Discrimination and Quantitation of Biologically Relevant Carboxylate Anions Using A [Dye•PAMAM] Complex
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Binding of Dye to PAMAM G5 Dendrimer
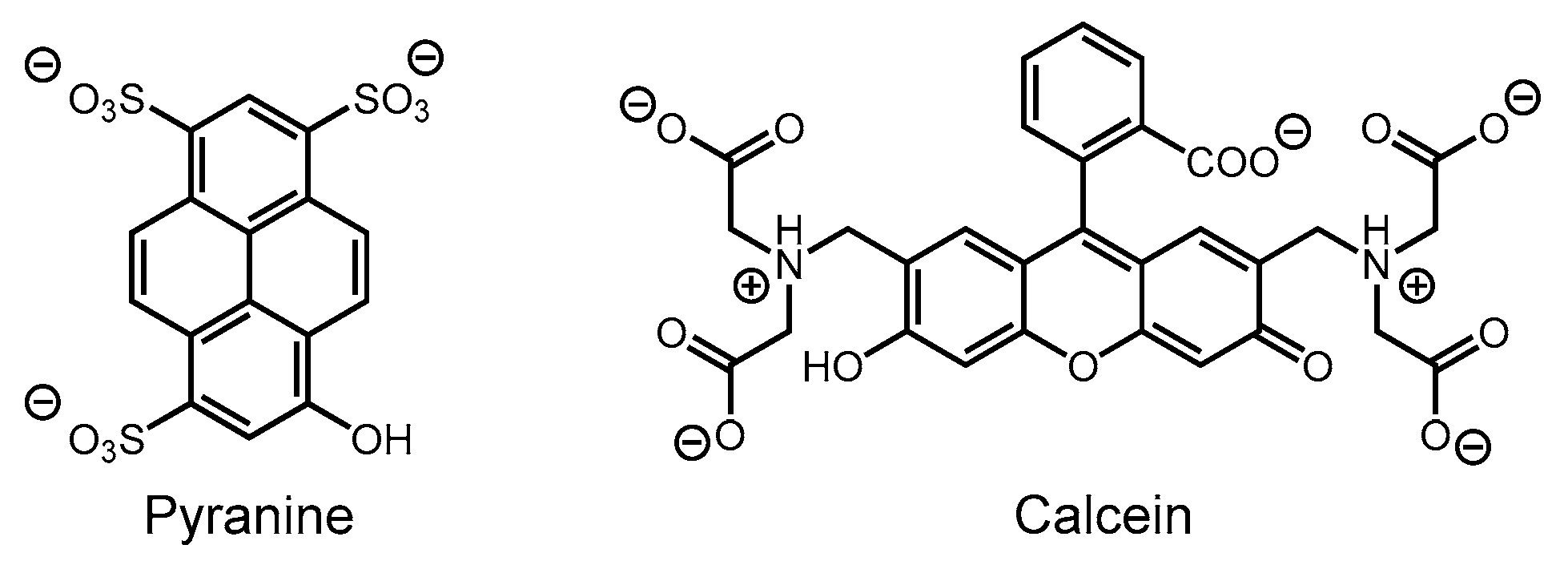
3.1.1. Binding of Pyranine
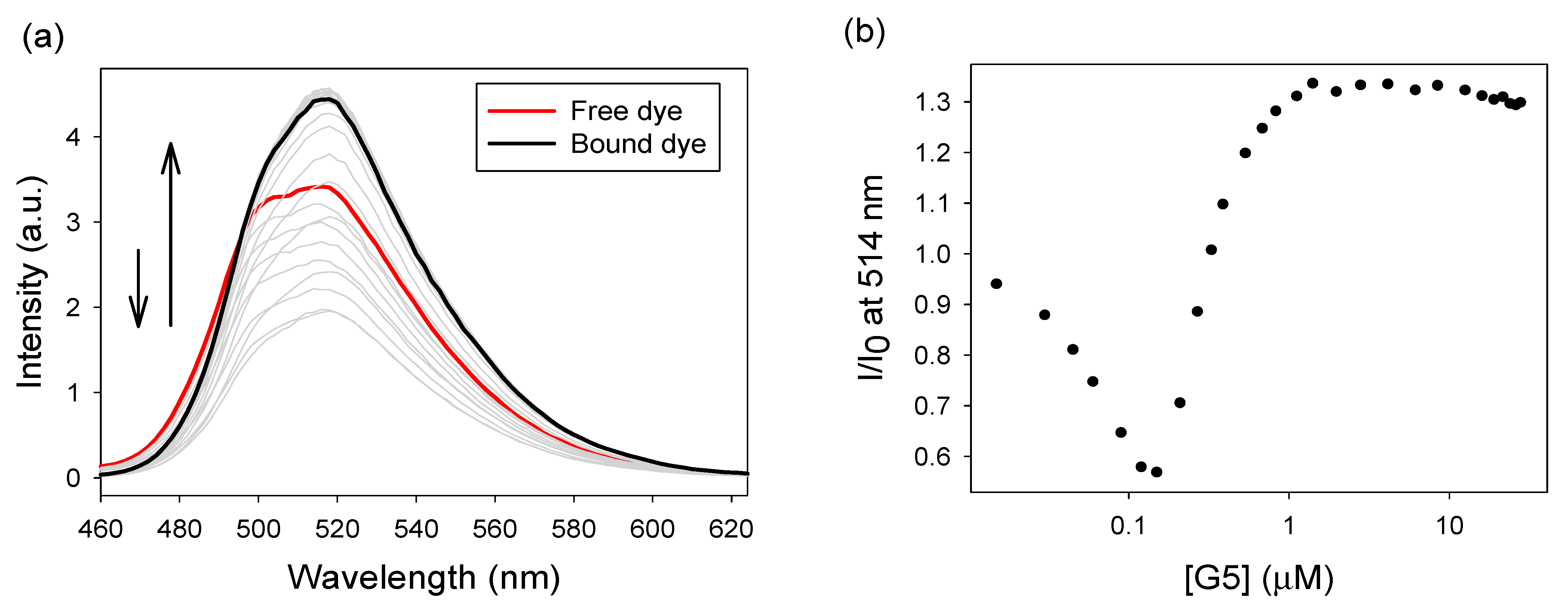
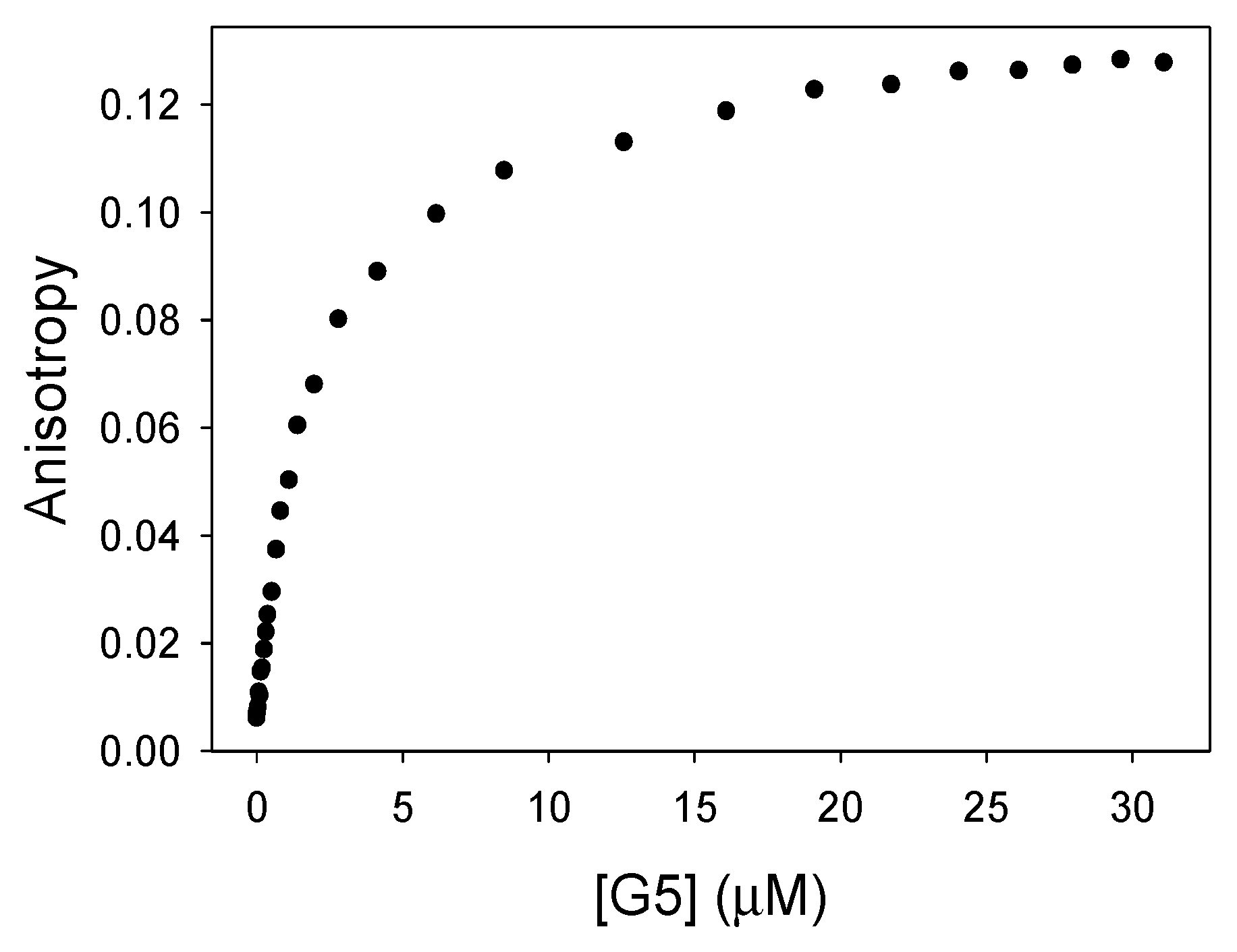
3.1.2. Binding of Calcein
3.2. Binding of Carboxylate to PAMAM G5 Dendrimer (Indicator Displacement Assay)
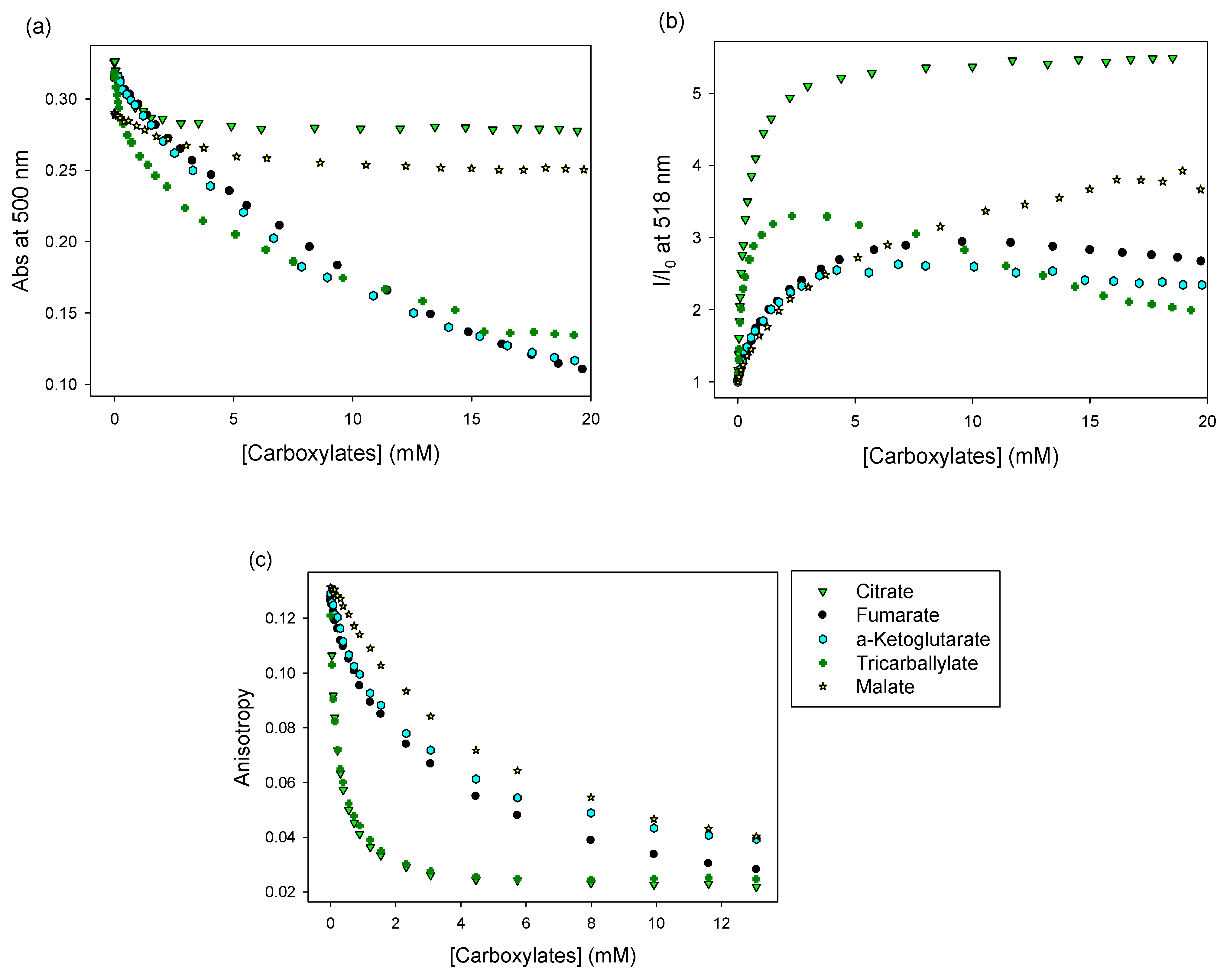
3.2.1. A Pyranine-Based IDA
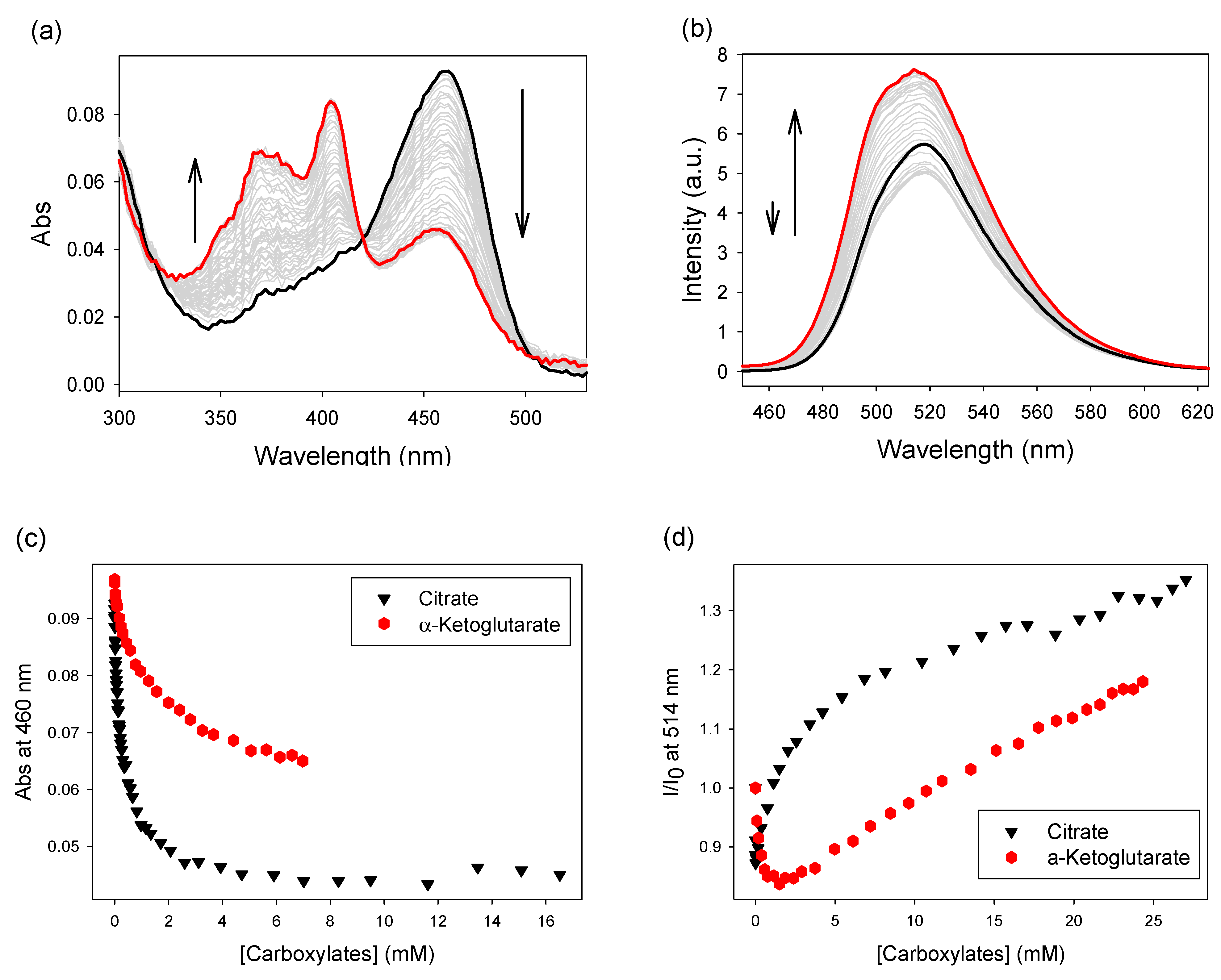
3.2.2. A Calcein-Based IDA
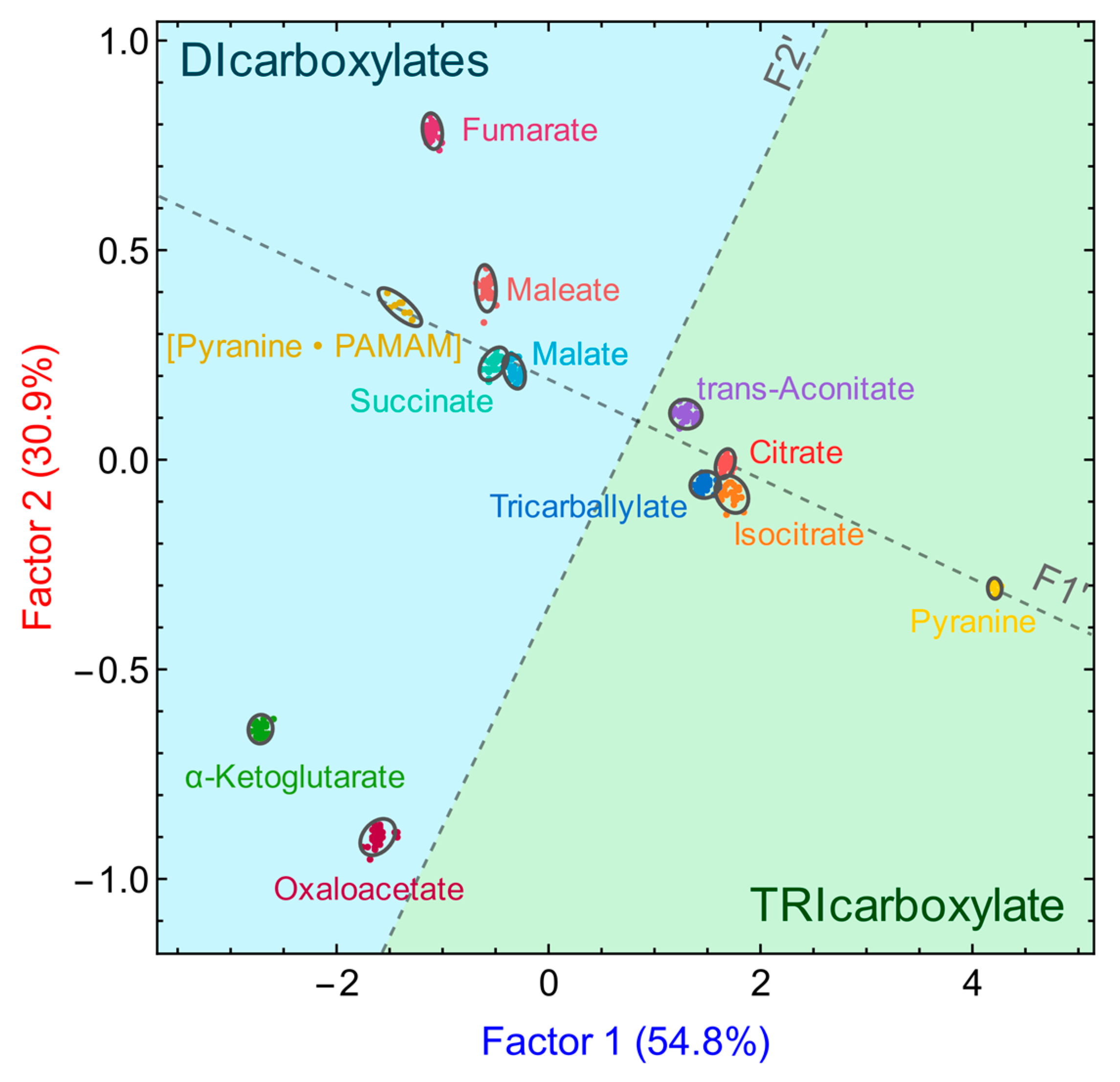
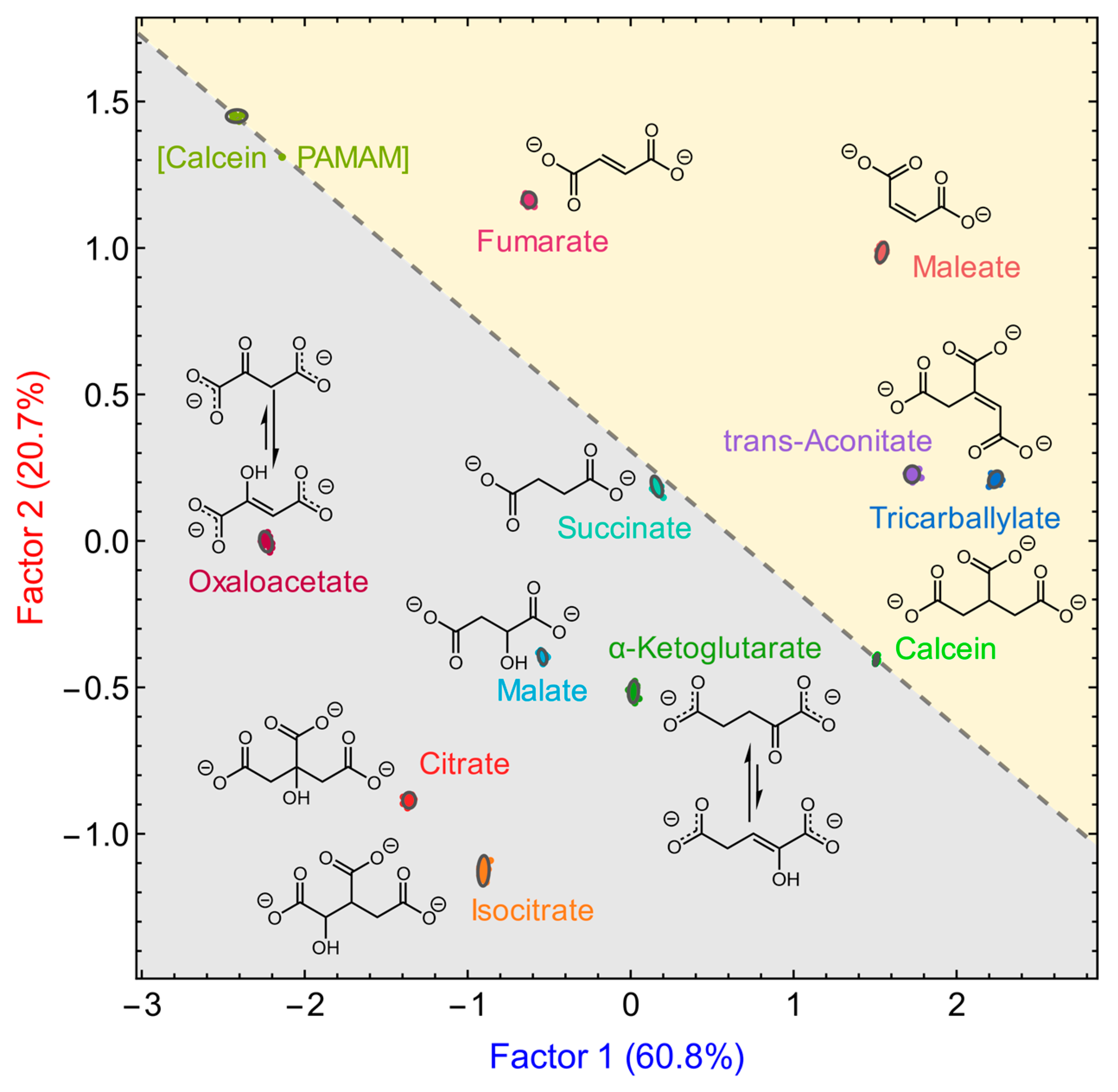
3.3. Anion Discrimination
3.3.1. Using the Pyranine-Based Sensor
3.3.2. Using the Calcein-Based Sensor
3.3.3. Limit of Discrimination
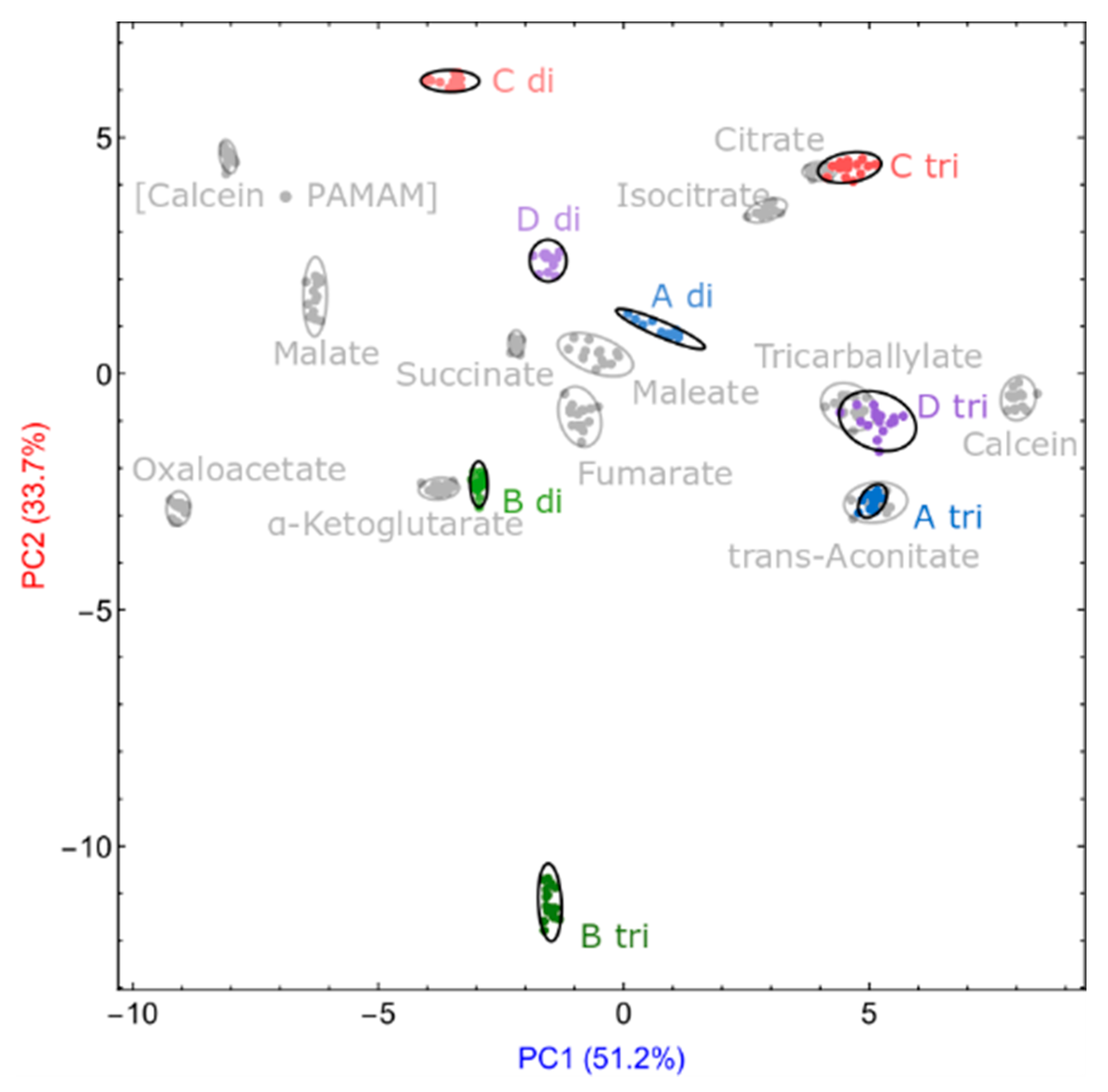
3.4. Unknown Quali-Quantitative Analysis
3.4.1. Quantification of Unknown Samples
