tawil, N.; Khan, R.; Aliakbar, A.M.; Hassan, M.H.; Chodavarapu, V.P.; Mandeville, R. CMOS Conductometric System for Growth Monitoring and Sensing of Bacteria. IEEE Trans. Biomed. Circuits Syst. 2011, 5, 223–230. [Google Scholar]
- Chouteau, C.; Dzyadevych, S.; Durrieu, C.; Chovelon, J.M. A bi-enzymatic whole cell conductometric biosensor for heavy metal ions and pesticides detection in water samples. Biosens. Bioelectron. 2005, 21, 273–281. [Google Scholar] [CrossRef]
- Furst, A.L.; Francis, M.B. Impedance-Based Detection of Bacteria. Chem. Rev. 2019, 119, 700–726. [Google Scholar] [CrossRef]
- Silley, P.; Forsythe, S. Impedance microbiology—A rapid change for microbiologists. J. Appl. Bacteriol. 1996, 80, 233–243. [Google Scholar] [CrossRef]
- Felice, C.J.; Valentinuzzi, M.E. Medium and interface components in impedance microbiology. IEEE Trans. Biomed. Eng. 1999, 46, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.N. The Charges Produced by the Growth of Bacteria in the Molecular Concentration and Electrical Conductivity of Culture Media. J. Exp. Med. 1899, 4, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawerla, M.; Stolle, A.; Schalch, B.; Eisgruber, H. Impedance microbiology: Applications in food hygiene. J. Food Prot. 1999, 62, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Felice, C.J.; Madrid, R.E.; Olivera, J.M.; Rotger, V.I.; Valentinuzzi, M.E. Impedance microbiology: Quantification of bacterial content in milk by means of capacitance growth curves. J. Microbiol. Methods 1999, 35, 37–42. [Google Scholar] [CrossRef]
- Strauss, W.M.; Malaney, G.W.; Tanner, R.D. The impedance method for monitoring total coliforms in wastewaters. Part I. Background and methodology. Folia Microbiol. 1984, 29, 162–169. [Google Scholar] [CrossRef]
- Colquhoun, K.O.; Timms, S.; Fricker, C.R. Detection of Escherichia coli in potable water using direct impedance technology. J. Appl. Bacteriol. 1995, 79, 635–639. [Google Scholar] [CrossRef]
- Clausen, C.H.; Dimaki, M.; Bertelsen, C.V.; Skands, G.E.; Rodriguez-Trujillo, R.; Thomsen, J.D.; Svendsen, W.E. Bacteria Detection and Differentiation Using Impedance Flow Cytometry. Sensors 2018, 18, 3496. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Lai, B.; Tang, X. Microbial Fuel Cell-Based Biosensors. Biosensors 2019, 9, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ElMekawy, A.; Hegab, H.M.; Pant, D.; Saint, C.P. Bio-analytical applications of microbial fuel cell-based biosensors for onsite water quality monitoring. J. Appl. Microbiol. 2018, 124, 302–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adekunle, A.; Raghavan, V.; Tartakovsky, B. A comparison of microbial fuel cell and microbial electrolysis cell biosensors for real-time environmental monitoring. Bioelectrochemistry 2019, 126, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Sik Hyun, M.; Gadd, G.M.; Joo Kim, H. A novel biomonitoring system using microbial fuel cells. J. Environ. Monit. 2007, 9, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lei, Y.; Li, B. A batch-mode cube microbial fuel cell based “shock” biosensor for wastewater quality monitoring. Biosens. Bioelectron. 2014, 62, 308–314. [Google Scholar] [CrossRef]
- Meighen, E.A. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 1991, 55, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Selifonova, O.; Burlage, R.; Barkay, T. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl. Environ. Microbiol. 1993, 59, 3083–3090. [Google Scholar] [CrossRef] [Green Version]
- Mancini, J.A.; Boylan, M.; Soly, R.R.; Graham, A.F.; Meighen, E.A. Cloning and expression of the Photobacterium phosphoreum luminescence system demonstrates a unique lux gene organization. J. Biol. Chem. 1988, 263, 14308–14314. [Google Scholar] [CrossRef]
