Discrimination of Different Breast Cell Lines on Glass Substrate by Means of Fourier Transform Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Preparation
2.2. FTIR Measurements and Spectral Analysis
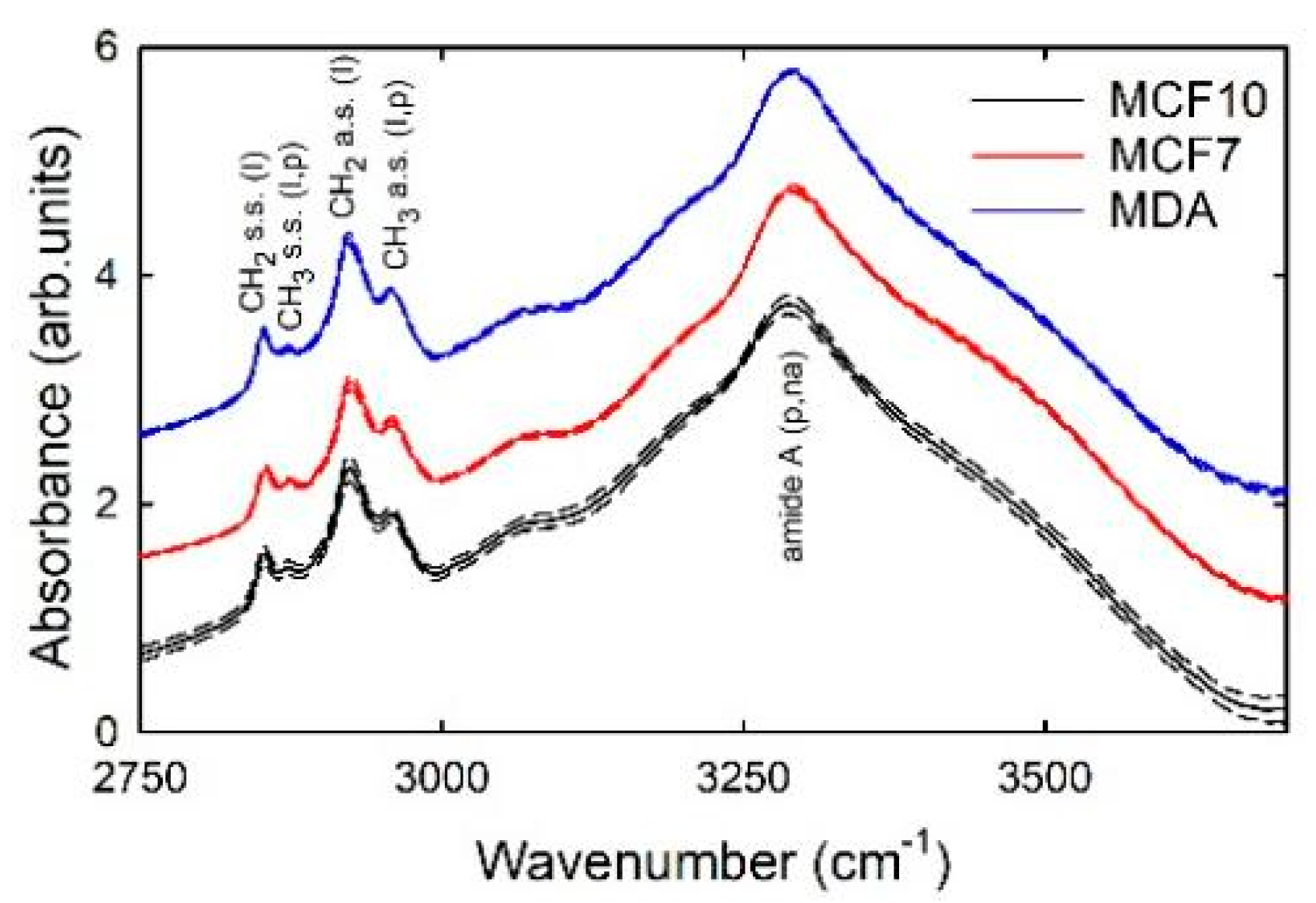
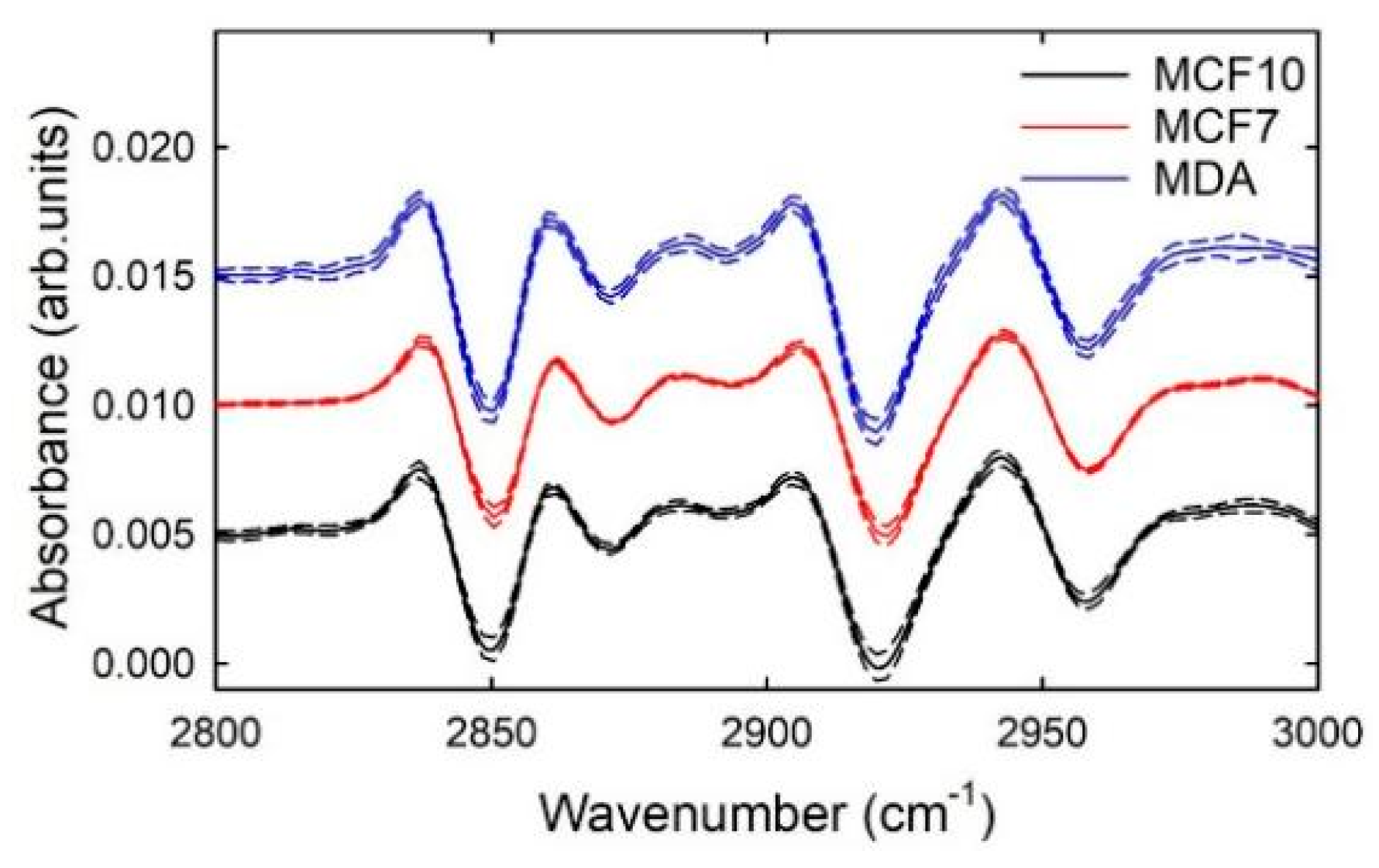
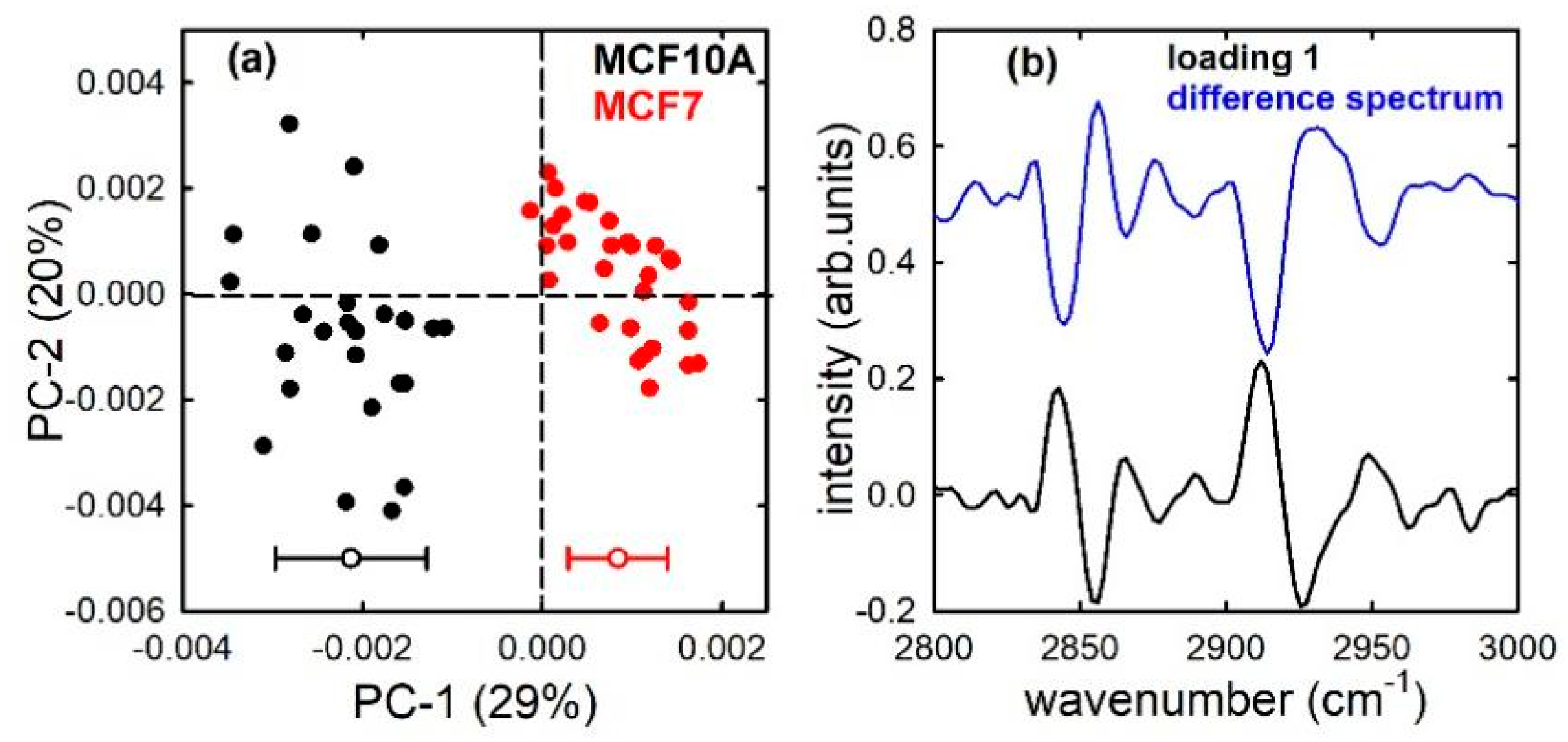
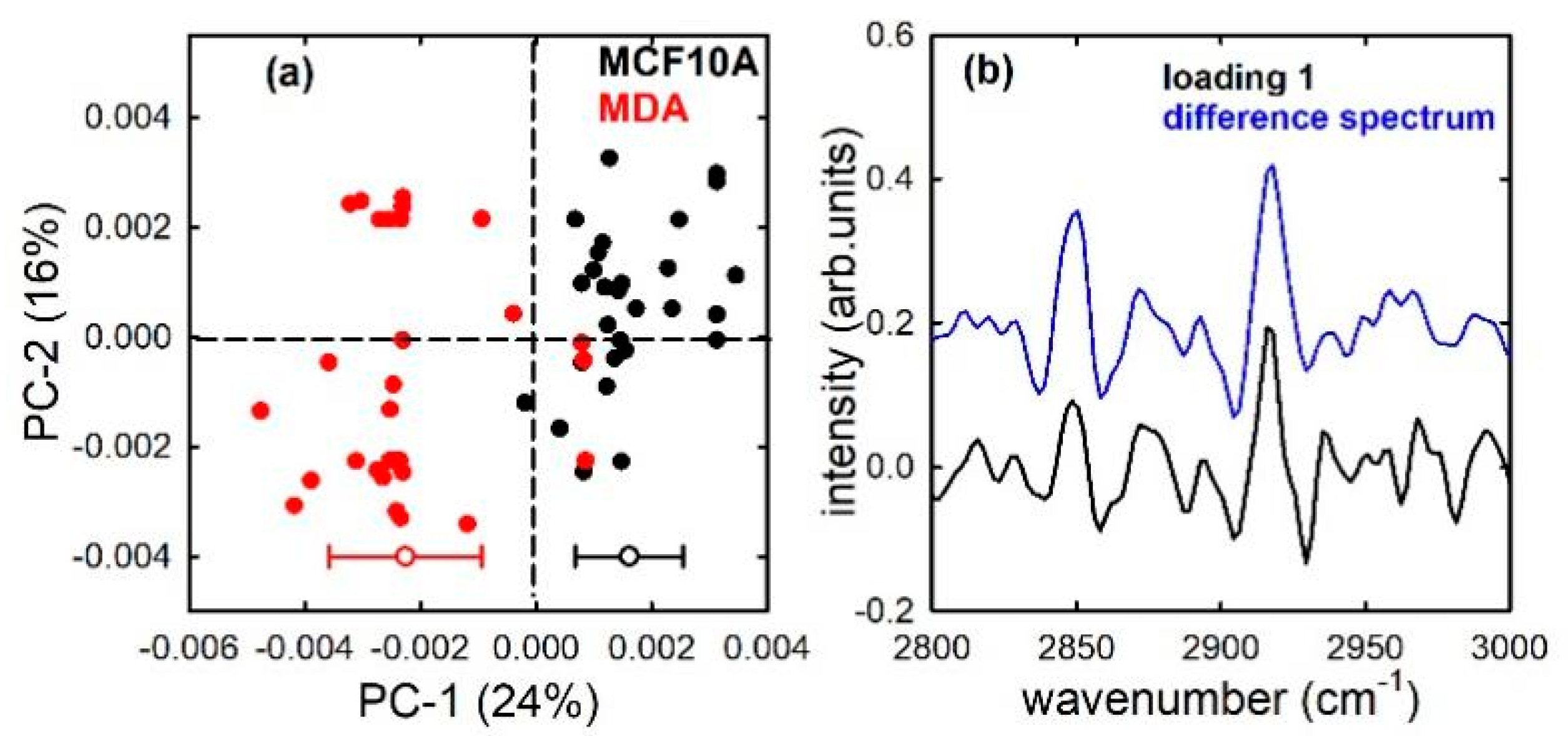
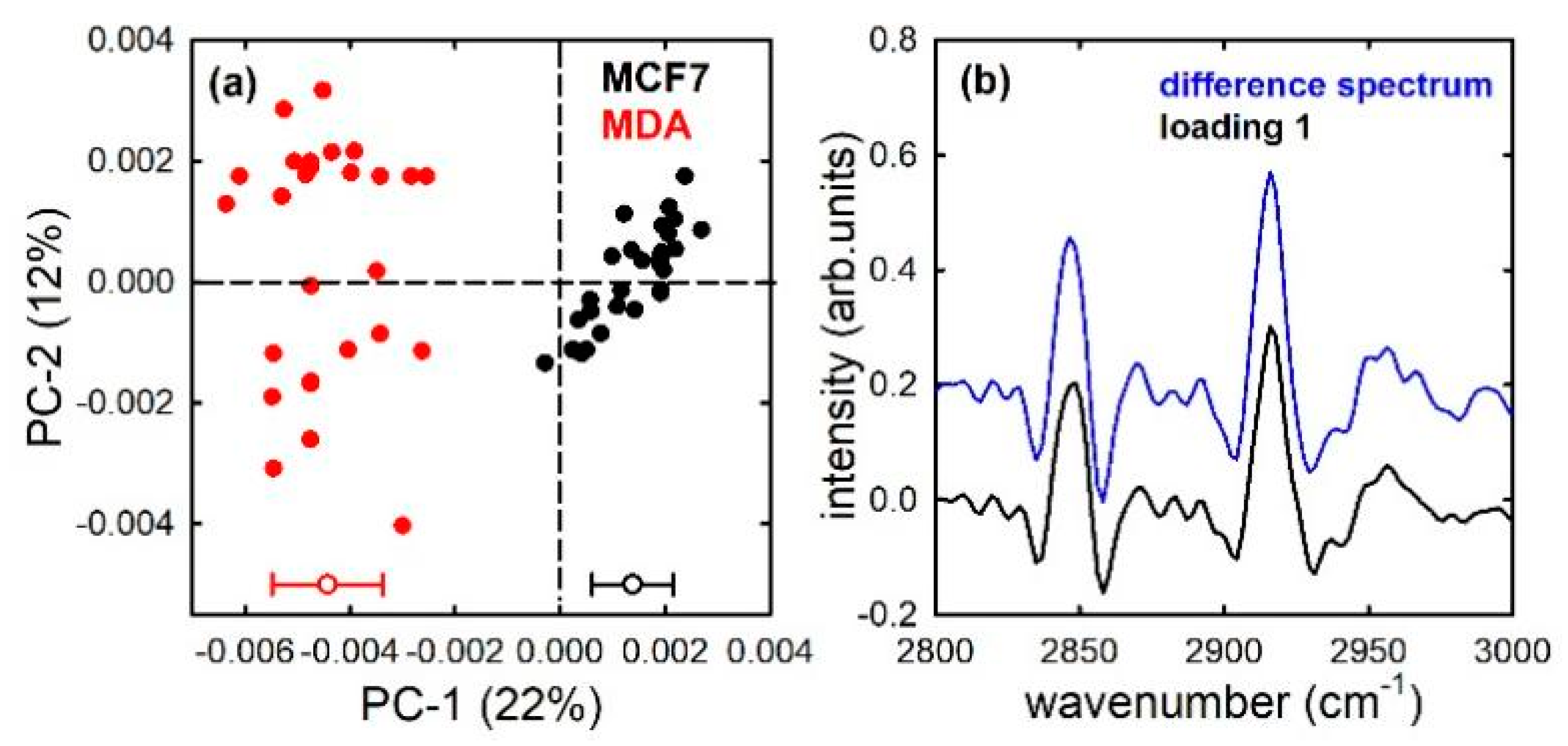
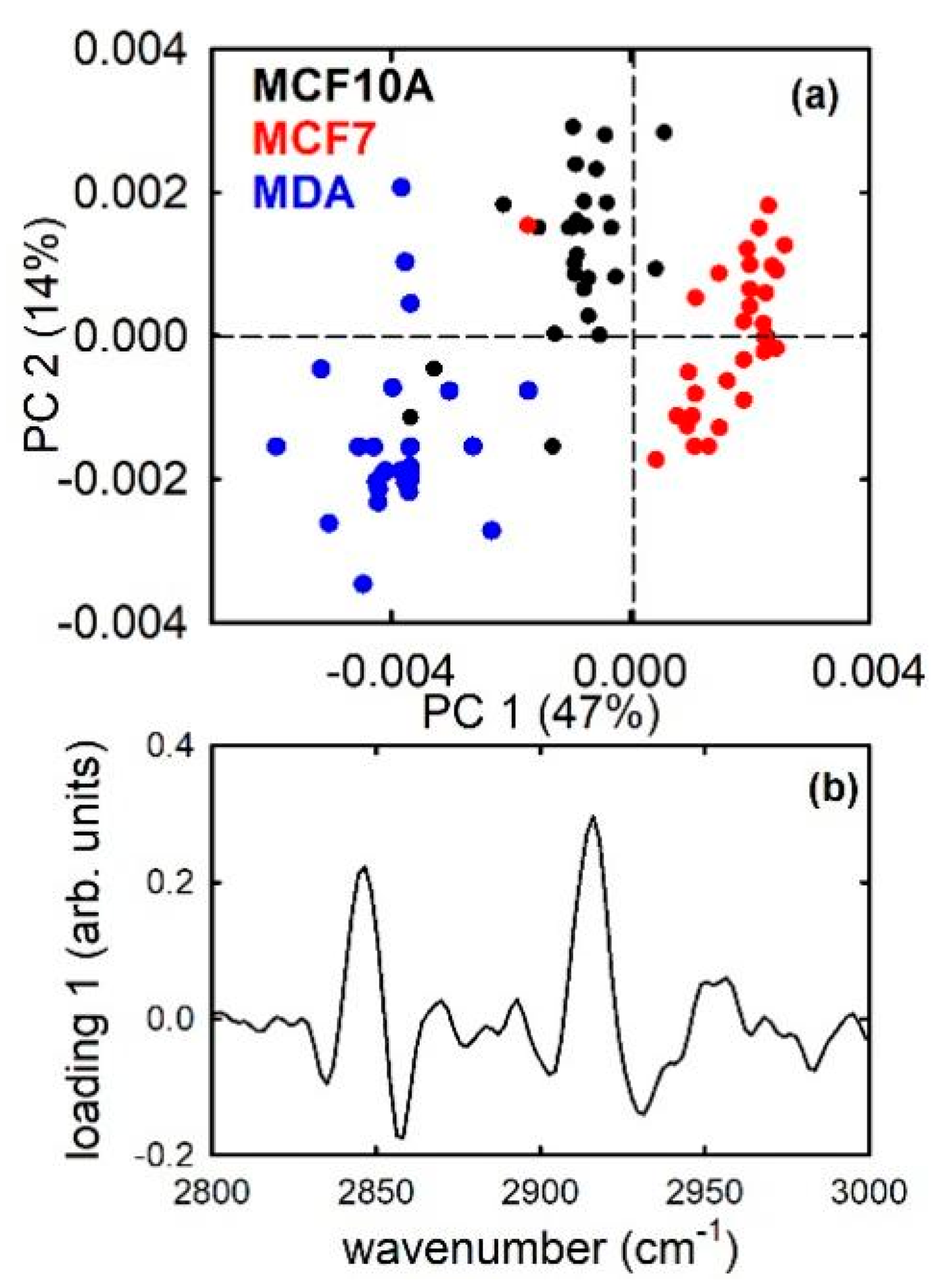
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Sheng, D.; Xu, F.; Yu, Q.; Fang, T.; Xia, J.; Li, S.; Wang, X. A study of structural differences between liver cancer cells and normal liver cells using FTIR spectroscopy. J. Mol. Struct. 2015, 1099, 18–23. [Google Scholar] [CrossRef]
