1. Introduction
Cancers are now responsible for the majority of global deaths and are expected to rank as the leading cause of death. Thus, cancer may be the most important barrier to increasing life expectancy in every country in the world in the 21st century [
1]. In the treatment of cancers, the correct diagnosis of the type and nature of tumors at as early a stage as possible is conducive to increased efficacy [
2]. The development of DNA microarray technology has made it possible to study the causes of cancers from the level of genes, which greatly improves the accuracy of diagnosis and the curative effect related to cancer. Although DNA microarray data are usually high-dimensional, with the number of genes in a sample often running into thousands or even tens of thousands, there are often only a few key genes that determine specific tumors [
3]. Since the original data contain excessive redundant genes and noise, directly using these data may lead to serious misclassification. Moreover, high-dimensional data also lead to a series of challenges such as a high storage cost and huge computation burden [
4]. Therefore, selecting the important genes related to cancer classification from the original huge number of genes is one of the key research areas with respect to gene data classification.
Currently, many effective methods of gene selection have been proposed. These methods can be roughly divided into three categories—i.e., filter, wrapper, and embedded—depending on their evaluation manner [
5]. The filter method employs the “certainty” metric to assign a score that reflects the ability of a gene to maintain the internal structure of data to determine the relevance between genes and specific cancers. However, as it neglects correlations among genes, this method may lose the important structural information underlying the original data. The wrapper method wraps genes into subsets and uses learning algorithms or predictive models to evaluate the importance of these subsets. However, the large number of subsets may induce a huge computational burden. The embedded method utilizes specific learning algorithm searching in the gene space for gene selection. In contrast with the other approaches, the embedded algorithm does not need to evaluate the classification ability of genes but only needs to select genes according to certain rules, leading to a lighter computational burden than the wrapped algorithm.
Embedded algorithms are divided into supervised feature (gene) selection (SFS) [
6,
7] and unsupervised feature selection (UFS) [
8,
9] approaches depending on whether they use label information. Typical SFS algorithms include SPFS [
10], LLFS [
11], mRMR [
12], L21RFS [
13], DLSR-FS [
14], feature selection through sparse guidance [
15,
16], and so on. Although the above methods use the sparsity of the graph structure or regression model to reduce the misclassification induced by noise, the expensive and laborious label cost limits the wide application of SFS approaches [
17,
18]. When attempting to discover characteristic patterns in data without labels, UFS is more challenging. Representative algorithms of the UFS type include SPEC [
19], FSSL [
20], JELSR [
21], EVSC [
22], Laplacian Score [
23], and so on.
Most of the above algorithms attempt to select features by uncovering the local manifold structure of data. More specifically, the above algorithms try to determine the embedding mapping which may reveal the low-dimensional manifold structure underlying the high-dimensional original gene data. Thus, the dimensionality reduction of the original gene data may be realized, and the inherent pattern of the data can even be found [
24]. Generally, the local manifold of the original data may be usually represented in the form of graphs such as a samples pair similarity graph [
10], k-NN graph [
23], local linear embedding [
25], and so on. In addition, besides the local structure of the original gene data, the global structure and the discriminant structure of the original gene data may also be explored to classify cancer [
26,
27,
28]. However, these methods merely focus on presenting the local structure, while they ignore the maintenance of the global structure of the original gene data [
29]; thus, their performance may be deteriorated by noise in the original data space.
