Cost-Effective Multiplex Fluorescence Detection System for PCR Chip †
Abstract
1. Introduction
2. Materials and Methods
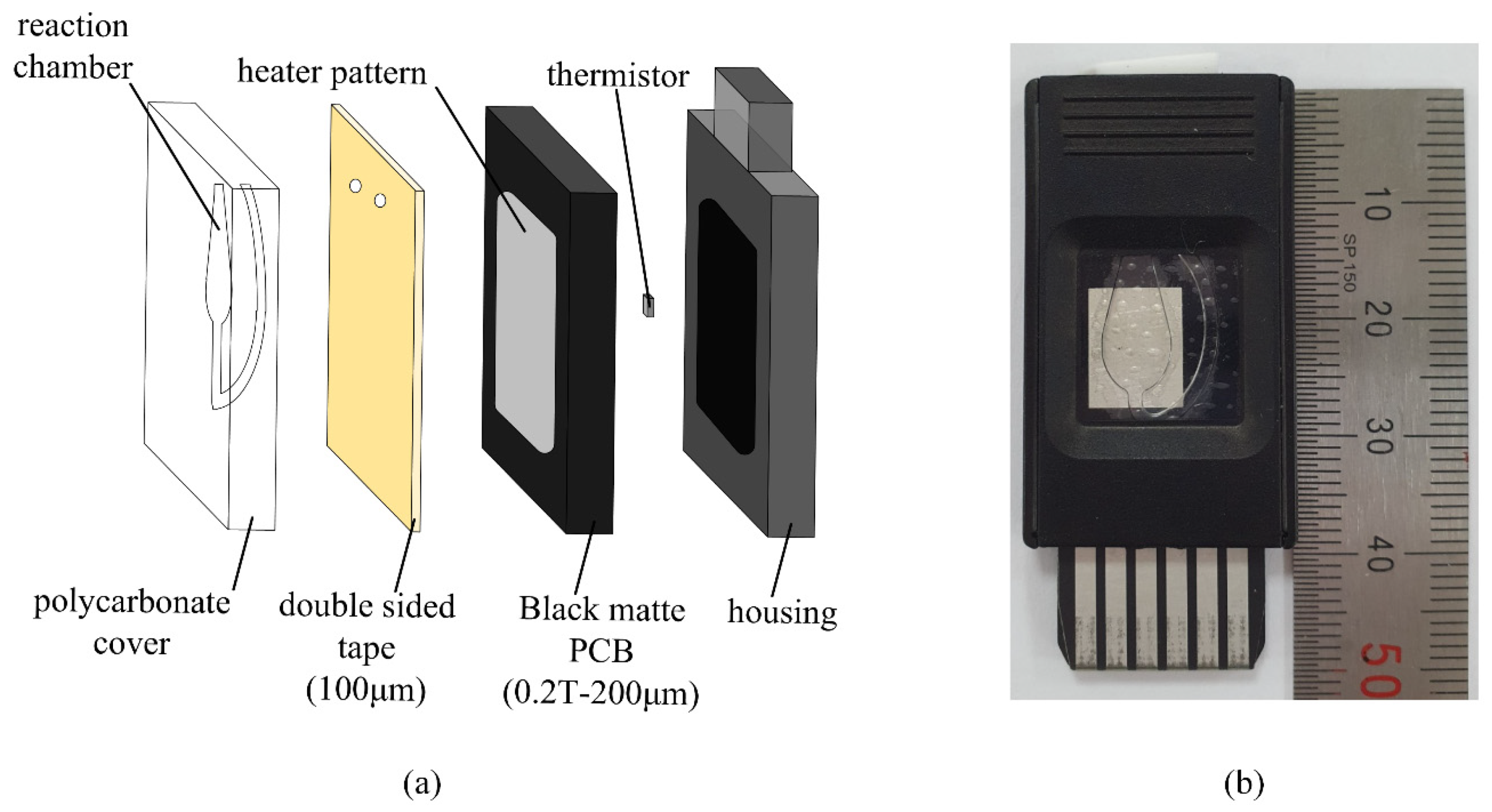
2.1. Overall System
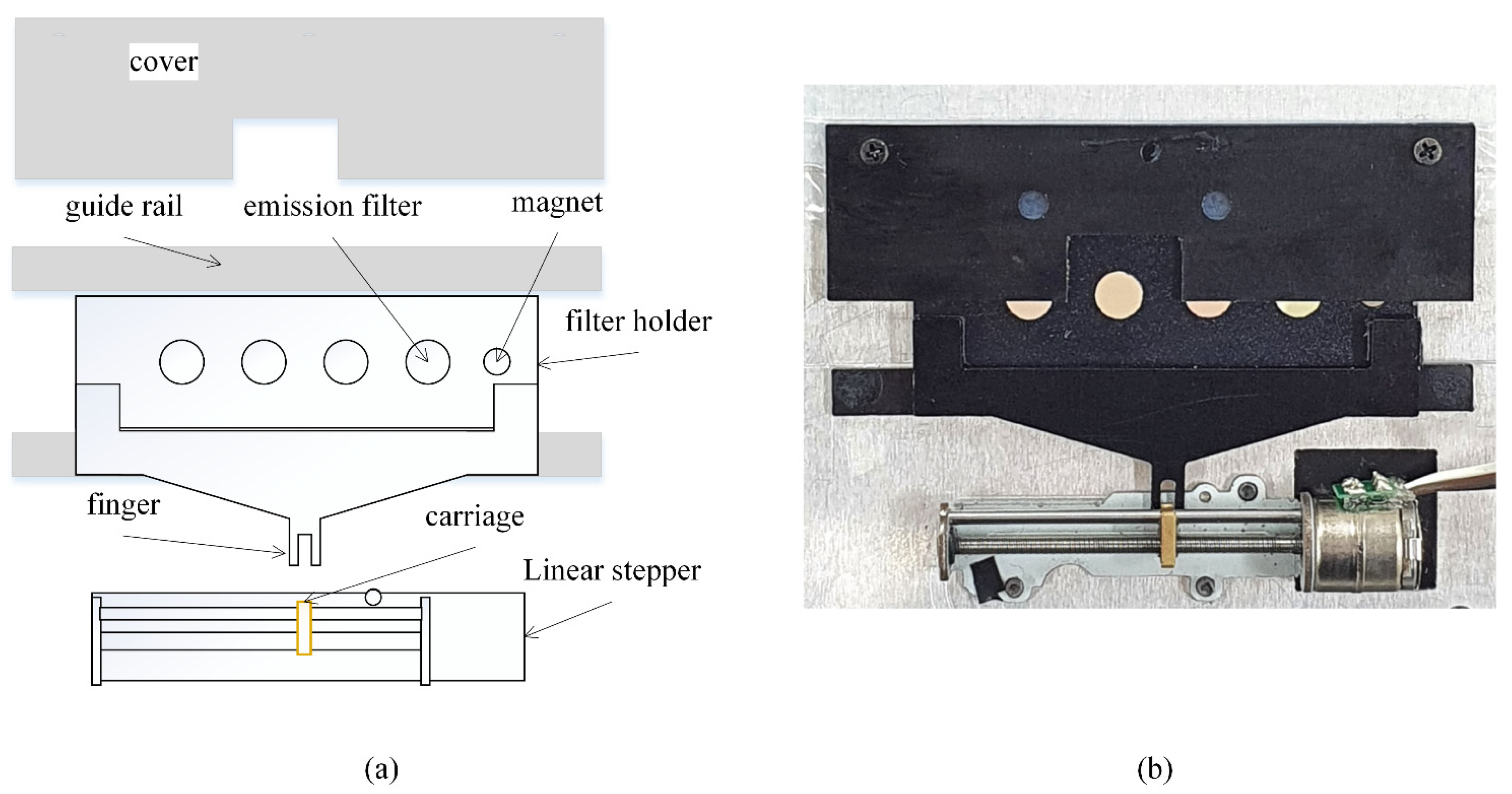
2.2. Excitation Unit and Emission Filter Wheel
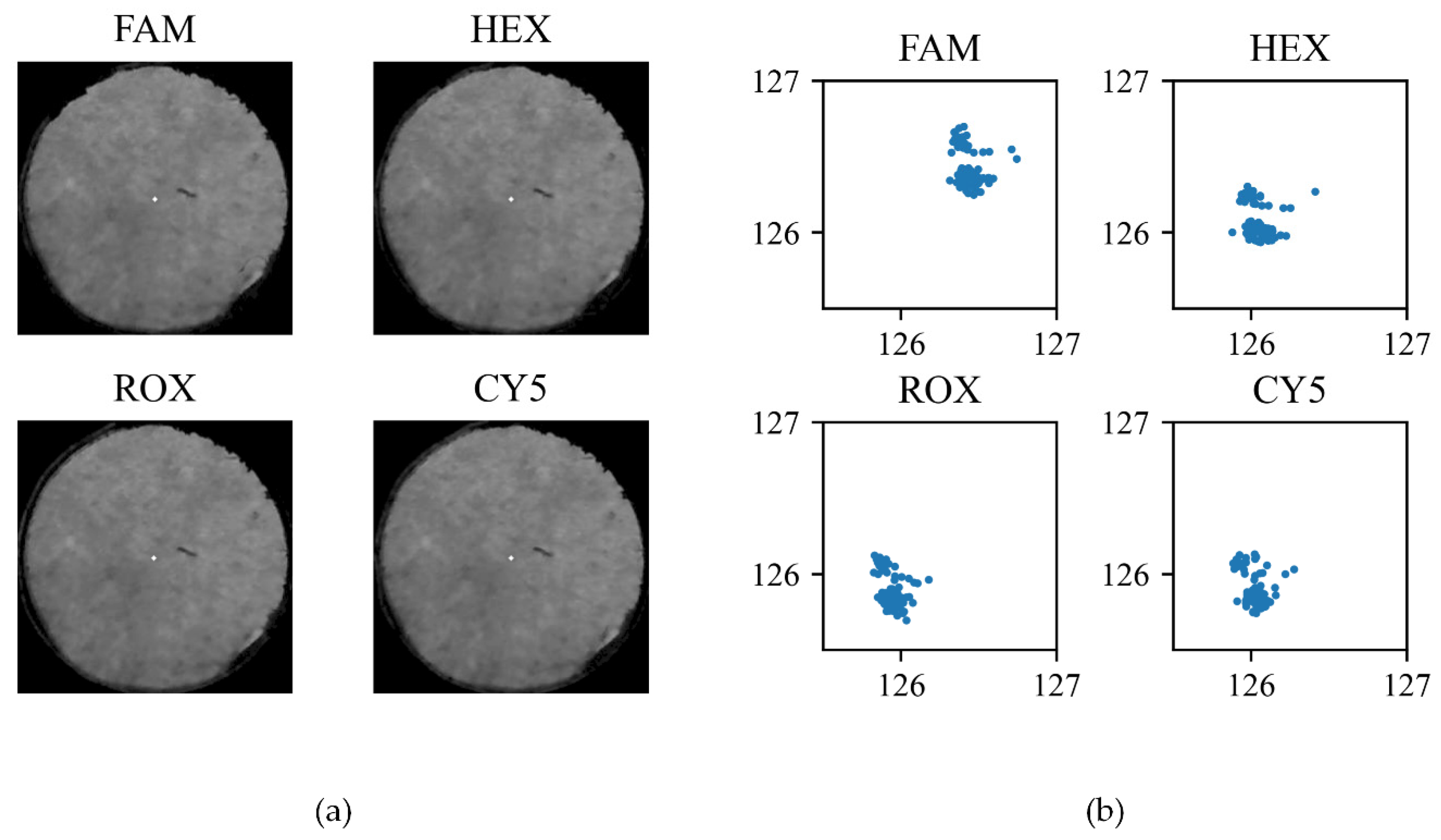
2.3. Flourescence Detection Performance Analysis
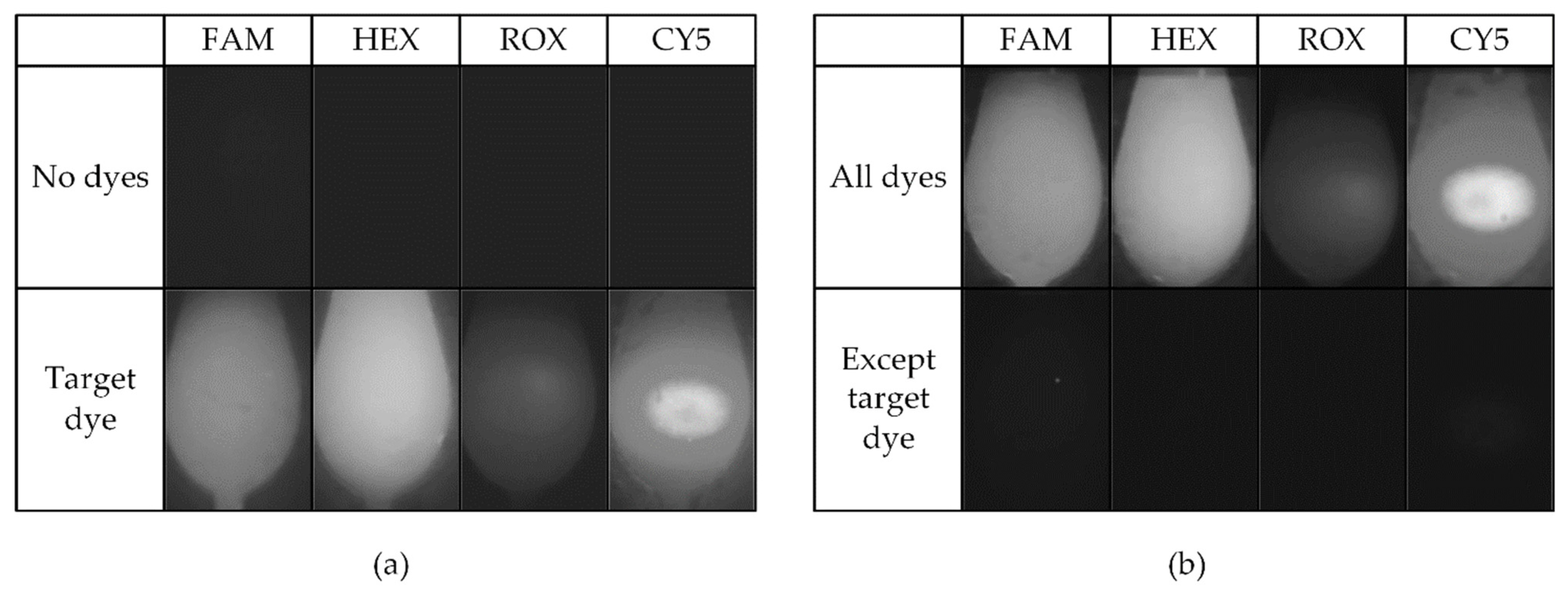
3. Experimental Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, Y.; Zhou, X.; Xing, D. Integrated paper-based detection chip with nucleic acid extraction and amplification for automatic and sensitive pathogen detection. Sens. Actuators B Chem. 2018, 261, 288–296. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.-Y.; Smith, P.W.; Diagana, T.T. Therapeutics for neglected infectious diseases: Progress and challenge. ACS Infect. Dis. 2015, 1, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Dominguez, D.C.; Li, X.; Sanchez, J.; Scott, G. A versatile PDMS/paper hybrid microfluidic platform for sensitive infectious disease diagnosis. Anal. Chem. 2014, 86, 7978–7986. [Google Scholar] [CrossRef]
- Zhang, H.; Miller, B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art. Biosens. Bioelectron. 2019, 141, 111476. [Google Scholar] [CrossRef]
- Abanses, J.C.; Dowd, M.D.; Simon, S.D.; Sharma, V. Impact of rapid influenza testing at triage on management of febrile infants and young children. Pediatr. Emerg. Care 2006, 22, 145–149. [Google Scholar] [CrossRef]
- Benito-Fernández, J.; Vázquez-Ronco, M.A.; Morteruel-Aizkuren, E.; Mintegui-Raso, S.; Sánchez-Etxaniz, J.; Fernández-Landaluce, A. Impact of rapid viral testing for influenza A and B viruses on management of febrile infants without signs of focal infection. Pediatr. Infect. Dis. J. 2006, 25, 1153–1157. [Google Scholar] [CrossRef]