- Butler, H.; Cameron, J.M.; Jenkins, C.A.; Hithell, G.; Hume, S.; Hunt, N.T.; Baker, M.J. Shining a light on clinical spectroscopy: Translation of diagnostic IR, 2D-IR and Raman spectroscopy towards the clinic. Clin. Spectrosc. 2019, 1, 100003. [Google Scholar] [CrossRef]
- Finlayson, D.; Rinaldi, C.; Baker, M.J. Is Infrared Spectroscopy Ready for the Clinic? Anal. Chem. 2019, 91, 12117–12128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, H.J.; Behl, I.; Calado, G.; Ibrahim, O.; Toner, M.; Galvin, S.; Healy, C.M.; Flint, S.; Lyng, F.M. Biomedical applications of vi-brational spectroscopy: Oral cancer diagnostics. Spectrochim Acta A Mol. Biomol. Spectrosc. 2021, 252, 119470. [Google Scholar] [CrossRef] [PubMed]
- Elshemey, W.M.; Ismail, A.M.; Elbialy, N.S. Molecular-Level Characterization of Normal, Benign, and Malignant Breast Tissues Using FTIR Spectroscopy. J. Med. Biol. Eng. 2016, 36, 369–378. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Depciuch, J.; Tołpa, B.; Witek, P.; Szmuc, K.; Kaznowska, E.; Osuchowski, M.; Król, P.; Cebulski, J. Raman and FTIR spec-troscopy in determining the chemical changes in healthy brain tissues and glioblastoma tumor tissues. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 225, 117526. [Google Scholar] [CrossRef] [PubMed]
- Seredin, P.; Goloshchapov, D.; Ippolitov, Y.; Vongsvivut, J. Spectroscopic signature of the patho-logical processes of carious dentine based on FTIR investigations of the oral biological fluids. Biomed. Opt. Express 2019, 10, 4050–4058. [Google Scholar] [CrossRef]
- Byrne, H.J.; Bonnier, F.; McIntyre, J.; Parachalil, D.R. Quantitative analysis of human blood serum using vibrational spectroscopy. Clin. Spectrosc. 2020, 2, 100004. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.; Lima, K.M.; Ashton, K.M.