Rheumatic Heart Disease Screening Based on Phonocardiogram
Abstract
1. Introduction
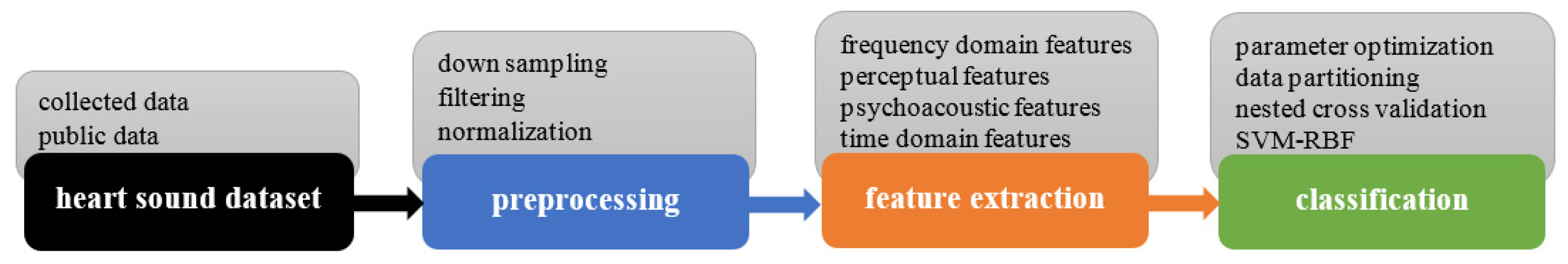
2. Materials and Methods
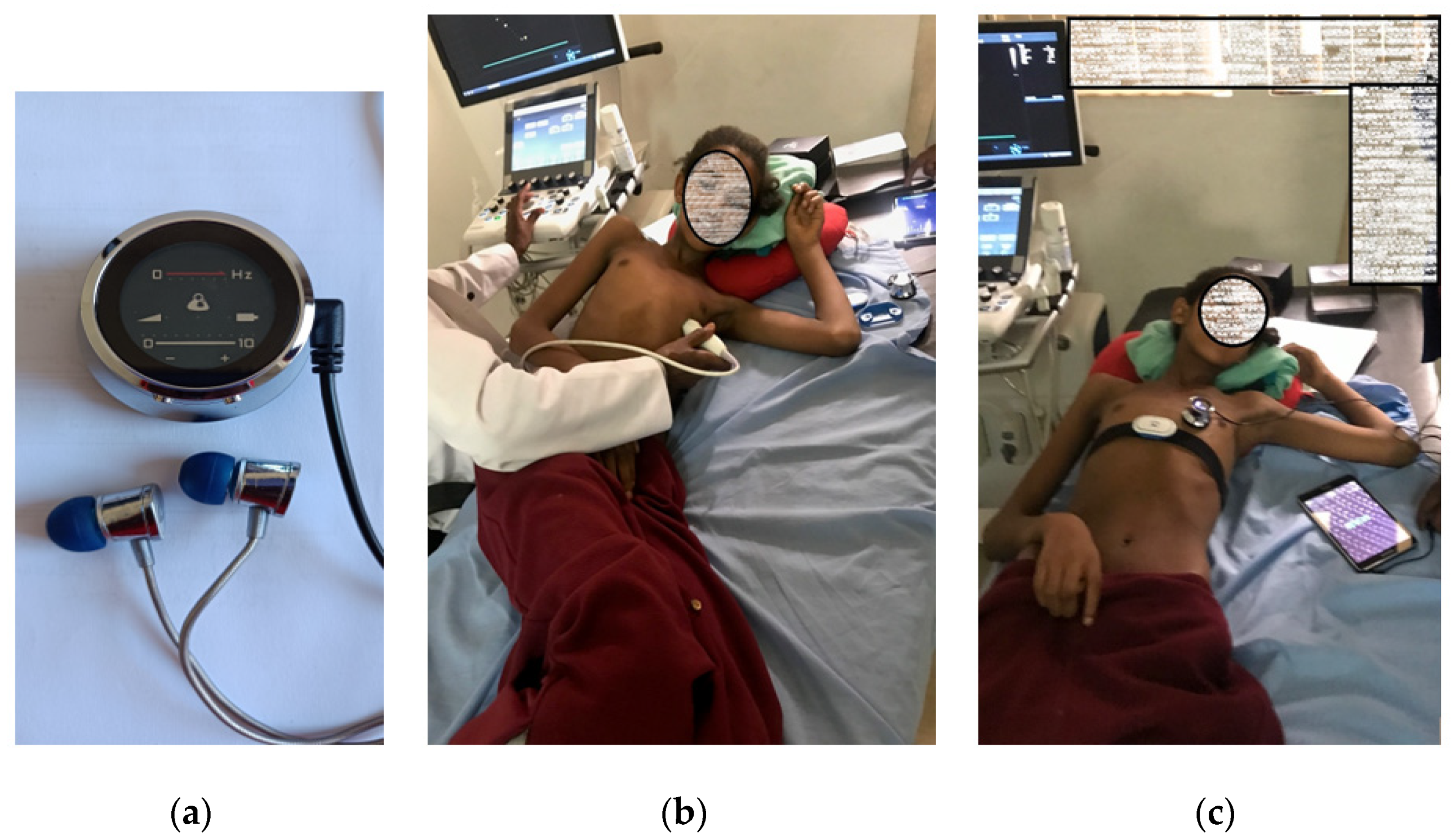
2.1. Heart Sound Dataset
2.1.1. RHD Dataset
2.1.2. Additional Heart Sound Dataset
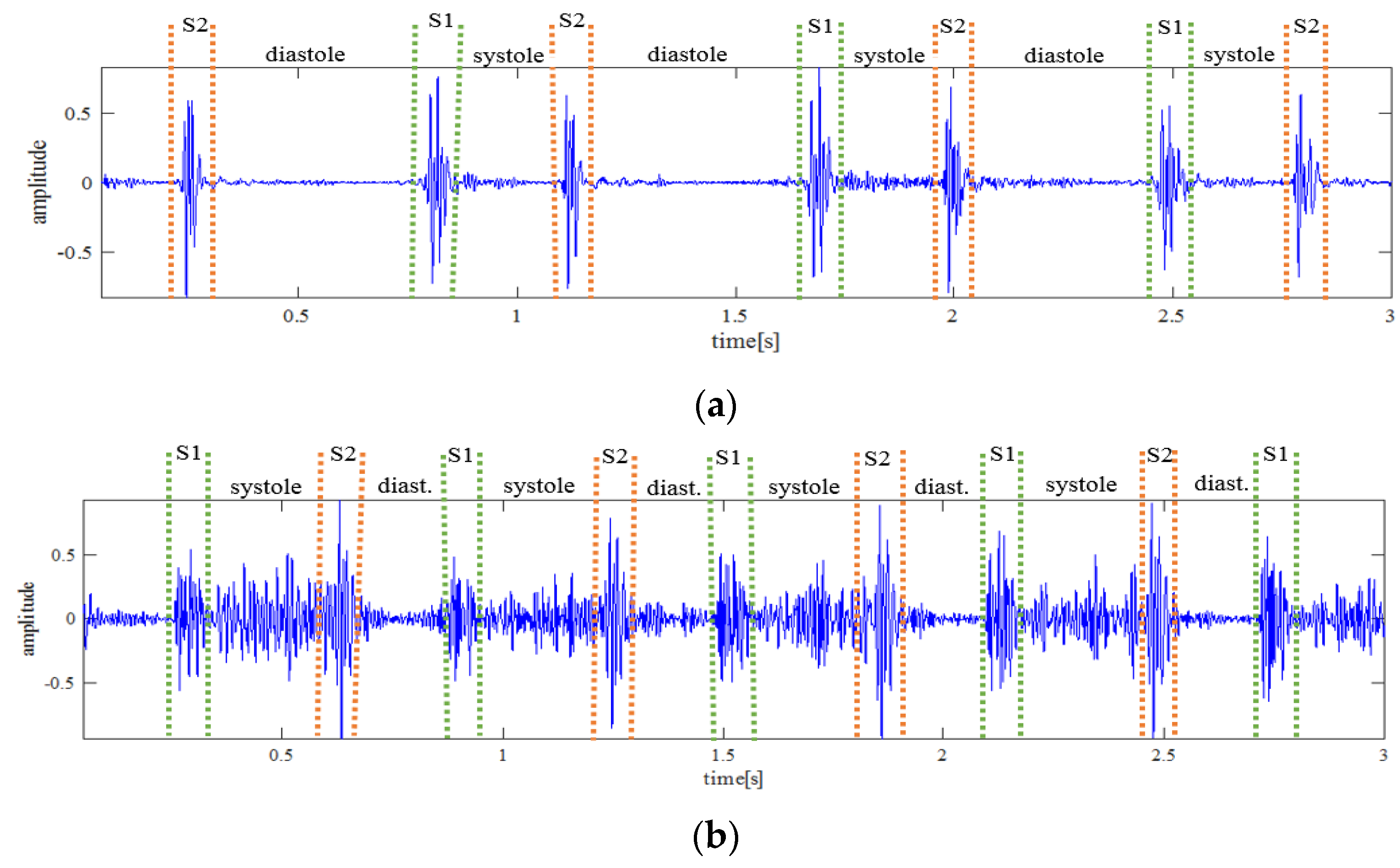
2.2. Preprocessing
2.3. Feature Extraction
2.4. Classification
2.4.1. SVMs
2.4.2. Nested Cross-Validation Approach
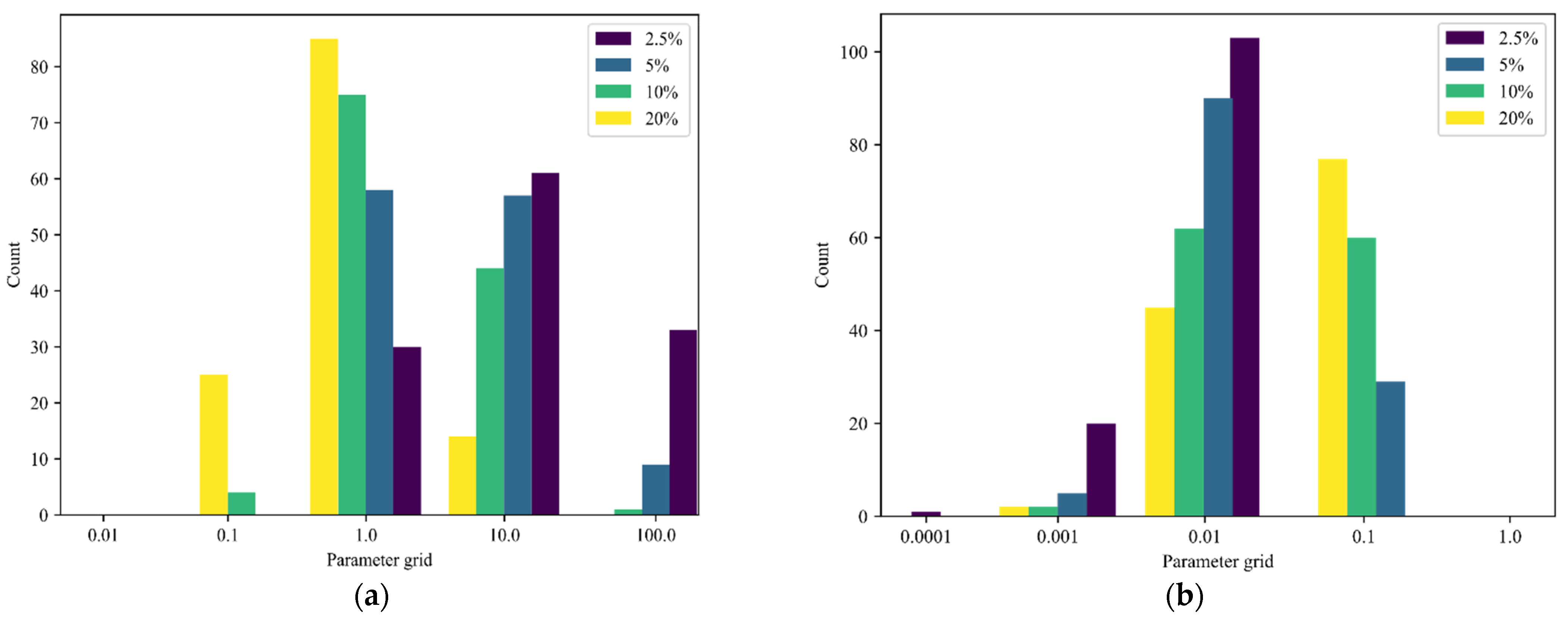
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Peters, F.; Karthikeyan, G.; Abrams, J.; Muhwava, L.; Zühlke, L. Rheumatic heart disease: Current status of diagnosis and therapy. Cardiovasc. Diagn. Ther. 2020, 10, 305–315. [Google Scholar] [CrossRef]
- Dylan, V.; Miller, M.; Revelo, P. Rheumatic Heart Disease. In Diagnostic Pathology, Diagnostic Pathology: Cardio-Vascular, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 204–207. ISBN 9780323595605. [Google Scholar]
- Dougherty, S.; Beaton, A.; Nascimento, B.R.; Zühlke, L.J.; Khorsandi, M.; Wilson, N. Prevention and control of rheumatic heart disease: Overcoming core challenges in resource-poor environments. Ann. Pediatr. Cardiol. 2018, 11, 68–78. [Google Scholar] [CrossRef]
- Rothenbühler, M.; O’Sullivan, C.J.; Stortecky, S.; Stefanini, G.G.; Spitzer, E.; Estill, J.; Shrestha, N.R.; Keiser, O.; Jüni, P.; Pilgrim, T. Active surveillance for rheumatic heart disease in endemic regions: A systematic review and meta-analysis of prevalence among children and adolescents. Lancet Glob. Health 2014, 2, e717–e726. [Google Scholar] [CrossRef]
- Carapetis, J.R.; Beaton, A.; Cunningham, M.W.; Guilherme, L.; Karthikeyan, G.; Mayosi, B.M.; Sable, C.; Steer, A.; Wilson, N.; Wyber, R.; et al. Acute rheumatic fever and rheumatic heart disease. Nat. Rev. Dis. Prim. 2016, 2, 15084. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Carapetis, J.R.; Griffiths, K.; Edwards, K.; Condon, J.R. Reply to Letter Regarding Article, “Acute Rheumatic Fever and Rheumatic Heart Disease: Incidence and Progression in the Northern Territory of Australia, 1997 to 2010”. Circulation 2014, 129, e397. [Google Scholar] [CrossRef][Green Version]