- Heitzer, A.; Malachowsky, K.; Thonnard, J.E.; Bienkowski, P.R.; White, D.C.; Sayler, G.S. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol. 1994, 60, 1487–1494. [Google Scholar] [CrossRef] [Green Version]
- Ivask, A.; Rolova, T.; Kahru, A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Robinson, G.M.; Tonks, K.M.; Thorn, R.M.; Reynolds, D.M. Application of bacterial bioluminescence to assess the efficacy of fast-acting biocides. Antimicrob. Agents Chemother. 2011, 55, 5214–5220. [Google Scholar] [CrossRef] [Green Version]
- Layton, A.C.; Muccini, M.; Ghosh, M.M.; Sayler, G.S. Construction of a bioluminescent reporter strain To detect polychlorinated biphenyls. Appl. Environ. Microbiol. 1998, 64, 5023–5026. [Google Scholar] [CrossRef] [Green Version]
- Leedjarv, A.; Ivask, A.; Virta, M.; Kahru, A. Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere 2006, 64, 1910–1919. [Google Scholar] [CrossRef]
- Biran, A.; Yagur-Kroll, S.; Pedahzur, R.; Buchinger, S.; Reifferscheid, G.; Ben-Yoav, H.; Shacham-Diamand, Y.; Belkin, S. Bacterial genotoxicity bioreporters. Microb. Biotechnol. 2010, 3, 412–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niazi, J.H.; Kim, B.C.; Ahn, J.M.; Gu, M.B. A novel bioluminescent bacterial biosensor using the highly specific oxidative stress-inducible pgi gene. Biosens. Bioelectron. 2008, 24, 670–675. [Google Scholar] [CrossRef]
- Van Dyk, T.K.; Majarian, W.R.; Konstantinov, K.B.; Young, R.M.; Dhurjati, P.S.; LaRossa, R.A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl. Environ. Microbiol. 1994, 60, 1414–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Youn, C.H.; Kim, B.C.; Gu, M.B. An oxidative stress-specific bacterial cell array chip for toxicity analysis. Biosens. Bioelectron. 2007, 22, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Southward, C.M.; Surette, M.G. The dynamic microbe: Green fluorescent protein brings bacteria to light. Mol. Microbiol. 2002, 45, 1191–1196. [Google Scholar] [CrossRef] [Green Version]
- Gireesh-Babu, P.; Chaudhari, A. Development of a broad-spectrum fluorescent heavy metal bacterial biosensor. Mol. Biol. Rep. 2012, 39, 11225–11229. [Google Scholar] [CrossRef]
- Stiner, L.; Halverson, L.J. Development and characterization of a green fluorescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl. Environ. Microbiol. 2002, 68, 1962–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biran, I.; Rissin, D.M.; Ron, E.Z.; Walt, D.R. Optical imaging fiber-based live bacterial cell array biosensor. Anal. Biochem. 2003, 315, 106–113. [Google Scholar] [CrossRef]
- Stocker, J.; Balluch, D.; Gsell, M.; Harms, H.; Feliciano, J.; Daunert, S.; Malik, K.A.; van der Meer, J.R. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 2003, 37, 4743–4750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Campbell, R.E.; Ting, A.Y.; Tsien, R.Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002, 3, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Mirasoli, M.; Feliciano, J.; Michelini, E.; Daunert, S.; Roda, A. Internal response correction for fluorescent whole-cell biosensors. Anal. Chem. 2002, 74, 5948–5953. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Gu, M.B. An integrated mini biosensor system for continuous water toxicity monitoring. Biosens. Bioelectron. 2005, 20, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Thouand, G.; Horry, H.; Durand, M.J.; Picart, P.; Bendriaa, L.; Daniel, P.; DuBow, M.S. Development of a biosensor for on-line detection of tributyltin with a recombinant bioluminescent Escherichia coli strain. Appl. Microbiol. Biotechnol. 2003, 62, 218–225. [Google Scholar] [CrossRef]
