EEG-Based Emotion Recognition by Convolutional Neural Network with Multi-Scale Kernels
Abstract
:1. Introduction
- We construct the network with multi-scale kernel convolution blocks to learn the local and global patterns of the preprocessed EEG features. The network could learn the salient connectivity patterns that have variation in structure.
- Utilizing the channel and band interaction information is effective for emotion recognition based on EEG signals. The time domain-based features along with the correlation information clearly enhance the performance of this task.
2. Related Works
3. Proposed Method
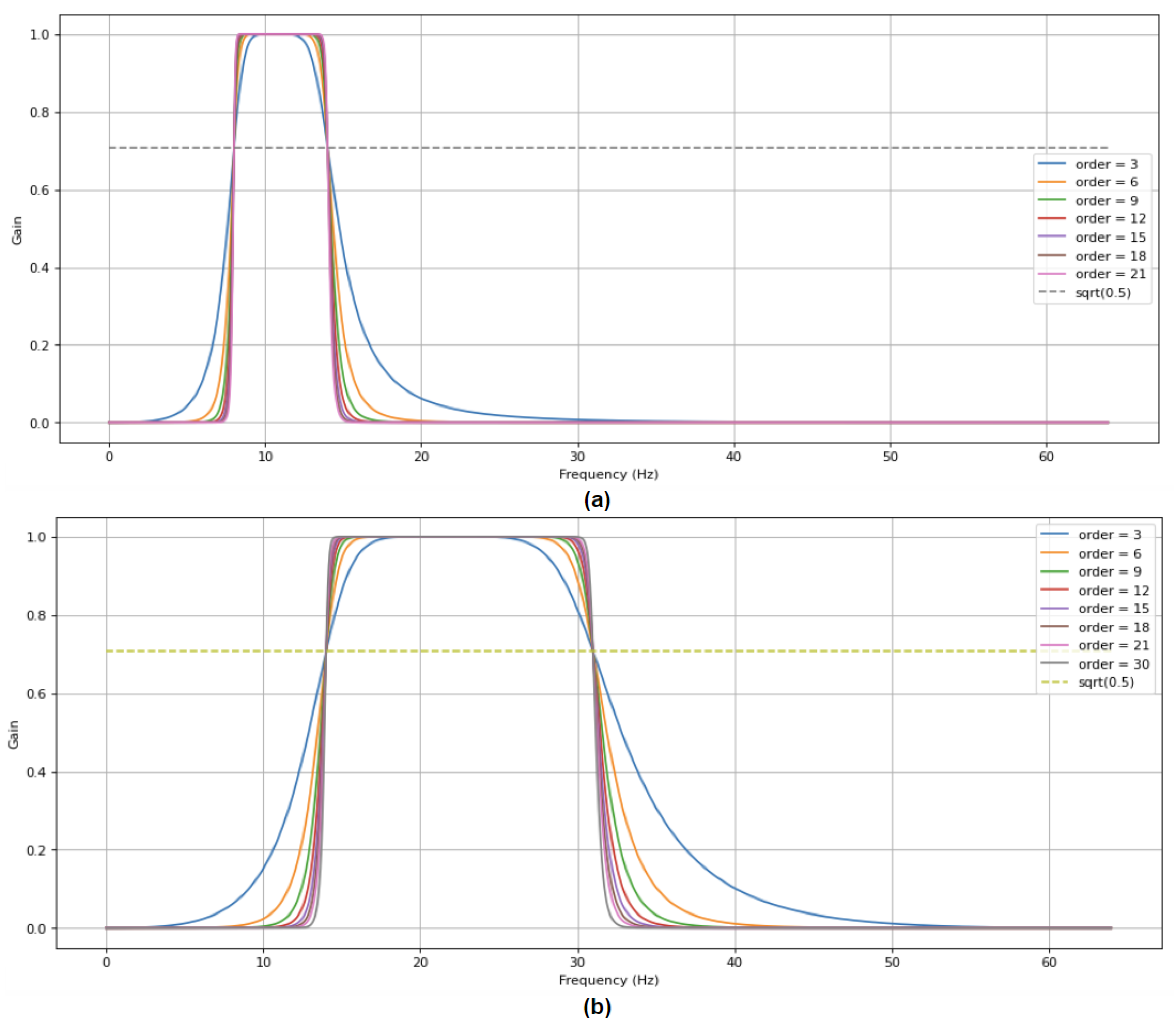
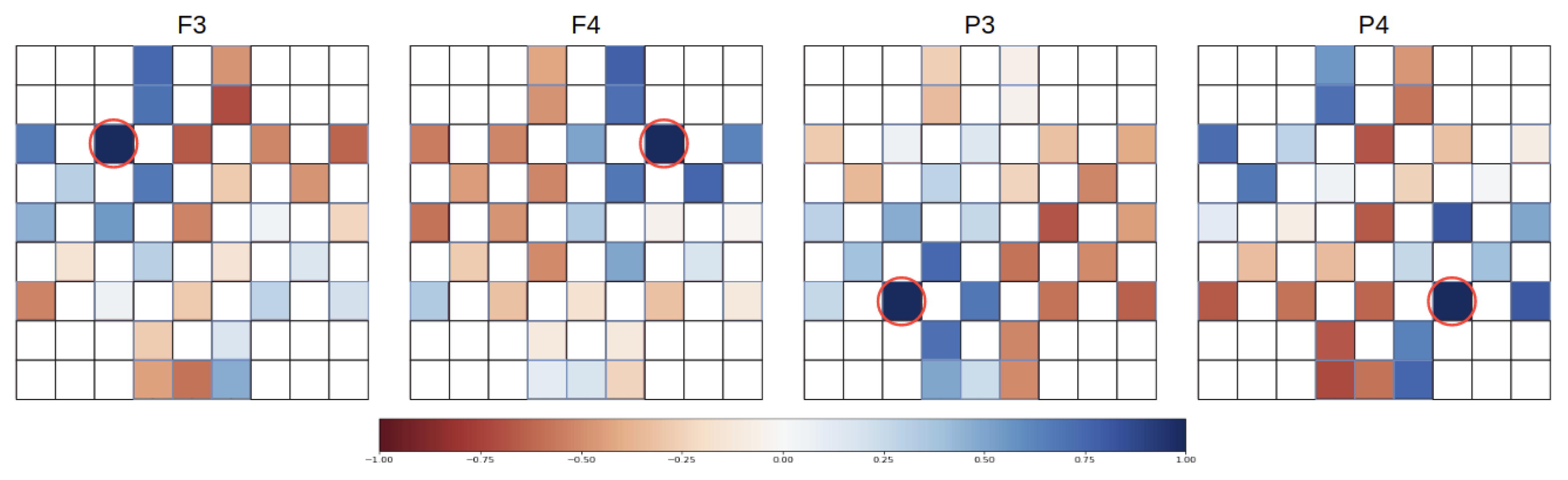
3.1. Dataset and Preprocessing
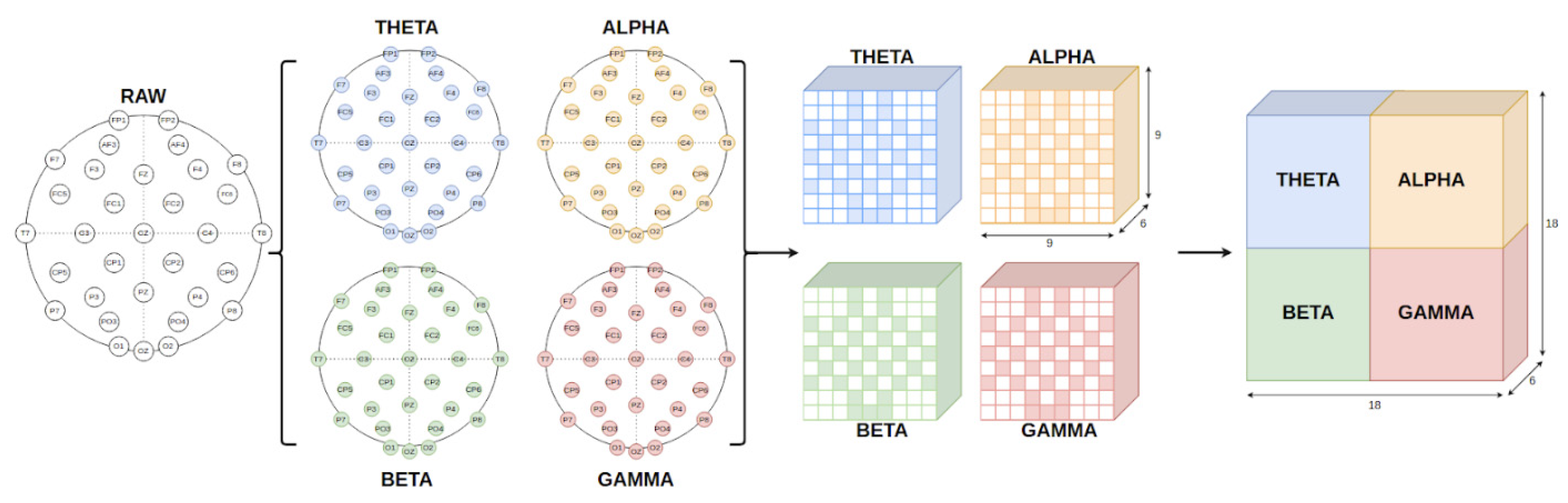
3.2. Multi Feature Extraction and Feature Representation
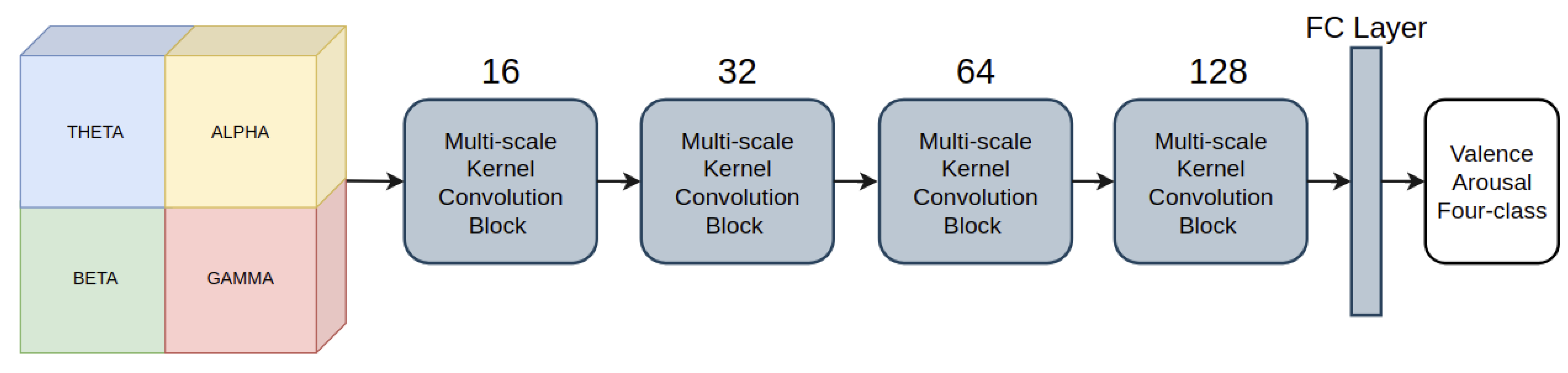
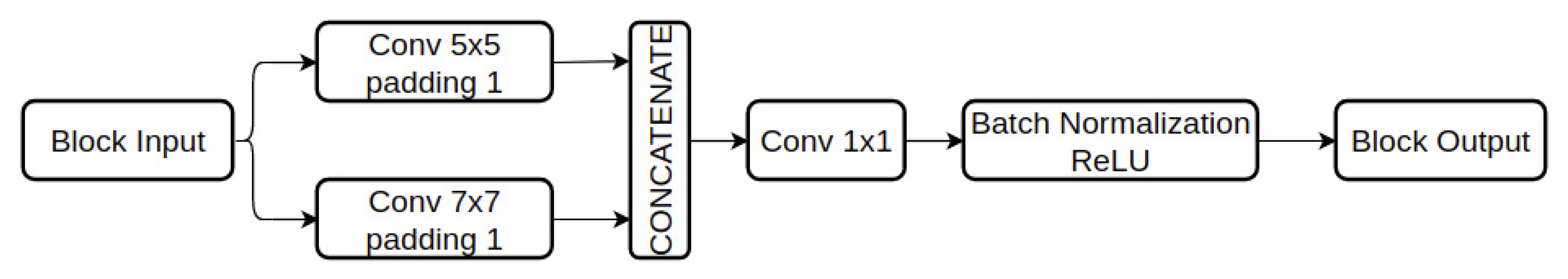
3.3. Deep Learning Model for Emotion Recognition
4. Experiments and Results
4.1. Experiemental Setup
4.2. Results
4.3. Ablation Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Deng, W. Deep facial expression recognition: A survey. IEEE Trans. Affect. Comput. 2020. [Google Scholar] [CrossRef] [Green Version]
- Fahad, M.S.; Ranjan, A.; Yadav, J.; Deepak, A. A survey of speech emotion recognition in natural environment. Digit. Signal Process. 2020, 110, 102951. [Google Scholar] [CrossRef]
- Noroozi, F.; Kaminska, D.; Corneanu, C.; Sapinski, T.; Escalera, S.; Anbarjafari, G. Survey on emotional body gesture recognition. IEEE Trans. Affect. Comput. 2018. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Liu, G.; Cheng, N.; Wei, J.; Shangguan, P.; Huang, W. Emotion recognition based on multi-variant correlation of physiological signals. IEEE Trans. Affect. Comput. 2014, 5, 126–140. [Google Scholar] [CrossRef]