- Hojat, K.; Duppenthaler, A.; Aebi, C. Impact of the availability of an influenza virus rapid antigen test on diagnostic decision making in a pediatric emergency department. Pediatr. Emerg. Care 2013, 29, 696–698. [Google Scholar] [CrossRef]
- Poehling, K.A.; Zhu, Y.; Tang, Y.-W.; Edwards, K. Accuracy and impact of a point-of-care rapid influenza test in young children with respiratory illnesses. Arch. Pediatr. Adolesc. Med. 2006, 160, 713–718. [Google Scholar] [CrossRef]
- Chin, C.D.; Linder, V.; Sia, S.K. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip 2007, 7, 41–57. [Google Scholar] [CrossRef]
- Faix, D.J.; Sherman, S.S.; Waterman, S.H. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009, 361, 728–729. [Google Scholar] [CrossRef] [PubMed]
- Drain, P.K.; Hyle, E.P.; Noubary, F.; Freedberg, K.A.; Wilson, D.; Bishai, W.R.; Rodriguez, W.; Bassett, I.V. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect. Dis. 2014, 14, 239–249. [Google Scholar] [CrossRef]
- Hansen, G.T. Point-of-care testing in microbiology: A mechanism for improving patient outcomes. Clin. Chem. 2020, 66, 124–137. [Google Scholar] [CrossRef]
- Zarei, M. Infectious pathogens meet point-of-care diagnostics. Biosens. Bioelectron. 2018, 106, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Batt, C.A. Food pathogen detection. Science 2007, 316, 1579–1580. [Google Scholar] [CrossRef]
- Park, S.; Zhang, Y.; Lin, S.; Wang, T.-H.; Yang, S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011, 29, 830–839. [Google Scholar] [CrossRef]