3.4.2. Identity and Concentration of Unknown Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brewster, J.L.; McKellar, J.L.O.; Finn, T.J.; Newman, J.; Peat, T.S.; Gerth, M.L. Structural basis for ligand recognition by a cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci. Rep. 2016, 6, 35198. [Google Scholar] [CrossRef]
- Compton, K.K.; Hildreth, S.B.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemoreceptor McpV senses short-chain carboxylates via direct binding. J. Bacteriol. 2018, 200, e00519. [Google Scholar] [CrossRef]
- Davey, E.A.; Zucchero, A.J.; Trapp, O.; Bunz, U.H.F. Discrimination of organic acids using a three molecule array based upon cruciform fluorophores. J. Am. Chem. Soc. 2011, 133, 7716–7718. [Google Scholar] [CrossRef]
- Takeuchi, T.; Montenegro, J.; Hennig, A.; Matile, S. Pattern generation with synthetic sensing systems in lipid bilayer membranes. Chem. Sci. 2011, 2, 303–307. [Google Scholar] [CrossRef]
- Umali, A.P.; LeBoeuf, S.E.; Newberry, R.W.; Kim, S.; Tran, L.; Rome, W.A.; Tian, T.; Taing, D.; Hong, J.; Kwan, M.; et al. Discrimination of flavonoids and red wine varietals by arrays of differential peptidic sensors. Chem. Sci. 2011, 2, 439–445. [Google Scholar] [CrossRef]