- Damasceno, A.; Mayosi, B.; Sani, M.; Ogah, O.; Mondo, C.; Ojji, D.B.; Dzudie, A.; Kouam, C.K.; Suliman, A.; Schrueder, N.; et al. The Causes, Treatment, and Outcome of Acute Heart Failure in 1006 Africans from 9 Countries. Arch. Intern. Med. 2012, 172, 1386–1394. [Google Scholar] [CrossRef]
- Okello, E.; Wanzhu, Z.; Musoke, C.; Twalib, A.; Kakande, B.; Lwabi, P.; Wilson, N.B.; Mondo, C.K.; Odoi-Adome, R.; Freers, J. Cardiovascular complications in newly diagnosed rheumatic heart disease patients at Mulago Hospital, Uganda: Cardiovascular topics. Cardiovasc. J. Afr. 2013, 24, 76–79. [Google Scholar] [CrossRef]
- Steer, A.C.; Carapetis, J. Prevention and treatment of rheumatic heart disease in the developing world. Nat. Rev. Cardiol. 2009, 6, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Reményi, B.; Wilson, N.; Steer, A.; Ferreira, B.; Kado, J.; Kumar, K.; Lawrenson, J.; Maguire, G.; Marijon, E.; Mirabel, M.; et al. World Heart Federation criteria for echocardiographic diagnosis of rheumatic heart disease—An evidence-based guideline. Nat. Rev. Cardiol. 2012, 9, 297–309. [Google Scholar] [CrossRef] [PubMed]
- RHD Australia. The 2020 Australian Guideline for Prevention, Diagnosis and Management of Acute Rheumatic Fever and Rheumatic Heart DISease, 3rd ed. 2020; Available online: https://www.rhdaustralia.org.au/system/files/fileuploads/arf_rhd_guidelines_3rd_edition_web_updated.pdf (accessed on 21 June 2021).
- Remenyi, B.; Carapetis, J.; Wyber, R.; Taubert, K.; Mayosi, B. Position statement of the World Heart Federation on the prevention and control of rheumatic heart disease. Nat. Rev. Cardiol. 2013, 10, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Noubiap, J.J.; Agbor, V.N.; Bigna, J.J.; Kaze, A.D.; Nyaga, U.F.; Mayosi, B.M. Prevalence and progression of rheumatic heart disease: A global systematic review and meta-analysis of population-based echocardiographic studies. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Oli, K.; Tekle-Haimanot, R.; Forsgren, L.; Ekstedt, J. Rheumatic Heart Disease Prevalence among Schoolchildren of an Ethiopian Rural Town. Cardiology 1992, 80, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Günther, G.; Asmera, J.; Parry, E. Death from rheumatic heart disease in rural Ethiopia. Lancet 2006, 367, 391. [Google Scholar] [CrossRef]
- Engel, M.E.; Haileamlak, A.; Zuhlke, L.; Lemmer, C.E.; Nkepu, S.; van de Wall, M.; Daniel, W.; King, M.S.; Mayosi, B.M. Prevalence of rheumatic heart disease in 4720 asymptomatic scholars from South Africa and Ethiopia. Heart 2015, 101, 1389–1394. [Google Scholar] [CrossRef]
- Yadeta, D.; Hailu, A.; Haileamlak, A.; Gedlu, E.; Guteta, S.; Tefera, E.; Tigabu, Z.; Tesfaye, H.; Daniel, W.; Mekonnen, D.; et al. Prevalence of rheumatic heart disease among school children in Ethiopia: A multisite echocardiography-based screening. Int. J. Cardiol. 2016, 221, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, D.; Guteta, S.; Alemayehu, B.; Mekonnen, D.; Gedlu, E.; Benti, H.; Tesfaye, H.; Berhane, S.; Hailu, A.; Luel, A.; et al. Spectrum of cardiovascular diseases in six main referral hospitals of Ethiopia. Hear. Asia 2017, 9, e010829. [Google Scholar] [CrossRef]
- Gemechu, T.; Mahmoud-Elsayed, H.; Parry, E.H.; Phillips, D.I.; Yacoub, M.H. Community-based prevalence study of rheumatic heart disease in rural Ethiopia. Eur. J. Prev. Cardiol. 2017, 24, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Barberato, S.H.; Romano, M.M.D.; Beck, A.L.S.; Rodrigues, A.C.T.; Almeida, A.L.C.; Assunção, B.M.B.L.; Gripp, E.D.A.; Guimarães, F.V.; Abensur, H.; Castillo, J.M.D.; et al. Position Statement on Indications of Echocardiography in Adults—2019. Arq Bras Cardiol. 2019, 113, 135–181. [Google Scholar]
- Godown, J.; Lu, J.C.; Beaton, A.; Sable, C.; Mirembe, G.; Sanya, R.; Aliku, T.; Yu, S.; Lwabi, P.; Webb, C.L.; et al. Handheld Echocardiography Versus Auscultation for Detection of Rheumatic Heart Disease. Pediatrics 2015, 135, e939–e944. [Google Scholar] [CrossRef]