- Yoo, S.K.; Lee, J.H.; Yun, S.S.; Gu, M.B.; Lee, J.H. Fabrication of a bio-MEMS based cell-chip for toxicity monitoring. Biosens. Bioelectron. 2007, 22, 1586–1592. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.R.; Choi, S.J.; Jeong, H.D.; Hong, S. Development of QCM biosensor to detect a marine derived pathogenic bacteria Edwardsiella tarda using a novel immobilisation method. Biosens. Bioelectron. 2009, 24, 1635–1640. [Google Scholar] [CrossRef]
- Woutersen, M.; van der Gaag, B.; Abrafi Boakye, A.; Mink, J.; Marks, R.S.; Wagenvoort, A.J.; Ketelaars, H.A.M.; Brouwer, B.; Heringa, M.B. Development and Validation of an On-Line Water Toxicity Sensor with Immobilized Luminescent Bacteria for On-Line Surface Water Monitoring. Sensors 2017, 17, 2682. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhou, T.; Guo, J.; Li, Y. Dark variants of luminous bacteria whole cell bioluminescent optical fiber sensor to genotoxicants. J. Huazhong Univ. Sci. Technol. Med. Sci. 2004, 24, 507–509. [Google Scholar]
- Hakkila, K.; Green, T.; Leskinen, P.; Ivask, A.; Marks, R.; Virta, M. Detection of bioavailable heavy metals in EILATox-Oregon samples using whole-cell luminescent bacterial sensors in suspension or immobilized onto fibre-optic tips. J. Appl. Toxicol. 2004, 24, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Charrier, T.; Durand, M.J.; Jouanneau, S.; Dion, M.; Pernetti, M.; Poncelet, D.; Thouand, G. A multi-channel bioluminescent bacterial biosensor for the on-line detection of metals and toxicity. Part I: Design and optimization of bioluminescent bacterial strains. Anal. Bioanal. Chem. 2011, 400, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Mitchell, R.J.; Kim, B.C.; Cullen, D.C.; Gu, M.B. A cell array biosensor for environmental toxicity analysis. Biosens. Bioelectron. 2005, 21, 500–507. [Google Scholar] [CrossRef]
- Campana, O.; Wlodkowic, D. The undiscovered country: Ecotoxicology meets microfluidics. Sensor. Actuat. B-Chem. 2018, 257, 692–704. [Google Scholar] [CrossRef]
- Campana, O.; Wlodkowic, D. Ecotoxicology Goes on a Chip: Embracing Miniaturized Bioanalysis in Aquatic Risk Assessment. Environ. Sci. Technol. 2018, 52, 932–946. [Google Scholar] [CrossRef]
- Bachmann, T. Transforming cyanobacteria into bioreporters of biological relevance. Trends Biotechnol. 2003, 21, 247–249. [Google Scholar] [CrossRef]
- Shao, C.Y.; Howe, C.J.; Porter, A.J.; Glover, L.A. Novel cyanobacterial biosensor for detection of herbicides. Appl. Environ. Microbiol. 2002, 68, 5026–5033. [Google Scholar] [CrossRef] [Green Version]
- Shing, W.L.; Heng, L.Y.; Surif, S. Performance of a cyanobacteria whole cell-based fluorescence biosensor for heavy metal and pesticide detection. Sensors 2013, 13, 6394–6404. [Google Scholar]
- Lefevre, F.; Chalifour, A.; Yu, L.; Chodavarapu, V.; Juneau, P.; Izquierdo, R. Algal fluorescence sensor integrated into a microfluidic chip for water pollutant detection. Lab Chip 2012, 12, 787–793. [Google Scholar] [CrossRef]
- Yuce, M.; Nazir, H.; Donmez, G. An advanced investigation on a new algal sensor determining Pb(II) ions from aqueous media. Biosens. Bioelectron. 2010, 26, 321–326. [Google Scholar] [CrossRef]
- Mbeunkui, F.; Richaud, C.; Etienne, A.L.; Schmid, R.D.; Bachmann, T.T. Bioavailable nitrate detection in water by an immobilized luminescent cyanobacterial reporter strain. Appl. Microbiol. Biotechnol. 2002, 60, 306–312. [Google Scholar]
- Schreiter, P.P.; Gillor, O.; Post, A.; Belkin, S.; Schmid, R.D.; Bachmann, T.T. Monitoring of phosphorus bioavailability in water by an immobilized luminescent cyanobacterial reporter strain. Biosens. Bioelectron. 2001, 16, 811–818. [Google Scholar] [CrossRef]