- Bicker, K.L.; Sun, J.; Harrell, M.; Zhang, Y.; Pena, M.M.; Thompson, P.R.; Lavigne, J.J. Synthetic lectin arrays for the detection and discrimination of cancer associated glycans and cell lines. Chem. Sci. 2012, 3, 1147–1156. [Google Scholar] [CrossRef]
- Hall, A.J.; Achilli, L.; Manesiotis, P.; Quaglia, M.; de Lorenzi, E.; Sellergren, B. A substructure approach toward polymeric receptors targeting dihydrofolate reductase inhibitors. 2. Molecularly imprinted polymers against Z-L-glutamic acid showing affinity for larger molecules. J. Org. Chem. 2003, 68, 9132–9135. [Google Scholar] [CrossRef]
- Linton, B.R.; Goodman, M.S.; Fan, E.; van Arman, S.A.; Hamilton, A.D. Thermodynamic aspects of dicarboxylate recognition by simple artificial receptors. J. Org. Chem. 2001, 66, 7313–7319. [Google Scholar] [CrossRef]
- Bourrellier, A.B.F.; Valot, B.; Guillot, A.; Ambard-Bretteville, F.; Vidal, J.; Hodges, M. Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc. Natl. Acad. Sci. USA 2010, 107, 502–507. [Google Scholar] [CrossRef]