Another challenge related to gene data classification is the dimension reduction of the original data. Since the original gene data are high-dimensional and have a complex topological structure, localizing the key genes related to cancer classification in the huge amount of original gene data is also challenging. Nie, Xu et al. [
30] proposed a unified UFS framework of dimensionality reduction, which uses a minimization regression residual criterion to linearize project data into a low-dimensional subspace. However, similar to the above-mentioned methods, maintaining the global structure of the original gene data is not included in their work. Inspired by Nie’s work, in this paper, we propose a unified UFS framework with characteristics including gene selection, global and local structure learning from original gene data. In the proposed UFS framework, we design a regression function composed of three parts which satisfies the requirement of embedding mapping including dimensionality reduction and the maintenance of the global and local structures of the original gene data. Specifically, the multi-dimensional scaling (MDS) method is first used to project the original gene data from the high-dimensional space into a low-dimensional space on the constraint of the Euclidean distance invariant. Then, the sparse regression method is employed based on the minimized regression residual criterion to learn the reconstruction coefficient in the low-dimensional space, meaning that the global structure of the original data can be maintained in the course of the dimensionality reduction of the original data. Finally, a probabilistic neighborhood graph model based on sample genes is used to maintain the local manifold structure of the data. The contributions of this article are summarized as follows.