- Mullis, K.B.; Faloona, F.A. [21] Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar]
- Ishmael, F.T.; Stellato, C. Principles and applications of polymerase chain reaction: Basic science for the practicing physician. Ann. Allergy Asthma Immunol. 2008, 101, 437–443. [Google Scholar] [CrossRef]
- de Mello Malta, F.; Amgarten, D.; Val, F.C.; Cervato, M.C.; de Azevedo, B.M.C.; de Souza Basqueira, M.; dos Santos Alves, C.O.; Nobrega, M.S.; de Souza Reis, R.; Sebe, P. Mass molecular testing for COVID19 using NGS-based technology and a highly scalable workflow. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Goudouris, E.S. Laboratory diagnosis of COVID-19. J. Pediatr. 2021, 97, 7–12. [Google Scholar] [CrossRef]
- Quesada-González, D.; Merkoçi, A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015, 73, 47–63. [Google Scholar] [CrossRef]
- Al-Soud, W.A.; Rådström, P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001, 39, 485–493. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Jiang, Y.; Shi, B.; Wu, D.; Wu, W. Low-cost battery-powered and user-friendly real-time quantitative PCR system for the detection of multigene. Micromachines 2020, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; He, G.; Wu, W. A PCR microreactor machinery with passive micropump and battery-powered heater for thermo-cycled amplifications of clinical-level and multiplexed DNA targets. Microchim. Acta 2018, 185, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Kang, K.-T.; Lee, N.Y. Bubble-free on-chip continuous-flow polymerase chain reaction: Concept and application. Analyst 2011, 136, 2287–2293. [Google Scholar] [CrossRef] [PubMed]