- Lee, H.R.; Jung, S.M.; Yoon, S.; Yoon, W.H.; Park, T.H.; Kim, S.; Shin, H.W.; Hwang, D.S.; Jung, S. Immobilization of planktonic algal spores by inkjet printing. Sci. Rep. 2019, 9, 12357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtson Nash, S.M.; Quayle, P.A.; Schreiber, U.; Muller, J.F. The selection of a model microalgal species as biomaterial for a novel aquatic phytotoxicity assay. Aquat. Toxicol. 2005, 72, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Franqueira, D.; Orosa, M.; Torres, E.; Herrero, C.; Cid, A. Potential use of flow cytometry in toxicity studies with microalgae. Sci. Total Environ. 2000, 247, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Campanella, L.; Cubadda, F.; Sammartino, M.P.; Saoncella, A. An algal biosensor for the monitoring of water toxicity in estuarine environments. Water Res. 2001, 35, 69–76. [Google Scholar] [CrossRef]
- Cho, C.W.; Pham, T.P.; Jeon, Y.C.; Min, J.; Jung, H.Y.; Lee, D.S.; Yun, Y.S. Microalgal photosynthetic activity measurement system for rapid toxicity assessment. Ecotoxicology 2008, 17, 455–463. [Google Scholar] [CrossRef]
- Shitanda, I.; Takada, K.; Sakai, Y.; Tatsuma, T. Amperometric biosensing systems based on motility and gravitaxis of flagellate algae for aquatic risk assessment. Anal. Chem. 2005, 77, 6715–6718. [Google Scholar] [CrossRef] [PubMed]
- Endo, R.; Omasa, K. Chlorophyll fluorescence imaging of individual algal cells: Effects of herbicide on Spirogyra distenta at different growth stages. Environ. Sci. Technol. 2004, 38, 4165–4168. [Google Scholar] [CrossRef]
- Hassannejad, S.; Lotfi, R.; Ghafarbi, S.P.; Oukarroum, A.; Abbasi, A.; Kalaji, H.M.; Rastogi, A. Early Identification of Herbicide Modes of Action by the Use of Chlorophyll Fluorescence Measurements. Plants 2020, 9, 529. [Google Scholar] [CrossRef]
- Anandraj, A.; White, S.; Naidoo, D.; Mutanda, T. Monitoring the acclimatization of a Chlorella sp. From freshwater to hypersalinity using photosynthetic parameters of pulse amplitude modulated fluorometry. Bioresour. Technol. 2020, 309, 123380. [Google Scholar] [CrossRef]
- Miao, A.J.; Wang, W.X.; Juneau, P. Comparison of Cd, Cu, and Zn toxic effects on four marine phytoplankton by pulse-amplitude-modulated fluorometry. Environ. Toxicol. Chem. 2005, 24, 2603–2611. [Google Scholar] [CrossRef]
- Muller, R.; Schreiber, U.; Escher, B.I.; Quayle, P.; Bengtson Nash, S.M.; Mueller, J.F. Rapid exposure assessment of PSII herbicides in surface water using a novel chlorophyll a fluorescence imaging assay. Sci. Total Environ. 2008, 401, 51–59. [Google Scholar] [CrossRef]
- Schreiber, U.; Muller, J.F.; Haugg, A.; Gademann, R. New type of dual-channel PAM chlorophyll fluorometer for highly sensitive water toxicity biotests. Photosynth. Res. 2002, 74, 317–330. [Google Scholar] [CrossRef]
- Tatsuma, T.; Yoshida, Y.; Shitanda, I.; Notsu, H. Algal biosensor array on a single electrode. Analyst 2009, 134, 223–225. [Google Scholar] [CrossRef]
- Umar, L.; Alexander, F.A.; Wiest, J. Application of algae-biosensor for environmental monitoring. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 7099–7102. [Google Scholar] [PubMed]
- Naessens, M.; Leclerc, J.C.; Tran-Minh, C. Fiber optic biosensor using Chlorella vulgaris for determination of toxic compounds. Ecotoxicol. Environ. Saf. 2000, 46, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Vedrine, C.; Leclerc, J.C.; Durrieu, C.; Tran-Minh, C. Optical whole-cell biosensor using Chlorella vulgaris designed for monitoring herbicides. Biosens. Bioelectron. 2003, 18, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Sekli Belaidi, F.; Farouil, L.; Salvagnac, L.; Temple-Boyer, P.; Seguy, I.; Heully, J.L.; Alary, F.; Bedel-Pereira, E.; Launay, J. Towards integrated multi-sensor platform using dual electrochemical and optical detection for on-site pollutant detection in water. Biosens. Bioelectron. 2019, 132, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Perez-Bueno, M.L.; Pineda, M.; Baron, M. Phenotyping Plant Responses to Biotic Stress by Chlorophyll Fluorescence Imaging. Front. Plant Sci. 2019, 10, 1135. [Google Scholar] [CrossRef]