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Potential of mid-infrared spectroscopy as a non-invasive diagnostic test in urine for endometrial or ovarian cancer. Analyst 2018, 143, 3156–3163. [Google Scholar] [CrossRef]
- Meade, A.D.; Howe, O.; Unterreiner, V.; Sockalingum, G.D.; Byrne, H.J.; Lyng, F.M. Vibrational spectroscopy in sensing radiobi-ological effects: Analyses of targeted and non-targeted effects in human keratinocytes. Faraday Discuss. 2016, 187, 213–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depciuch, J.; Stanek-Widera, A.; Warchulska, M.; Lange, D.; Sarwa, K.; Koziorowska, A.; Kula, M.; Cebulski, J. Identification of chemical changes in healthy breast tissue caused by chemotherapy using Raman and FTIR spectroscopy: A preliminary study. Infrared Phys. Technol. 2019, 102, 102989. [Google Scholar] [CrossRef]
- Monici, M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [CrossRef]
- Butler, H.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [Green Version]
- Traynor, D.; Behl, I.; O’Dea, D.; Bonnier, F.; Nicholson, S.; O’Connell, F.; Maguire, A.; Flint, S.; Galvin, S.; Healy, C.M.; et al. Raman spectral cytopathology for cancer diagnostic applications. Nat. Protoc. 2021, 16, 1–20. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Ding, R.; Shen, L.; Gao, P.; Xu, H.; Xiu, C.; Zhang, H.; Song, D.; Han, B. Characterization and imaging of surgical specimens of invasive breast cancer and normal breast tissues with the application of raman spec-tral mapping: A feasibility study and comparison with randomized single-point detection method. Oncol. Lett. 2020, 20, 2969. [Google Scholar] [CrossRef] [PubMed]