- Priye, A.; Wong, S.; Bi, Y.; Carpio, M.; Chang, J.; Coen, M.; Cope, D.; Harris, J.; Johnson, J.; Keller, A. Lab-on-a-drone: Toward pinpoint deployment of smartphone-enabled nucleic acid-based diagnostics for mobile health care. Anal. Chem. 2016, 88, 4651–4660. [Google Scholar] [CrossRef]
- Tseng, Y.-T.; Wang, C.-H.; Chang, C.-P.; Lee, G.-B. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens. Bioelectron. 2016, 82, 105–111. [Google Scholar] [CrossRef]
- Loo, J.; Kwok, H.; Leung, C.; Wu, S.; Law, I.; Cheung, Y.; Cheung, Y.; Chin, M.; Kwan, P.; Hui, M. Sample-to-answer on molecular diagnosis of bacterial infection using integrated lab-on-a-disc. Biosens. Bioelectron. 2017, 93, 212–219. [Google Scholar] [CrossRef]
- Kim, S.H.; Shin, J.H. Point-of-care diagnostics for infectious diseases: Present and future. Korean J. Med. 2018, 93, 181–187. [Google Scholar] [CrossRef][Green Version]
- Kettler, H.; White, K.; Hawkes, S.J. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommendations; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed point-of-care testing–xPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef]
- Zarei, M. Advances in point-of-care technologies for molecular diagnostics. Biosens. Bioelectron. 2017, 98, 494–506. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Quantification strategies in real-time polymerase chain reaction. Quant. Real-Time PCR Appl. Microbiol. 2012, 53–62. [Google Scholar]
- Navarro, E.; Serrano-Heras, G.; Castaño, M.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef]