We combine structure learning and feature selection to propose a new feature selection framework. Since the MDS method is employed in the proposed framework to preserve the original space structure, which is reconstructed in a low-dimensional space, the proposed framework can preserve both the global structure and local structure underlying the original gene data;
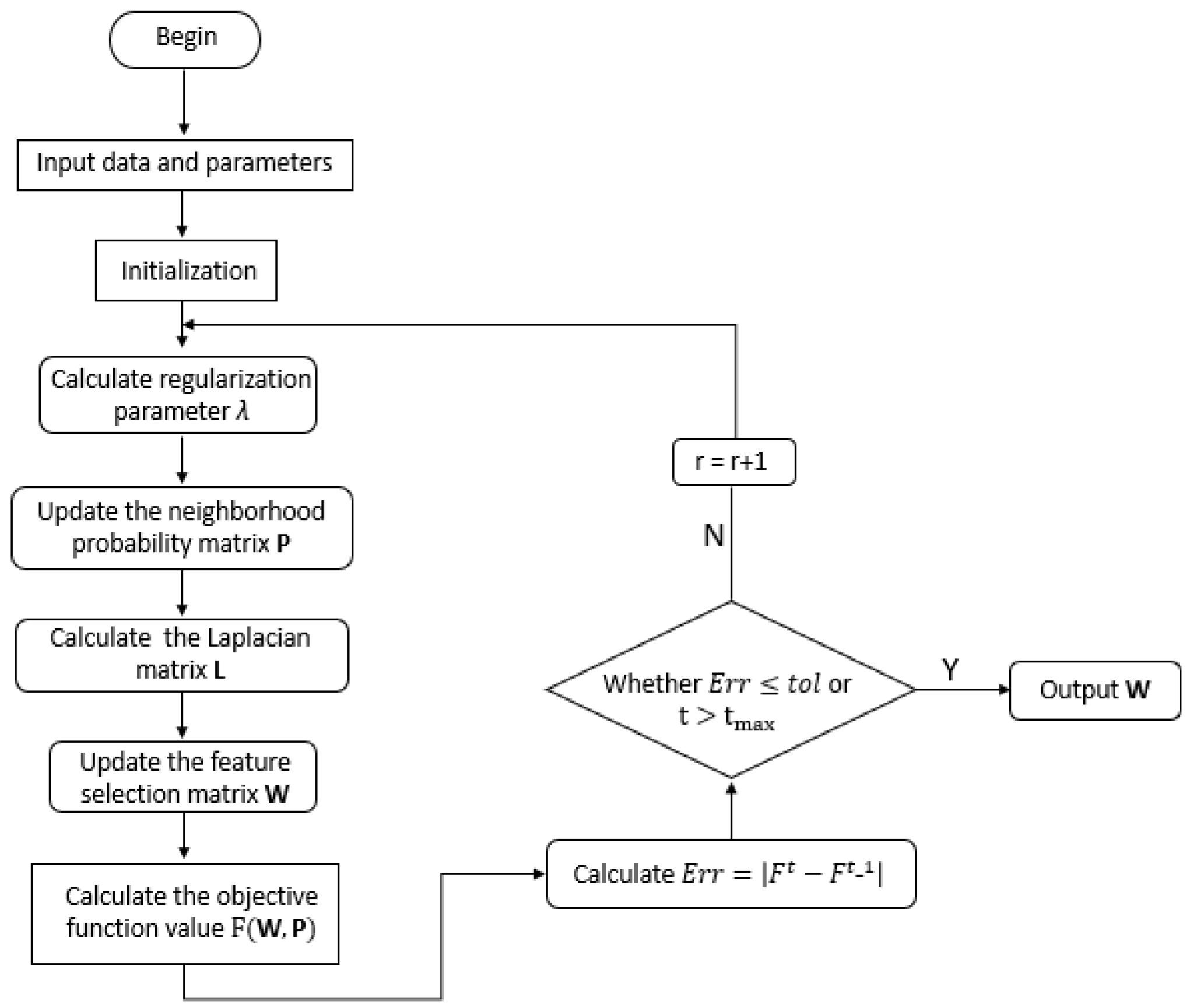
The alternating direction method of multipliers (ADMM) is proposed to handle non-convex optimization related to the proposed framework. In addition, an efficient strategy related to the inverse of the high-dimensional matrix is also included in the proposed method;
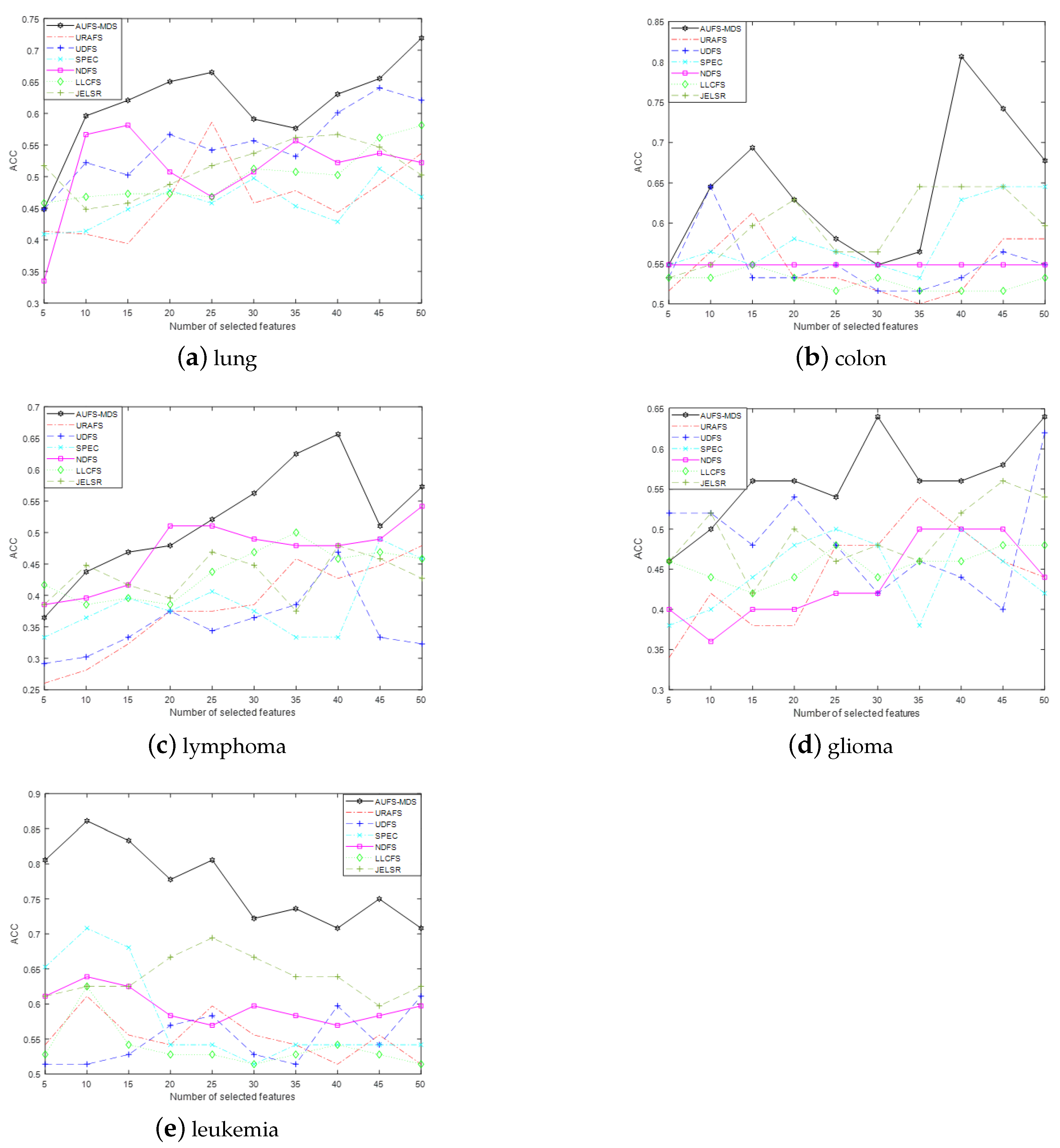
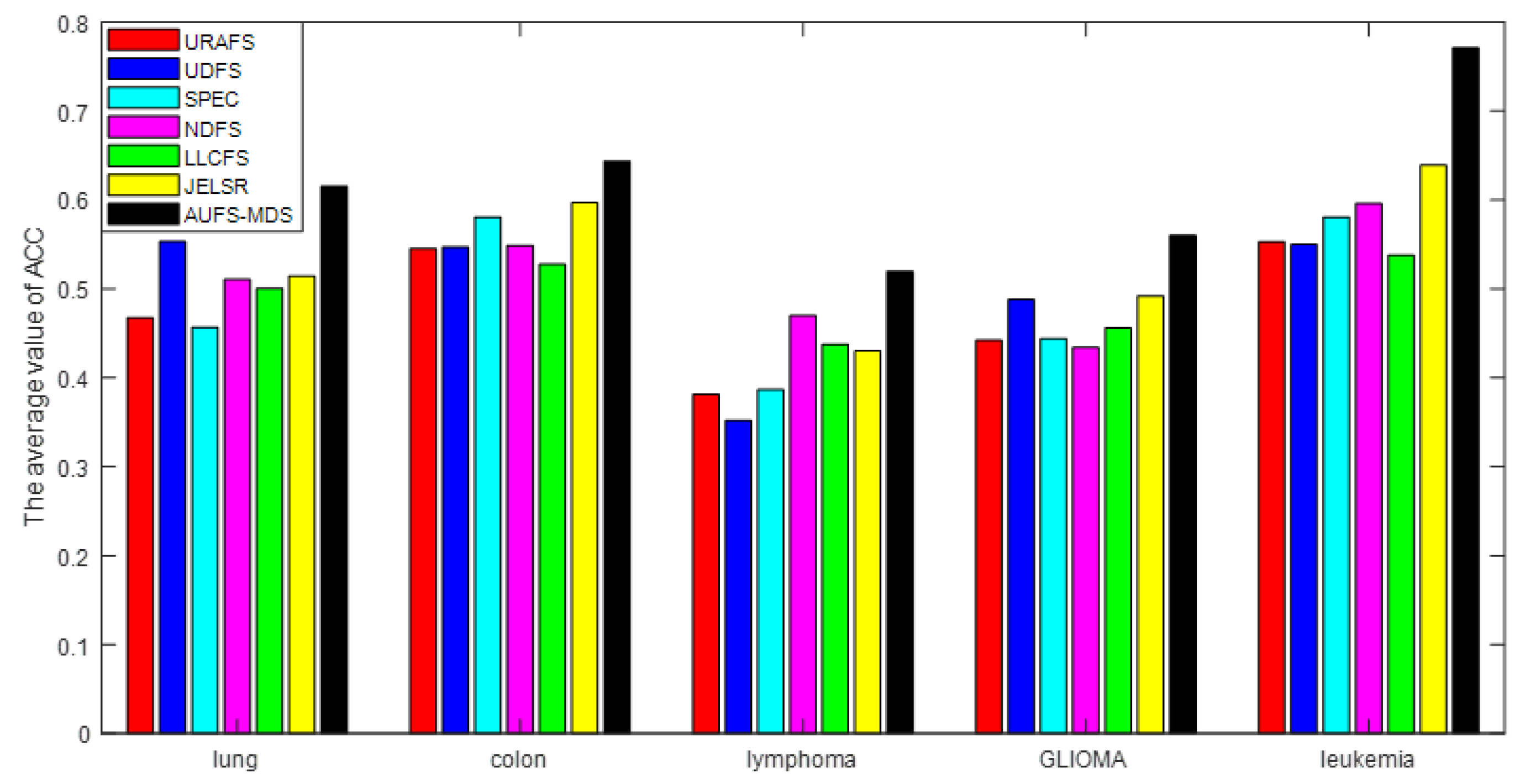
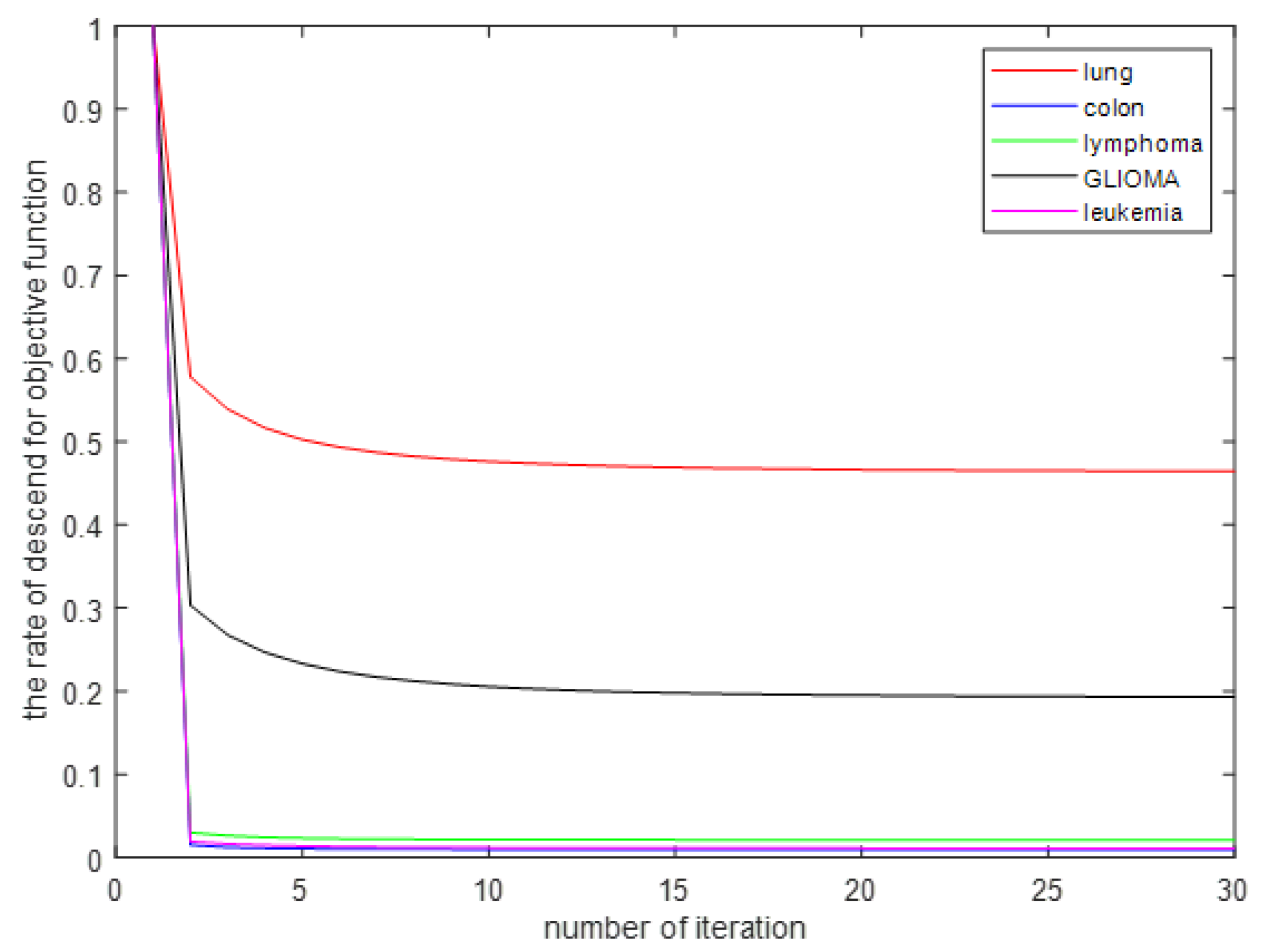
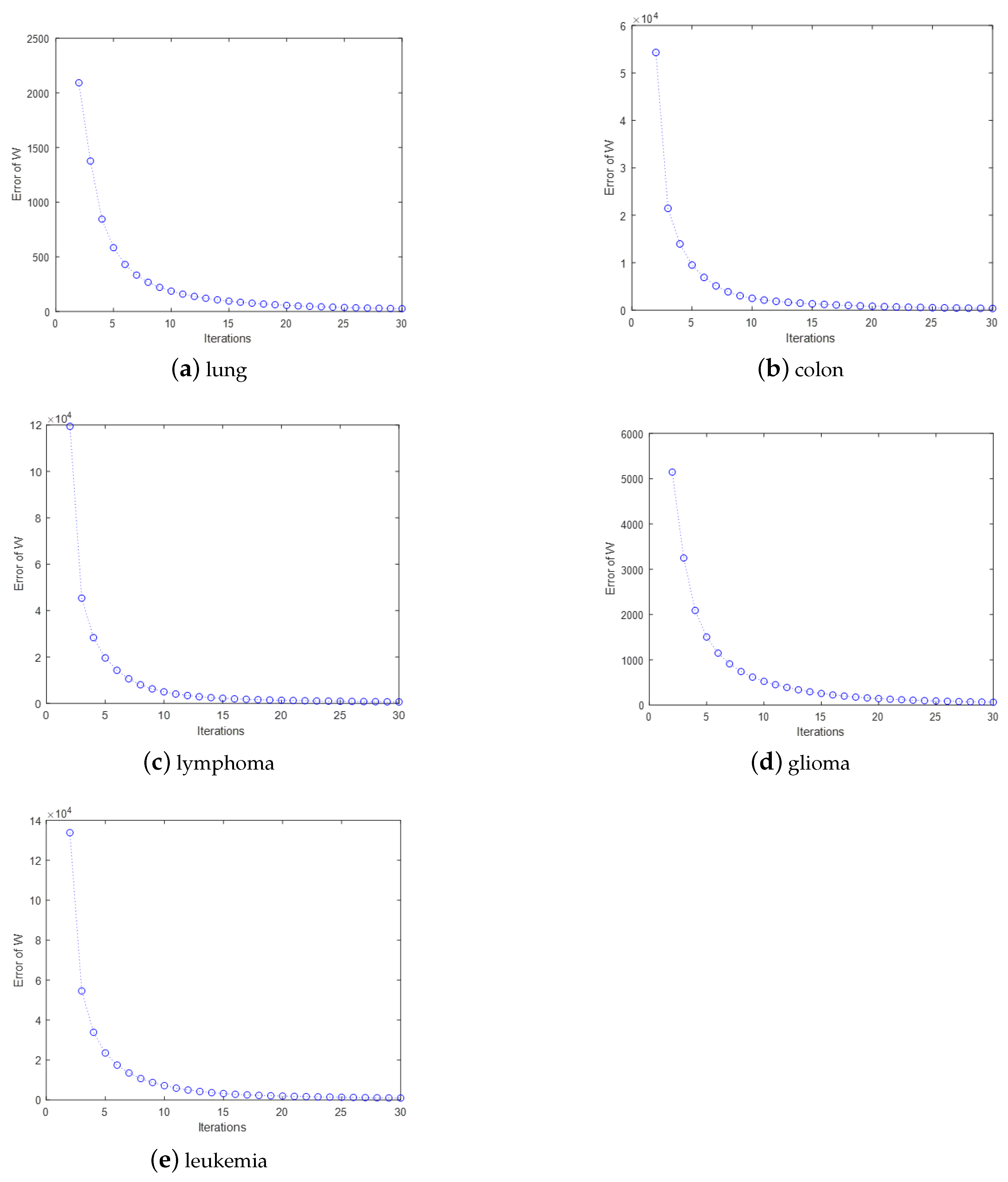
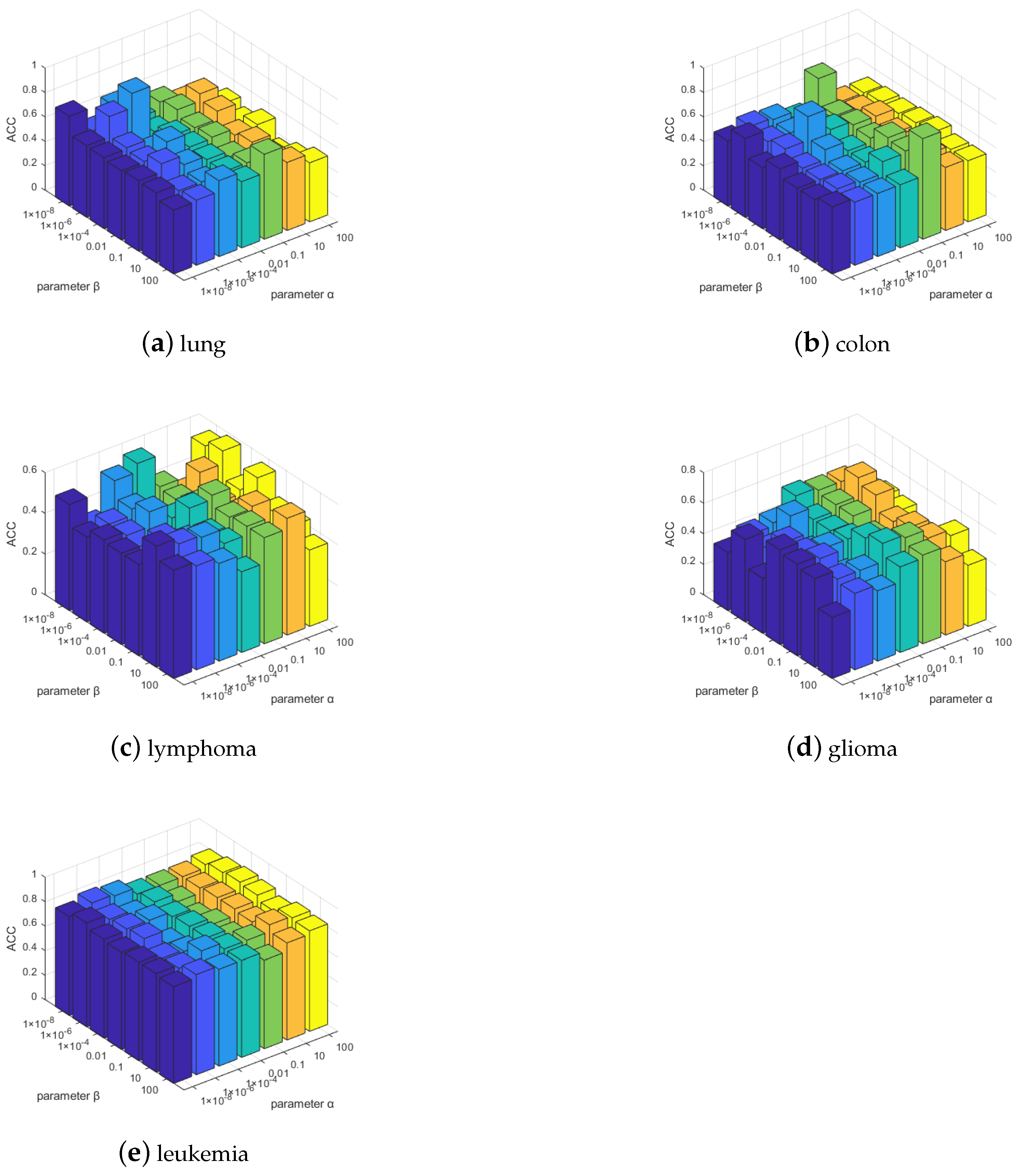