- Staritzbichler, R.; Hunold, P.; Estrela-Lopis, I.; Hildebrand, P.W.; Isermann, B.; Kaiser, T. Raman spectros-copy on blood serum samples of patients with end-stage liver disease. PLoS ONE 2021, 16, e0256045. [Google Scholar] [CrossRef]
- Hardy, M.; Kelleher, L.; de Carvalho Gomes, P.; Buchan, E.; Chu, H.O.M.; Goldberg Oppenheimer, P. Hin On Martin Chu & Pola Gold-berg Oppenheimer Methods in Raman spectroscopy for saliva studies—A review. Appl. Spectrosc. Rev. 2021, 1–57. [Google Scholar]
- Perna, G.; Capozzi, V.; Lasalvia, M. A Comparison between FTIR Spectra from HUKE and SH-SY5Y Cell Lines Grown on Different Substrates. Appl. Sci. 2020, 10, 8825. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Rutter, A.V.; Crees, J.; Wright, H.; van Pittius, D.G.; Yousef, I.; Sulé-Suso, J. Fourier transform infrared spectra of cells on glass coverslips. A further step in spectral pathology. Analyst 2018, 143, 5711–5717. [Google Scholar] [CrossRef]
- Rutter, A.V.; Crees, J.; Wright, H.; Raseta, M.; Van Pittius, D.G.; Roach, P.; Sulé-Suso, J. Identification of a Glass Substrate to Study Cells Using Fourier Transform Infrared Spectroscopy: Are We Closer to Spectral Pathology? Appl. Spectrosc. 2020, 74, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Zeaiter, M.; Rutledge, D. Preprocessing methods. In Comprehensive Chemometrics: Chemical and Biochemical Data Analysis Vol. 3; Brown, S.D., Tauler, R., Walczak, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 121–231. [Google Scholar]
- Hanson, B.A. ChemoSpec: Exploratory Chemometrics for Spectroscopy. R Package Version 4.4.97. 2017. Available online: https://CRAN.R-project.org/package=ChemoSpec (accessed on 20 July 2021).