- Gardezi, S.; Myerson, S.G.; Chambers, J.; Coffey, S.; D’Arcy, J.; Hobbs, F.D.R.; Holt, J.; Kennedy, A.; Loudon, M.; Prendergast, A.; et al. Cardiac auscultation poorly predicts the presence of valvular heart disease in asymptomatic primary care patients. Heart 2018, 104, 1832–1835. [Google Scholar] [CrossRef]
- Ploutz, M.; Lu, J.; Scheel, J.; Webb, C.L.; Ensing, G.J.; Aliku, T.; Lwabi, P.; Sable, C.; Beaton, A. Handheld echocardiographic screening for rheumatic heart disease by non-experts. Heart 2016, 102, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.V.; Brown, A.; Maguire, G.P.; Atkinson, D.; Carapetis, J. Utility of auscultatory screening for detecting rheumatic heart disease in high-risk children in Australia’s Northern Territory. Med. J. Aust. 2013, 199, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.J.; Mackie, A.; Finley, J.P. Cardiac Auscultation in the Modern Era. Cardiol. Rev. 2017, 25, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Montinari, M.R.; Minelli, S. The first 200 years of cardiac auscultation and future perspectives. J. Multidiscip. Healthc. 2019, 12, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Gerbarg, D.S.; Taranta, A.; Spagnuolo, M.; Hofler, J.J. Computer analysis of phonocardiograms. Prog. Cardiovasc. Dis. 1963, 5, 393–405. [Google Scholar] [CrossRef]
- Springer, D.B. Mobile Phone-Based Rheumatic Heart Disease Detection. Ph.D. Thesis, University of Oxford, Oxford, UK, 2015. [Google Scholar]
- Jadhav, A.R.; Ghontale, A.G.; Ganesh, A. Heart sounds segmentation and classification using adaptive learning neural networks. In Proceedings of the 2017 International Conference on Signal Processing and Communication (ICSPC), Coimbatore, India, 28–29 July 2017; pp. 33–38. [Google Scholar] [CrossRef]
- Rubin, J.; Abreu, R.; Ganguli, A.; Nelaturi, S.; Matei, I.; Sricharan, K. Recognizing Abnormal Heart Sounds Using Deep Learning. arXiv 2017, arXiv:1707.04642. [Google Scholar]