The convergence and computational complexity of the proposed algorithm is discussed. Extensive experiments on multiple gene data demonstrate the superiority of our framework and method.
The rest of the paper is organized as follows.
Section 2 briefly recalls the existing unsupervised embedded feature selection algorithms and introduces the MDS algorithm.
Section 3 introduces the proposed approach and the optimization process. In
Section 4, we analyze the convergence and parameter selection of the proposed algorithm. In
Section 5, we conduct extensive experiments on multiple datasets and discuss and analyze several experimental results. In the last section, we present the conclusion and future prospects.
2. Related Work
In this section, we review several typical UFS algorithms.
In the past few years, UFS based on the spectral analysis technique has shown outstanding performance. Zhao and Liu [
19] proposed the spectrum feature selection (SPEC) algorithm, which employs spectral analysis based on graph theory to select features with correlation. Due to the lack of an embedded learning process and the low sparsity of the graph caused by excessive samples, SPEC may be susceptible to noise and irrelevant features. Li, Yang et al. [
28] proposed a non-negative discriminant feature selection algorithm (NDFS), which uses the correlation between discriminant information and features to select features. Specifically, NDFS first uses the spectral clustering technique to detect the structure underlying the original gene data and then learns the clustering label to construct the feature selection matrix, finally selecting features with discriminant information. Although the influence on the graph structure of noise is reduced by the structure learning and graph