- Torres, E.P.; Torres, E.A.; Hernández-Álvarez, M.; Yoo, S.G. EEG-based BCI emotion recognition: A survey. Sensors 2020, 20, 5083. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sourina, O. Real-time fractal-based valence level recognition from EEG. In Transactions on Computational Science XVIII; Springer: Berlin/Heidelberg, Germany, 2013; pp. 101–120. [Google Scholar]
- Shaw, L.; Routray, A. Statistical features extraction for multivariate pattern analysis in meditation EEG using PCA. In Proceedings of the 2016 IEEE EMBS International Student Conference (ISC), Ottawa, ON, Canada, 29–31 May 2016; pp. 1–4. [Google Scholar]
- Xing, X.; Li, Z.; Xu, T.; Shu, L.; Hu, B.; Xu, X. SAE+ LSTM: A New framework for emotion recognition from multi-channel EEG. Front. Neurorobot. 2019, 13, 37. [Google Scholar] [CrossRef]
- Karahan, E.; Rojas-Lopez, P.A.; Bringas-Vega, M.L.; Valdés-Hernández, P.A.; Valdes-Sosa, P.A. Tensor analysis and fusion of multimodal brain images. Proc. IEEE 2015, 103, 1531–1559. [Google Scholar] [CrossRef]
- Atkinson, J.; Campos, D. Improving BCI-based emotion recognition by combining EEG feature selection and kernel classifiers. Expert Syst. Appl. 2016, 47, 35–41. [Google Scholar] [CrossRef]
- Al-Fahoum, A.S.; Al-Fraihat, A.A. Methods of EEG signal features extraction using linear analysis in frequency and time-frequency domains. Int. Sch. Res. Not. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Qiu, S.; Du, C.; Wang, Y.; He, H. Domain adaptation for EEG emotion recognition based on latent representation similarity. IEEE Trans. Cogn. Dev. Syst. 2019, 12, 344–353. [Google Scholar] [CrossRef]
- Li, P.; Liu, H.; Si, Y.; Li, C.; Li, F.; Zhu, X.; Huang, X.; Zeng, Y.; Yao, D.; Zhang, Y.; et al. EEG based emotion recognition by combining functional connectivity network and local activations. IEEE Trans. Biomed. Eng. 2019, 66, 2869–2881. [Google Scholar] [CrossRef]