- Tatke, G.; Kumari, H.; Silva-Herzog, E.; Ramirez, L.; Mathee, K. Pseudomonas aeruginosa MifS-MifR two-component system is specific for α-ketoglutarate utilization. PLoS ONE 2015, 10, e0129629. [Google Scholar] [CrossRef]
- Eng, C.; Kiuru, M.; Fernandez, M.J.; Aaltonen, L.A. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat. Rev. Cancer 2003, 3, 193–202. [Google Scholar] [CrossRef]
- de Silva, F.R.O.; Nabeshima, C.T.; Bellini, H.; Courrol, L.C. Early diagnosis of prostate cancer by citrate determination in urine with europium–oxytetracycline complex. Appl. Spectrosc. 2012, 66, 958–961. [Google Scholar] [CrossRef]
- Selnaes, K.M.; Gribbestad, I.S.; Bertilsson, H.; Wright, A.; Angelsen, A.; Heerschap, A.; Tessem, M.B. Spatially matched in vivo and ex vivo MR metabolic profiles of prostate cancer–Investigation of a correlation with Gleason score. NMR Biomed. 2013, 26, 600–606. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. Prostatic fluid electrolyte composition for the screening of prostate cancer: A potential solution to a major problem. Prostate Cancer Prostatic Dis. 2009, 12, 17–24. [Google Scholar] [CrossRef][Green Version]