- Putignani, L.; Mancinelli, L.; Del Chierico, F.; Menichella, D.; Adlerstein, D.; Angelici, M.; Marangi, M.; Berrilli, F.; Caffara, M.; di Regalbono, D.F. Investigation of Toxoplasma gondii presence in farmed shellfish by nested-PCR and real-time PCR fluorescent amplicon generation assay (FLAG). Exp. Parasitol. 2011, 127, 409–417. [Google Scholar] [CrossRef]
- Demeke, T.; Jenkins, G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 2010, 396, 1977–1990. [Google Scholar] [CrossRef]
- Li, X.; Wu, W.; Manz, A. Thermal gradient for fluorometric optimization of droplet PCR in virtual reaction chambers. Microchim. Acta 2017, 184, 3433–3439. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bai, H.; Mauk, M.G.; Saif, L.; Bau, H.H. A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab Chip 2021, 21, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Li, Y.; Wu, D.; Wu, W. A handheld continuous-flow real-time fluorescence qPCR system with a PVC microreactor. Analyst 2020, 145, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Gallegos, R.A.; Rios, A.; Garcia-Cordero, J.L. An affordable and portable thermocycler for real-time PCR made of 3D-printed parts and off-the-shelf electronics. Anal. Chem. 2018, 90, 5563–5568. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Weaver, S.C.; Wong, P.-Y.; Lie, S.; Wang, E.; Guerbois, M.; Vayugundla, S.P.; Wong, S. Rapid, affordable and portable medium-throughput molecular device for Zika virus. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Obahiagbon, U.; Smith, J.T.; Zhu, M.; Katchman, B.A.; Arafa, H.; Anderson, K.S.; Christen, J.M.B. A compact, low-cost, quantitative and multiplexed fluorescence detection platform for point-of-care applications. Biosens. Bioelectron. 2018, 117, 153–160. [Google Scholar] [CrossRef]