- Murugappan, M.; Ramachandran, N.; Sazali, Y. Classification of human emotion from EEG using discrete wavelet transform. J. Biomed. Sci. Eng. 2010, 3, 390–396. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jiang, D.; Zhang, Y. A common spatial pattern and wavelet packet decomposition combined method for EEG-based emotion recognition. J. Adv. Comput. Intell. Intell. Inform. 2019, 23, 274–281. [Google Scholar] [CrossRef]
- Shahabi, H.; Moghimi, S. Toward automatic detection of brain responses to emotional music through analysis of EEG effective connectivity. Comput. Hum. Behav. 2016, 58, 231–239. [Google Scholar] [CrossRef]
- Ackermann, P.; Kohlschein, C.; Bitsch, J.Á.; Wehrle, K.; Jeschke, S. EEG-based Automatic Emotion Recognition: Feature Extraction, Selection and Classification Methods. In Proceedings of the 2016 IEEE 18th International Conference on E-Health Networking, Applications and Services (Healthcom), Munich, Germany, 14–17 September 2016; pp. 1–6. [Google Scholar]
- Mohammadi, Z.; Frounchi, J.; Amiri, M. Wavelet-based emotion recognition system using EEG signal. Neural Comput. Appl. 2017, 28, 1985–1990. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Kwon, Y.H.; Shin, S.B.; Kim, S.D. Electroencephalography based fusion two-dimensional (2D)-convolution neural networks (CNN) model for emotion recognition system. Sensors 2018, 18, 1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Chen, S.; Deng, S. A EEG-based emotion recognition model with rhythm and time characteristics. Brain Inform. 2019, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Salama, E.S.; El-Khoribi, R.A.; Shoman, M.E.; Shalaby, M.A.W. EEG-based emotion recognition using 3D convolutional neural networks. Int. J. Adv. Comput. Sci. Appl 2018, 9, 329–337. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Hsieh, S. Classifying different emotional states by means of EEG-based functional connectivity patterns. PLoS ONE 2014, 9, e95415. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; He, H. Hierarchical convolutional neural networks for EEG-based emotion recognition. Cogn. Comput. 2018, 10, 368–380. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, M.; Li, C.; Liu, Y.; Song, R.; Liu, A.; Chen, X. Emotion recognition from multi-channel eeg via deep forest. IEEE J. Biomed. Health Inform. 2020, 25, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zheng, X.; Hu, B.; Zhang, Y.; Cui, X. EEG emotion recognition using fusion model of graph convolutional neural networks and LSTM. Appl. Soft Comput. 2021, 100, 106954. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG emotion recognition using dynamical graph convolutional neural networks. IEEE Trans. Affect. Comput. 2018, 11, 532–541. [Google Scholar] [CrossRef] [Green Version]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. Deap: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W. Multichannel EEG-based emotion recognition via group sparse canonical correlation analysis. IEEE Trans. Cogn. Dev. Syst. 2016, 9, 281–290. [Google Scholar] [CrossRef]
- Rozgi’c, V.; Vitaladevuni, S.N.; Prasad, R. Robust EEG emotion classification using segment level decision fusion. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 1286–1290. [Google Scholar]
- Zhuang, X.; Rozgi’c, V.; Crystal, M. Compact unsupervised EEG response representation for emotion recognition. In Proceedings of the IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), Valencia, Spain, 1–4 June 2014; pp. 736–739. [Google Scholar]