- Potes, C.; Saman, P.; Asif, R.; Bryan, C. Ensemble of feature-based and deep learning-based classifiers for detection of abnormal heart sounds. In Proceedings of the 2016 Computing in Cardiology Conference, Vancouver, BC, Canada, 11–14 September 2016; pp. 621–624. [Google Scholar]
- Gupta, C.; Palaniappan, R.; Rajan, S.; Swaminathan, S.; Krishnan, S. Segmentation and classification of heart sounds. In Proceedings of the Canadian Conference on Electrical and Computer Engineering, Saskatoon, SK, Canada, 1–4 May 2005; pp. 1674–1677. [Google Scholar]
- Yuenyong, S.; Nishihara, A.; Kongprawechnon, W.; Tungpimolrut, K. A framework for automatic heart sound analysis without segmentation. Biomed. Eng. Online 2011, 10, 13. [Google Scholar] [CrossRef]
- Zhang, W.; Han, J. Towards Heart Sound Classification Without Segmentation Using Convolutional Neural Network. In Proceedings of the 2017 Computing in Cardiology, Rennes, France, 24–27 September 2017; Volume 44. [Google Scholar]
- Khan, N.M.; Khan, M.S.; Khan, G.M. Automated Heart Sound Classification from Unsegmented Phonocardiogram Signals Using Time Frequency Features. Int. J. Comput. Inf. Eng. 2018, 12, 598–603. [Google Scholar]
- Langley, P.; Murray, A. Heart sound classification from unsegmented phonocardiograms. Physiol. Meas. 2017, 38, 1658–1670. [Google Scholar] [CrossRef]
- Banerjee, P.; Mondal, A. An Irregularity Measurement Based Cardiac Status Recognition Using Support Vector Machine. J. Med. Eng. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Careena, P.; Preetha, M.; Arun, P. Research on Murmur from Time Domain Features of Heart Sounds. Int. J. Recent Technol. Eng. 2019, 8, 736–743. [Google Scholar]
- Sun, S. An innovative intelligent system based on automatic diagnostic feature extraction for diagnosing heart diseases. Knowl.-Based Syst. 2015, 75, 224–238. [Google Scholar] [CrossRef]
- Sengur, A.; Turkoglu, I. A hybrid method based on artificial immune system and fuzzy k-NN algorithm for diagnosis of heart valve diseases. Expert Syst. Appl. 2008, 35, 1011–1020. [Google Scholar] [CrossRef]