- Song, J.; Liu, C.; Mauk, M.G.; Rankin, S.C.; Lok, J.B.; Greenberg, R.M.; Bau, H.H. Two-stage isothermal enzymatic amplification for concurrent multiplex molecular detection. Clin. Chem. 2017, 63, 714–722. [Google Scholar] [CrossRef]
- Hwang, J.-S.; Kim, J.-D.; Kim, Y.-S.; Song, H.-J.; Park, C.-Y. Performance evaluation of optimal real-time polymerase chain reaction achieved with reduced voltage. Biomed. Eng. Online 2018, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-J.; Kim, J.-D.; Kim, Y.-S.; Song, H.-J.; Park, C.-Y. Evaluation-independent system for DNA section amplification. Biomed. Eng. Online 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-D.; Park, C.-Y.; Kim, Y.-S.; Hwang, J.-S. Quantitative Analysis of Fluorescence Detection Using a Smartphone Camera for a PCR Chip. Sensors 2021, 21, 3917. [Google Scholar] [CrossRef] [PubMed]







| Fluorescence | Excitation Filter | Emission Filter | ||
|---|---|---|---|---|
| CWL (nm) | FWHM (nm) | CWL (nm) | FWHM (nm) | |
| FAM | 470 | 30 | 520 | 20 |
| HEX | 530 | 20 | 565 | 22 |
| ROX | 570 | 20 | 615 | 40 |
| CY5 | 630 | 20 | 665 | 20 |
| Fluorescence | Part Number | Manufacturer | Dominant Wavelength (nm) |
|---|---|---|---|
| FAM | C503B-BAS-CY0C0461 | CreeLED, Inc. | 470 |
| HEX | C503B-GAN-CB0F0791 | CreeLED, Inc. | 527 |
| ROX | LTL2P3KGKNN | LITEON | 572 |
| CY5 | VLCS5830 | Vishay Semiconductor Opto Division | 624 |
| Reagents | Concentration | Volume (µL) |
|---|---|---|
| DNA (Chlamydia trachomatis) | 106 copy | 5.4 µL |
| Master mix | - | 18 µL |
| Primer mix | 0.28 pmole/µL | 9 µL |
| DDW | - | 3.6 µL |