- Ghosh, K.; Sarkar, A.R. Pyridinium-based symmetrical diamides as chemosensors in visual sensing of citrate through indicator displacement assay (IDA) and gel formation. Org. Biomol. Chem. 2011, 9, 6551–6558. [Google Scholar] [CrossRef]
- Pourreza, N.; Sharifi, H.; Golmohammadi, H. Curcumin nanoparticles combined with cloud point extraction for citrate determination in food and drug samples. Microchem. J. 2016, 129, 213–218. [Google Scholar] [CrossRef]
- Stepinski, J.; Pawlowska, D.; Angielski, S. Effect of lithium on renal gluconeogenesis. Acta Biochim. Pol. 1984, 31, 229–240. [Google Scholar]
- Ghosh, I.K.; Saha, R.; Frohlich, A. Patra, Selective sensing of fumarate over maleate by benzimidazolium–based fluororeceptors. Mini Rev. Org. Chem. 2011, 8, 31–37. [Google Scholar]
- Costero, A.M.; Colera, M.; Gavina, P.; Gil, S. Fluorescent sensing of maleate versus fumarate by a neutral cyclohexane based thiourea receptor. Chem. Commun. 2006, 761–763. [Google Scholar] [CrossRef]
- Jolly, A.M.; Bonizzoni, M. Intermolecular forces driving encapsulation of small molecules by PAMAM dendrimers in water. Macromolecules 2014, 47, 6281–6288. [Google Scholar] [CrossRef]
- Xu, Y.; Bonizzoni, M. Disposable paper strips for carboxylate discrimination. Analyst 2020, 145, 3505–3516. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Redemann, C.T.; Szoka, F.C., Jr. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconj. Chem. 1996, 7, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A new class of polymers: Starburst-dendritic macromolecules. Polym. J. 2002, 34, 132–147. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, L.; Crooks, R.M. Determination of the intrinsic proton binding constants for poly(amidoamine) dendrimers via potentiometric pH titration. Macromolecules 2003, 36, 5725–5731. [Google Scholar] [CrossRef]
- Otto, M. Chemometrics: Statistics and Computer Application in Analytical Chemistry, 3rd ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016. [Google Scholar]