- Alqudah, A.M. Towards classifying non-segmented heart sound records using instantaneous frequency based features. J. Med. Eng. Technol. 2019, 43, 418–430. [Google Scholar] [CrossRef]
- Zhang, W.; Han, J.; Deng, S. Heart sound classification based on scaled spectrogram and partial least squares regression. Biomed. Signal Process. Control 2017, 32, 20–28. [Google Scholar] [CrossRef]
- Noponen, A.L.; Lukkarinen, S.; Angerla, A.; Sepponen, R. Phonospectrographic analysis of heart murmur in children. BMC Pediatrics 2007, 7, 23. [Google Scholar] [CrossRef]
- Asmare, M.H.; Woldehanna, F.; Janssens, L.; Vanrumste, B. Rheumatic Heart Disease Detection Using Deep Learning from Spectro-Temporal Representation of Un-segmented Heart Sounds. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; Volume 2020, pp. 168–171. [Google Scholar] [CrossRef]
- Deng, M.; Meng, T.; Cao, J.; Wang, S.; Zhang, J.; Fan, H. Heart sound classification based on improved MFCC features and convolutional recurrent neural networks. Neural Netw. 2020, 130, 22–32. [Google Scholar] [CrossRef]
- Nogueira, D.M.; Ferreira, C.A.; Gomes, E.; Jorge, A. Classifying Heart Sounds Using Images of Motifs, MFCC and Temporal Features. J. Med. Syst. 2019, 43, 168. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Agarwal, A. DropConnected neural networks trained on time-frequency and inter-beat features for classi-fying heart sounds. Physiol. Meas. 2017, 38, 1645–1647. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Valencia, L.D.; Godino-Llorente, J.I.; Blanco-Velasco, M.; Castellanos-Dominguez, G. Feature Extraction from Parametric Time–Frequency Representations for Heart Murmur Detection. Ann. Biomed. Eng. 2010, 38, 2716–2732. [Google Scholar] [CrossRef] [PubMed]
- Bernhard Suhm. Heart Sound Classifier. MATLAB Central File Ex-Change. 2021. Available online: https://www.mathworks.com/matlabcentral/fileexchange/65286-heart-sound-classifier (accessed on 21 June 2021).