| Total | - | 36 µL |
| Step | Temperature (°C) | Duration(s) | Cycles |
|---|---|---|---|
| Pre-incubation | 50 °C | 2 m | 1 |
| Pre-heating | 95 °C | 10 m | |
| Denaturation | 95 °C | 15 s | 40 |
| Annealing | 60 °C | 1 m |
| Individual Dye Experiment | Fluorescence Crosstalk Experiment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fluorescence | Fluorescence | ||||||||
| FAM | HEX | ROX | CY5 | FAM | HEX | ROX | CY5 | ||
| No dyes | 4.2 | 2.0 | 2.0 | 2.0 | All dyes | 75.8 | 96.7 | 22.5 | 80.5 |
| Target | 62.1 | 95.3 | 21.1 | 71.2 | Except target | 5.6 | 2.0 | 2.0 | 2.6 |
| Gap | 57.9 | 93.3 | 19.1 | 69.2 | Gap | 70.1 | 94.6 | 20.5 | 77.8 |
| Relative gap | 13.7 | 46.6 | 9.5 | 34.6 | Relative gap | 12.4 | 45.3 | 10.2 | 28.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, S.-H.; Park, J.-S.; Koo, S.-B.-N.; Park, C.-Y.; Kim, Y.-S.; Kim, J.-D. Cost-Effective Multiplex Fluorescence Detection System for PCR Chip. Sensors 2021, 21, 6945. https://doi.org/10.3390/s21216945
Yun S-H, Park J-S, Koo S-B-N, Park C-Y, Kim Y-S, Kim J-D. Cost-Effective Multiplex Fluorescence Detection System for PCR Chip. Sensors. 2021; 21(21):6945. https://doi.org/10.3390/s21216945
Chicago/Turabian StyleYun, Sung-Hun, Ji-Sung Park, Seul-Bit-Na Koo, Chan-Young Park, Yu-Seop Kim, and Jong-Dae Kim. 2021. "Cost-Effective Multiplex Fluorescence Detection System for PCR Chip" Sensors 21, no. 21: 6945. https://doi.org/10.3390/s21216945
APA StyleYun, S.-H., Park, J.-S., Koo, S.-B.-N., Park, C.-Y., Kim, Y.-S., & Kim, J.-D. (2021). Cost-Effective Multiplex Fluorescence Detection System for PCR Chip. Sensors, 21(21), 6945. https://doi.org/10.3390/s21216945