- Brereton, R.G. Chemometrics: Data Driven Extraction for Science; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006; pp. 364–365. [Google Scholar]
- Jablonski, A. Anisotropy of fluorescence of molecules excited by excitation transfer. Acta Phys. Pol. A 1970, 38, 453–458. [Google Scholar]
- Jolly, A.M.; Bonizzoni, M. PAMAM dendrimers as supramolecular hosts through non-covalent interactions. Supramol. Chem. 2015, 27, 151–160. [Google Scholar] [CrossRef]
- Bonizzoni, M.; Long, S.R.; Rainwater, C.; Anslyn, E.V. PAMAM dendrimer-induced aggregation of 5-carboxyfluorescein. J. Org. Chem. 2012, 77, 1258–1266. [Google Scholar] [CrossRef]
- Ihde, M.H.; Steelman, A.M.; Bonizzoni, M. Fluorescent probes for the supramolecular interactions responsible for binding of polycyclic aromatic hydrocarbons to hyperbranched polyelectrolytes in aqueous media. Isr. J. Chem. 2021, 61, 261–272. [Google Scholar] [CrossRef]
- Mallet, A.M.; Liu, Y.; Bonizzoni, M. An off-the-shelf sensing system for physiological phosphates. Chem. Commun. 2014, 50, 5003–5006. [Google Scholar] [CrossRef]







| Unk | Actual Identity | Concentration (mM) | % Error | |