sparsity, NDFS can only work in the situation in which a linear relationship between the features and the clustering pseudo tags exists; moreover, the clustering tag technique employed by NDFS cannot fully capture the local structure information underlying the original data.
As mentioned above, the graph of the original gene data is susceptible to noise and irrelevant features; thus, it is necessary to reveal the data relationship in the low-dimensional subspace of the original gene data. Hou, Nie et al. [
21] proposed the joint low-dimensional embedded learning and sparse regression (JELSR) feature selection method. However, their method merely focuses on low-dimensional manifold embedding, thus ignoring the maintenance of the global structure of original gene data, leading to some globally important information being missing. Ye and Zhang et al. [
18] incorporated linear discriminant analysis (LDA), an adaptive structure based on spectral analysis and
-norm sparse regression into the joint learning framework of UFS. Although, their method employs the
-norm to enforce the row sparsity of the feature selection matrix, leading the projection matrix based on the LDA method to have the capability of feature selection, limitations of the traditional LDA method, such as suboptimal solutions and ignoring local manifolds, are also inherited. In this paper, we employ the multi-dimensional scaling (MDS) algorithm to reduce the dimensions of the original gene data and to maintain the global structure of the original gene data. In contrast to LDA and principal component analysis (PCA), the goal of MDS is not to preserve the maximum divisibility of the original data but to pay more attention to maintaining the internal characteristics of features underlying high-dimensional data.