- Li, L.; Wang, X.; Du, X.; Liu, Y.; Liu, C.; Qin, C. Classification of heart sound signals with BP neural network and lo-gistic regression. In Proceedings of the 2017 Chinese Automation Congress, Jinan, China, 20–22 October 2017; pp. 7380–7383. [Google Scholar]
- Fahad, H.M.; Khan, M.U.G.; Saba, T.; Rehman, A.; Iqbal, S. Microscopic abnormality classification of cardiac murmurs using ANFIS and HMM. Microsc. Res. Tech. 2018, 81, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Asmare, M.; Woldehanna, F.; Janssens, L.; Vanrumste, B. Automated Rheumatic Heart Disease Detection from Phonocardiogram in Cardiology Ward. In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies—Cognitive Health IT, Valletta, Malta, 24–26 February 2020; pp. 839–844, ISBN 978-989-758-398-8, ISSN 2184-4305. [Google Scholar] [CrossRef]
- Sawant, N.K.; Patidar, S.; Nesaragi, N.; Acharya, U.R. Automated detection of abnormal heart sound signals using Fano-factor constrained tunable quality wavelet transform. Biocybern. Biomed. Eng. 2021, 41, 111–126. [Google Scholar] [CrossRef]
- Son, G.Y.; Kwon, S. Classification of Heart Sound Signal Using Multiple Features. Appl. Sci. 2018, 8, 2344. [Google Scholar] [CrossRef]
- Available online: https://www.thinklabs.com (accessed on 19 March 2021).
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Li, Q.; Moody, B.; Juan, R.A.; Chorro, F.J.; Castells, F.; Roig, J.M.; Silva, I.; Johnson, A.E.W.; et al. An open access database for the evaluation of heart sound algorithms. Physiol. Meas. 2016, 37, 2181–2213. [Google Scholar] [CrossRef] [PubMed]
- Breebaart, J.; McKinney, M.F. Features for Audio Classification. Philips Res. 2004, 2, 113–129. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence–Volume 2 (IJCAI’95); Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1995; pp. 1137–1143. [Google Scholar]
- Vapnik, V.N. An overview of statistical learning theory. In IEEE Transactions on Neural Networks; IEEE: Manhattan, NY, USA, 1999; Volume 10, pp. 988–999. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Burges, C.J. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Moss, H.; Leslie, D.; Rayson, P. Using J-K-fold cross-validation to reduce variance when tuning NLP models. In Proceedings of the COLING 2018. Conference on Computational Linguistics, Santa Fe, NM, USA, 20–26 August 2018. [Google Scholar]
- Dougherty, S.; Khorsandi, M.; Herbst, P. Rheumatic heart disease screening: Current concepts and challenges. Ann. Pediatr. Cardiol. 2017, 10, 39–49. [Google Scholar] [CrossRef]

| No. | Dataset | No. of Records | Female/Male | Age (Years) | Average Recording Duration |
|---|---|---|---|---|---|
| 1 | Eth_PwRHD | 124 | 74/50 | 22.9 ± 8.9 | 3.59 min |
| 2 | Eth_HC | 46 | 15/31 | 14.4 ± 9.4 | 4.88 min |
| 3 | Physionet_cHC | 3 | * n.a. | 29 ± 8 | 47 ± 25 s |
| 4 | Physionet_fHC | 78 | n.a. | 56 ± 16 | 33 ± 5 s |
| No. | Feature Group | Number of Features | List of Features |
|---|---|---|---|
| 1 | Acoustic domain | 4 | Acoustic Roughness |
| Acoustic Loudness | |||
| Acoustic Sharpness | |||
| Acoustic Fluctuation | |||
| 2 | Frequency domain | 5 | Spectral entropy |
| Dominant frequency value | |||
| Dominant frequency magnitude | |||
| Dominant frequency ratio | |||
| Bandwidth | |||
| 3 | Time domain | 9 | Median |
| Mean absolute deviation | |||
| The first quartile | |||
| The third quartile | |||
| Interquartile range | |||
| Skewness | |||
| Kurtosis | |||