- Schreiber, U.; Quayle, P.; Schmidt, S.; Escher, B.I.; Mueller, J.F. Methodology and evaluation of a highly sensitive algae toxicity test based on multiwell chlorophyll fluorescence imaging. Biosens. Bioelectron. 2007, 22, 2554–2563. [Google Scholar] [CrossRef]
- Oxborough, K. Imaging of chlorophyll a fluorescence: Theoretical and practical aspects of an emerging technique for the monitoring of photosynthetic performance. J. Exp. Bot. 2004, 55, 1195–1205. [Google Scholar] [CrossRef]
- Gavel, A.; Marsalek, B. A novel approach for phytotoxicity assessment by CCD fluorescence imaging. Environ. Toxicol. 2004, 19, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Hupp, S.; Rosenkranz, M.; Bonfig, K.; Pandey, C.; Roitsch, T. Noninvasive Phenotyping of Plant-Pathogen Interaction: Consecutive In Situ Imaging of Fluorescing Pseudomonas syringae, Plant Phenolic Fluorescence, and Chlorophyll Fluorescence in Arabidopsis Leaves. Front. Plant Sci. 2019, 10, 1239. [Google Scholar] [CrossRef] [Green Version]
- Tsopela, A.; Laborde, A.; Salvagnac, L.; Ventalon, V.; Bedel-Pereira, E.; Seguy, I.; Temple-Boyer, P.; Juneau, P.; Izquierdo, R.; Launay, J. Development of a lab-on-chip electrochemical biosensor for water quality analysis based on microalgal photosynthesis. Biosens. Bioelectron. 2016, 79, 568–573. [Google Scholar] [CrossRef] [Green Version]
- Skommer, J.; Akagi, J.; Takeda, K.; Fujimura, Y.; Khoshmanesh, K.; Wlodkowic, D. Multiparameter Lab-on-a-Chip flow cytometry of the cell cycle. Biosens. Bioelectron. 2013, 42, 586–591. [Google Scholar] [CrossRef]
- Wlodkowic, D.; Skommer, J.; Akagi, J.; Fujimura, Y.; Takeda, K. Multiparameter analysis of apoptosis using lab-on-a-chip flow cytometry. Curr. Protoc. Cytom. 2013, 66, 9–42. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Darzynkiewicz, Z. Rise of the Micromachines: Microfluidics and the Future of Cytometry. Recent Adv. Cytom. Part A Instrum. Methods 2011, 102, 105–125. [Google Scholar]
- Franklin, N.M.; Stauber, J.L.; Lim, R.P. Development of flow cytometry-based algal bioassays for assessing toxicity of copper in natural waters. Environ. Toxicol. Chem. 2001, 20, 160–170. [Google Scholar] [CrossRef]
- Rakers, S.; Imse, F.; Gebert, M. Real-time cell analysis: Sensitivity of different vertebrate cell cultures to copper sulfate measured by xCELLigence(A (R)). Ecotoxicology 2014, 23, 1582–1591. [Google Scholar] [CrossRef]
- Bohrn, U.; Mucha, A.; Werner, C.F.; Trattner, B.; Backer, M.; Krumbe, C.; Schienle, M.; Stutz, E.; Schmitt-Landsiedel, D.; Fleischer, M.; et al. A critical comparison of cell-based sensor systems for the detection of Cr(VI) in aquatic environment. Sens. Actuat B-Chem. 2013, 182, 58–65. [Google Scholar] [CrossRef]
- Yan, G.; Du, Q.; Wei, X.; Miozzi, J.; Kang, C.; Wang, J.; Han, X.; Pan, J.; Xie, H.; Chen, J.; et al. Application of Real-Time Cell Electronic Analysis System in Modern Pharmaceutical Evaluation and Analysis. Molecules 2018, 23, 3280. [Google Scholar] [CrossRef] [Green Version]
- Giaever, I.; Keese, C.R. A morphological biosensor for mammalian cells. Nature 1993, 366, 591–592. [Google Scholar] [CrossRef] [PubMed]
- Curtis, T.M.; Widder, M.W.; Brennan, L.M.; Schwager, S.J.; van der Schalie, W.H.; Fey, J.; Salazar, N. A portable cell-based impedance sensor for toxicity testing of drinking water. Lab Chip 2009, 9, 2176–2183. [Google Scholar] [CrossRef] [PubMed]