- Pandey, P.; Seeja, K. Subject independent emotion recognition from EEG using VMD and deep learning. J. King Saud Univ. Comput. Inf. Sci. 2019. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, W.; Cui, Z.; Zong, Y.; Li, Y. Spatial–temporal recurrent neural network for emotion recognition. IEEE Trans. Cybern. 2018, 49, 839–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Hu, B.; Xu, L.; Moore, P.; Su, Y. Feature-level fusion of multimodal physiological signals for emotion recognition. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 395–399. [Google Scholar]
- Zhang, J.; Chen, M.; Hu, S.; Cao, Y.; Kozma, R. PNN for EEG-based Emotion Recognition. In Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Budapest, Hungary, 9–12 October 2016; pp. 002319–002323. [Google Scholar]
- Kim, Y.; Choi, A. EEG-Based Emotion Classification Using Long Short-Term Memory Network with Attention Mechanism. Sensors 2020, 20, 6727. [Google Scholar] [CrossRef]
- Tao, W.; Li, C.; Song, R.; Cheng, J.; Liu, Y.; Wan, F.; Chen, X. EEG-based Emotion Recognition Via Channel-Wise Attention and Self Attention. IEEE Trans. Affect. Comput. 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, S.; Ma, X.; He, H. A prototype-based SPD matrix network for domain adaptation EEG emotion recognition. Pattern Recognit. 2021, 110, 107626. [Google Scholar] [CrossRef]
- Du, X.; Ma, C.; Zhang, G.; Li, J.; Lai, Y.K.; Zhao, G.; Deng, X.; Liu, Y.J.; Wang, H. An Efficient LSTM Network for Emotion Recognition from Multichannel EEG Signals. IEEE Trans. Affect. Comput. 2020. [Google Scholar] [CrossRef]
| Domain | Arousal | Valence | Four-Class |
|---|---|---|---|
| Time | 89.92 | 89.24 | 84.48 |
| Frequency | 68.77 | 68.05 | 54.91 |
| Time–Frequency | 79.59 | 80.05 | 69.84 |
| Correlation | 65.27 | 60.88 | 41.84 |
| Frequency Bands | Arousal | Valence | Four-Class |
|---|---|---|---|
| Theta | 74.10 | 71.74 | 59.19 |
| Alpha | 74.72 | 71.71 | 58.94 |
| Beta | 84.52 | 84.70 | 76.25 |
| Gamma | 87.39 | 86.91 | 81.10 |
| Raw | 89.92 | 89.24 | 84.48 |
| Beta-Gamma | 94.28 | 94.00 | 93.04 |
| All | 98.27 | 98.36 | 98.38 |
| Kernel Size | Arousal | Valence | Four-Class |