- Kar, S.; Katti, D.; Katti, K.S. Fourier transform infrared spectroscopy based spectral biomarkers of metastasized breast cancer progression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 208, 85–96. [Google Scholar] [CrossRef]
- Verdonck, M.; Wald, N.; Janssis, J.; Yan, P.; Meyer, C.; Legat, A.; Speiser, D.E.; Desmedt, C.; Larsimont, D.; Sotiriou, C.; et al. Breast cancer and melanoma cell line identification by FTIR imaging after formalin-fixation and paraffin-embedding. Analyst 2013, 138, 4083–4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Xiao, X.; Tan, J.; Cai, Q. In situ evaluation of breast cancer cell growth with 3D ATR-FTIR spectroscopy. Vib. Spectrosc. 2009, 49, 64–67. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Mohlenhoff, B.; Romeo, M.; Diem, M.; Wood, B.R. Mie-Type Scattering and Non-Beer-Lambert Absorption Behavior of Human Cells in Infrared Microspectroscopy. Biophys. J. 2005, 88, 3635–3640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnier, F.; Byrne, H. Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems. Analyst 2011, 137, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Nieva, C.; Marro, M.; Codina, N.S.; Rao, S.; Petrov, D.; Sierra, A. The Lipid Phenotype of Breast Cancer Cells Characterized by Raman Microspectroscopy: Towards a Stratification of Malignancy. PLoS ONE 2012, 7, e46456. [Google Scholar] [CrossRef] [PubMed]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogene 2016, 5, e189. [Google Scholar] [CrossRef]
- Corbet, C.; Feron, O. Emerging roles of lipid metabolism in cancer progression. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Eiriksson, F.F.; Nøhr, M.K.; Costa, M.; Bödvarsdottir, S.K.; Ögmundsdottir, H.M.; Thorsteinsdottir, M. Lipidomic study of cell lines reveals differences between breast cancer subtypes. PLoS ONE 2020, 15, e0231289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lasalvia, M.; Capozzi, V.; Perna, G. Discrimination of Different Breast Cell Lines on Glass Substrate by Means of Fourier Transform Infrared Spectroscopy. Sensors 2021, 21, 6992. https://doi.org/10.3390/s21216992
Lasalvia M, Capozzi V, Perna G. Discrimination of Different Breast Cell Lines on Glass Substrate by Means of Fourier Transform Infrared Spectroscopy. Sensors. 2021; 21(21):6992. https://doi.org/10.3390/s21216992
Chicago/Turabian StyleLasalvia, Maria, Vito Capozzi, and Giuseppe Perna. 2021. "Discrimination of Different Breast Cell Lines on Glass Substrate by Means of Fourier Transform Infrared Spectroscopy" Sensors 21, no. 21: 6992. https://doi.org/10.3390/s21216992
APA StyleLasalvia, M., Capozzi, V., & Perna, G. (2021). Discrimination of Different Breast Cell Lines on Glass Substrate by Means of Fourier Transform Infrared Spectroscopy. Sensors, 21(21), 6992. https://doi.org/10.3390/s21216992