|---|---|---|---|---|
| Calc’d | Actual | |||
| A | trans-Aconitate | 54.3 | 36.9 | 47.2 |
| B | α-Ketoglutarate | 23.1 | 25.4 | 9.06 |
| C | Citrate | 34.4 | 38.5 | 10.6 |
| D | Tricarballylate | 2.27 | 3.05 | 25.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Bonizzoni, M. Discrimination and Quantitation of Biologically Relevant Carboxylate Anions Using A [Dye•PAMAM] Complex. Sensors 2021, 21, 3637. https://doi.org/10.3390/s21113637
Xu Y, Bonizzoni M. Discrimination and Quantitation of Biologically Relevant Carboxylate Anions Using A [Dye•PAMAM] Complex. Sensors. 2021; 21(11):3637. https://doi.org/10.3390/s21113637
Chicago/Turabian StyleXu, Yifei, and Marco Bonizzoni. 2021. "Discrimination and Quantitation of Biologically Relevant Carboxylate Anions Using A [Dye•PAMAM] Complex" Sensors 21, no. 11: 3637. https://doi.org/10.3390/s21113637
APA StyleXu, Y., & Bonizzoni, M. (2021). Discrimination and Quantitation of Biologically Relevant Carboxylate Anions Using A [Dye•PAMAM] Complex. Sensors, 21(11), 3637. https://doi.org/10.3390/s21113637