|---|---|---|---|
| 3 × 3 | 88.82 | 87.07 | 84.12 |
| 5 × 5 | 94.45 | 93.75 | 93.30 |
| 7 × 7 | 97.23 | 97.36 | 97.31 |
| 3 × 3 + 5 × 5 | 96.63 | 96.16 | 96.38 |
| 3 × 3 + 7 × 7 | 98.38 | 98.15 | 98.12 |
| 5 × 5 + 7 × 7 | 98.26 | 98.36 | 98.38 |
| 3 × 3 + 5 × 5 + 7 × 7 | 98.29 | 98.38 | 98.12 |
| Kernel Size | Year | Arousal | Valence |
|---|---|---|---|
| Kwon et al. [21] | 2018 | 76.56 | 80.46 |
| Zhang et al. [37] | 2016 | 81.21 | 81.76 |
| Yin et al. [28] | 2021 | 85.27 | 84.81 |
| Salama et al. [23] | 2018 | 88.49 | 87.44 |
| Kim et al. [38] | 2020 | 88.30 | 90.10 |
| Cheng et al. [27] | 2021 | 97.53 | 97.69 |
| The proposed method | 2021 | 98.27 | 98.36 |
| Arousal | Valence | Four-Class | |
|---|---|---|---|
| Shift 0 s | 98.26 | 98.36 | 98.38 |
| Shift 1 s | 98.94 | 98.72 | 98.23 |
| Shift 2 s | 98.37 | 98.28 | 97.80 |
| Shift 3 s | 98.23 | 98.30 | 97.97 |
| Shift 4 s | 98.34 | 98.25 | 97.88 |
| Shift 5 s | 98.40 | 98.26 | 97.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, T.-D.-T.; Kim, S.-H.; Yang, H.-J.; Lee, G.-S. EEG-Based Emotion Recognition by Convolutional Neural Network with Multi-Scale Kernels. Sensors 2021, 21, 5092. https://doi.org/10.3390/s21155092
Phan T-D-T, Kim S-H, Yang H-J, Lee G-S. EEG-Based Emotion Recognition by Convolutional Neural Network with Multi-Scale Kernels. Sensors. 2021; 21(15):5092. https://doi.org/10.3390/s21155092
Chicago/Turabian StylePhan, Tran-Dac-Thinh, Soo-Hyung Kim, Hyung-Jeong Yang, and Guee-Sang Lee. 2021. "EEG-Based Emotion Recognition by Convolutional Neural Network with Multi-Scale Kernels" Sensors 21, no. 15: 5092. https://doi.org/10.3390/s21155092
APA StylePhan, T.-D.-T., Kim, S.-H., Yang, H.-J., & Lee, G.-S. (2021). EEG-Based Emotion Recognition by Convolutional Neural Network with Multi-Scale Kernels. Sensors, 21(15), 5092. https://doi.org/10.3390/s21155092