- van der Schalie, W.H.; James, R.R.; Gargan, T.P. Selection of a battery of rapid toxicity sensors for drinking water evaluation. Biosens. Bioelectron. 2006, 22, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Wegener, J.; Keese, C.R.; Giaever, I. Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp. Cell Res. 2000, 259, 158–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, N.; Wang, X.; Xu, X.; Abassi, Y.A. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 2011, 740, 33–43. [Google Scholar] [PubMed]
- Brennan, L.M.; Widder, M.W.; McAleer, M.K.; Mayo, M.W.; Greis, A.P.; van der Schalie, W.H. Preparation and Testing of Impedance-based Fluidic Biochips with RTgill-W1 Cells for Rapid Evaluation of Drinking Water Samples for Toxicity. J. Vis. Exp. 2016, 109, 53555. [Google Scholar] [CrossRef] [Green Version]
- Kubisch, R.; Bohrn, U.; Fleischer, M.; Stutz, E. Cell-based sensor system using L6 cells for broad band continuous pollutant monitoring in aquatic environments. Sensors 2012, 12, 3370–3393. [Google Scholar] [CrossRef]
- Brennan, L.M.; Widder, M.W.; Lee, L.E.; van der Schalie, W.H. Long-term storage and impedance-based water toxicity testing capabilities of fluidic biochips seeded with RTgill-W1 cells. Toxicology 2012, 26, 736–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, T.M.; Tabb, J.; Romeo, L.; Schwager, S.J.; Widder, M.W.; van der Schalie, W.H. Improved cell sensitivity and longevity in a rapid impedance-based toxicity sensor. J. Appl. Toxicol. 2009, 29, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Bury, N.R.; Schnell, S.; Hogstrand, C. Gill cell culture systems as models for aquatic environmental monitoring. J. Exp. Biol. 2014, 217 Pt 5, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.E.; Dayeh, V.R.; Schirmer, K.; Bols, N.C. Applications and potential uses of fish gill cell lines: Examples with RTgill-W1. Cell Dev. Biol. Anim. 2009, 45, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, K.; Dixon, D.G.; Greenberg, B.M.; Bols, N.C. Ability of 16 priority PAHs to be directly cytotoxic to a cell line from the rainbow trout gill. Toxicology 1998, 127, 129–141. [Google Scholar] [CrossRef]
- Minghetti, M.; Schnell, S.; Chadwick, M.A.; Hogstrand, C.; Bury, N.R. A primary FIsh Gill Cell System (FIGCS) for environmental monitoring of river waters. Aquat. Toxicol. 2014, 154, 184–192. [Google Scholar] [CrossRef]
- Schnell, S.; Bawa-Allah, K.; Otitoloju, A.; Hogstrand, C.; Miller, T.H.; Barron, L.P.; Bury, N.R. Environmental monitoring of urban streams using a primary fish gill cell culture system (FIGCS). Ecotoxicol. Environ. Saf. 2015, 120, 279–285. [Google Scholar] [CrossRef]
- Maunder, R.J.; Baron, M.G.; Owen, S.F.; Jha, A.N. Investigations to extend viability of a rainbow trout primary gill cell culture. Ecotoxicology 2017, 26, 1314–1326. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wlodkowic, D.; Karpiński, T.M. Live-Cell Systems in Real-Time Biomonitoring of Water Pollution: Practical Considerations and Future Perspectives. Sensors 2021, 21, 7028. https://doi.org/10.3390/s21217028
Wlodkowic D, Karpiński TM. Live-Cell Systems in Real-Time Biomonitoring of Water Pollution: Practical Considerations and Future Perspectives. Sensors. 2021; 21(21):7028. https://doi.org/10.3390/s21217028
Chicago/Turabian StyleWlodkowic, Donald, and Tomasz M. Karpiński. 2021. "Live-Cell Systems in Real-Time Biomonitoring of Water Pollution: Practical Considerations and Future Perspectives" Sensors 21, no. 21: 7028. https://doi.org/10.3390/s21217028
APA StyleWlodkowic, D., & Karpiński, T. M. (2021). Live-Cell Systems in Real-Time Biomonitoring of Water Pollution: Practical Considerations and Future Perspectives. Sensors, 21(21), 7028. https://doi.org/10.3390/s21217028