| Shannon’s energy | |||
| Zero Crossing rate | |||
| 4 | Perceptual domain | 13 | MFCC1 to MFCC13 |
| Parameter | 2.5% | 5% | 10% | 20% | Stratified 10-Fold |
|---|---|---|---|---|---|
| f1-score | 59.0 ± 1.7 | 72.2 ± 0.8 | 80.1 ± 1.5 | 81.1 ± 1.5 | 96.0 ± 0.9 |
| Recall | 89.0 ± 1.2 | 92.3 ± 0.4 | 95.6 ± 1.0 | 93.9 ± 1.2 | 95.8 ± 1.5 |
| Precision | 44.2 ± 2.7 | 59.2 ± 3.6 | 68.9 ± 3.3 | 71.4 ± 2.0 | 96.2 ± 0.6 |
| specificity | 95.2 ± 0.5 | 94.8 ± 0.6 | 92.6 ± 0.8 | 86.9 ± 1.8 | 96.0 ± 0.6 |
| Authors | Features (Numbers) | Evaluation | f1-Score | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| Springer et al. [28] | Combination of undecimated wavelet transform (360) and MFCC (13) | 10-fold CV | 90.3 ± 2.0 | 86.3 ± 3.1 | 94.7 ± 0.9 | 94.6 ± 0.9 |
| nested CV at 5% | 63.3 ± 2.4 | 72.4 ± 1.4 | 56.2 ± 3.0 | 94.3 ± 0.2 | ||
| Careena P. et al. [38] | Time domain features (10) | 10-fold CV | 83.2 ± 0.8 | 82.0 ± 1.6 | 84.6 ± 1.7 | 84.4 ± 2.5 |
| nested CV at 5% | 0.40 ± 1.6 | 67.9 ± 2.4 | 28.3 ± 1.5 | 90.4 ± 0.5 | ||
| A. M. Alqudah et al. [41] | Frequency domain features (8) | 10-fold CV | 86.6 ± 1.2 | 89.7 ± 2.4 | 83.9 ± 3.6 | 82.1 ± 5.0 |
| nested CV at 5% | 37.5 ± 1.0 | 85.8 ± 1.7 | 24.0 ± 0.9 | 82.9 ± 0.6 | ||
| M. Deng et al. [45] | perceptual features (MFCC and its first and second derivatives) (26) | 10-fold CV | 89.4 ± 1.5 | 90.4 ± 1.9 | 88.5 ± 1.6 | 87.5 ± 1.9 |
| nested CV at 5% | 44.6 ± 1.0 | 90.6 ± 1.5 | 29.6 ± 0.7 | 86.1 ± 0.2 | ||
| Asmare et al. [52] | Combination of time (6), frequency (3) and perceptual features (13) | 10-fold CV | 93.9 ± 0.4 | 94.1 ± 1.1 | 93.7 ± 0.8 | 93.3 ± 0.9 |
| nested CV at 5% | 66.8 ± 2.9 | 91.5 ± 1.9 | 52.6 ± 3.3 | 93.5 ± 0.7 | ||
| N. K Sawant et al. [53] | Combination of time (3), frequency (4), and perceptual features (13) | 10-fold CV | 91.7 ± 1.3 | 90.8 ± 1.5 | 92.6 ± 1.2 | 92.6 ± 1.0 |
| nested CV at 5% | 62.4 ± 2.2 | 94.7 ± 1.6 | 46.6 ± 2.3 | 91.7 ± 0.7 | ||
| This paper | Combination of time (9), frequency (4), perceptual (13) and acoustic features (4) | 10-fold CV | 96.0 ± 0.9 | 95.8 ± 1.5 | 96.2 ± 0.6 | 96.0 ± 0.6 |
| nested CV at 5% | 72.2 ± 0.8 | 92.3 ± 0.4 | 59.2 ± 3.6 | 94.8 ± 0.6 |
| Parameter | 2.5% | 5% | 10% | 20% | Stratified 10-Fold |
|---|---|---|---|---|---|
| f1-score | 58.4 ± 1.4 | 68.7 ± 3.0 | 70.4 ± 2.1 | 73.8 ± 2.4 | 95.2 ± 0.8 |
| Recall | 85.5 ± 1.0 | 87.6 ± 2.6 | 84.8 ± 2.0 | 83.9 ± 1.9 | 96.1 ± 1.2 |
| Precision | 44.4 ± 2.5 | 56.5 ± 3.4 | 60.2 ± 2.2 | 65.9 ± 3.4 | 94.3 ± 0.7 |
| specificity | 95.2 ± 0.6 | 94.3 ± 0.5 | 92.1 ± 0.7 | 89.0 ± 1.5 | 93.7 ± 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asmare, M.H.; Filtjens, B.; Woldehanna, F.; Janssens, L.; Vanrumste, B. Rheumatic Heart Disease Screening Based on Phonocardiogram. Sensors 2021, 21, 6558. https://doi.org/10.3390/s21196558
Asmare MH, Filtjens B, Woldehanna F, Janssens L, Vanrumste B. Rheumatic Heart Disease Screening Based on Phonocardiogram. Sensors. 2021; 21(19):6558. https://doi.org/10.3390/s21196558
Chicago/Turabian StyleAsmare, Melkamu Hunegnaw, Benjamin Filtjens, Frehiwot Woldehanna, Luc Janssens, and Bart Vanrumste. 2021. "Rheumatic Heart Disease Screening Based on Phonocardiogram" Sensors 21, no. 19: 6558. https://doi.org/10.3390/s21196558
APA StyleAsmare, M. H., Filtjens, B., Woldehanna, F., Janssens, L., & Vanrumste, B. (2021). Rheumatic Heart Disease Screening Based on Phonocardiogram. Sensors, 21(19), 6558. https://doi.org/10.3390/s21196558







