Author Contributions
All authors contributed equally in preparing the manuscript. Conceptualization, T.B.A.A., S.F.M., and M.M.R. (Md. Motiur Rahman); methodology, T.B.A.A., S.F.M., M.M.R. (Md. Motiur Rahman), and M.M.R. (Md. Masud Rana); software, T.B.A.A., S.F.M., and M.M.R. (Md. Motiur Rahman); validation, T.B.A.A., M.M.R. (Md. Masud Rana), M.R.I., and M.A.P.M.; formal analysis, T.B.A.A., M.M.R. (Md. Masud Rana), M.R.I., and I.M.M.; investigation, T.B.A.A., M.M.R. (Md. Masud Rana), I.M.M., and M.A.P.M.; data curation, T.B.A.A. and M.M.R. (Md. Masud Rana); writing—original draft preparation, T.B.A.A.; writing—review and editing, M.M.R. (Md. Masud Rana), M.R.I., I.M.M., M.A.P.M., and A.Z.K.; supervision, M.M.R. (Md. Masud Rana), M.R.I., and A.Z.K. All authors have read and agreed to the published version of the manuscript.
Figure 1.
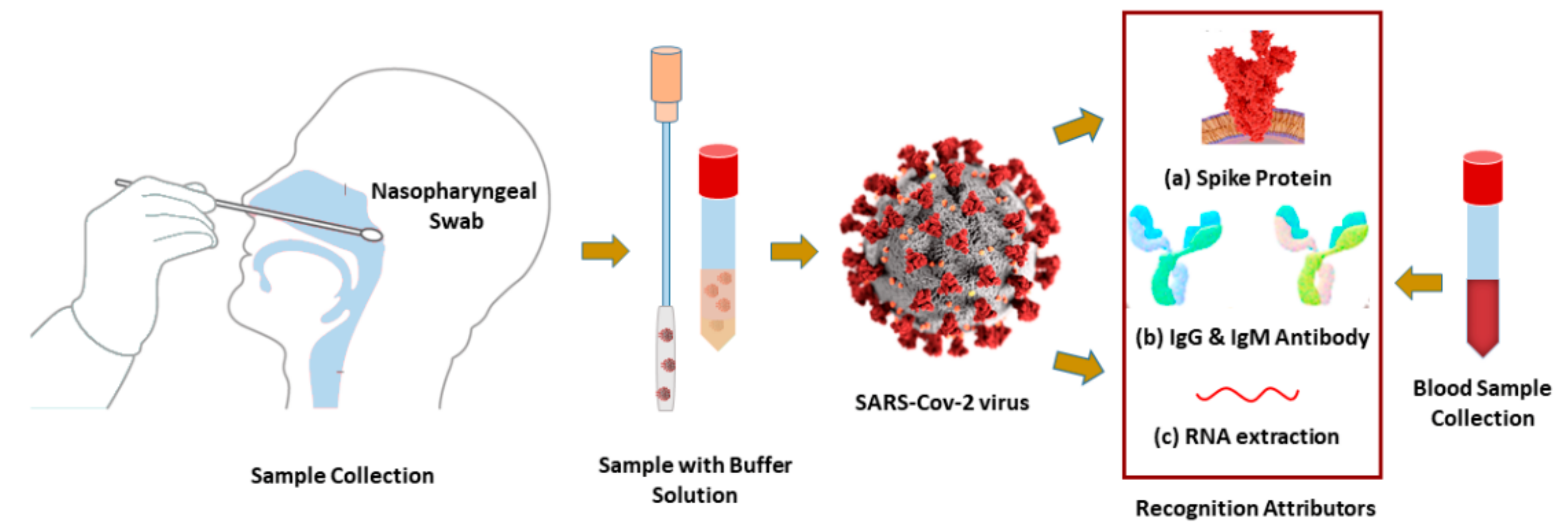
Schematic diagram of diagnosis biomarkers: (a) SARS-CoV-2 virus spike protein RBD, (b) IgG or IgM antibodies, and (c) or RNA-oligonucleotides are collected from nasopharyngeal swabs or human blood.
Figure 1.
Schematic diagram of diagnosis biomarkers: (a) SARS-CoV-2 virus spike protein RBD, (b) IgG or IgM antibodies, and (c) or RNA-oligonucleotides are collected from nasopharyngeal swabs or human blood.
Figure 2.
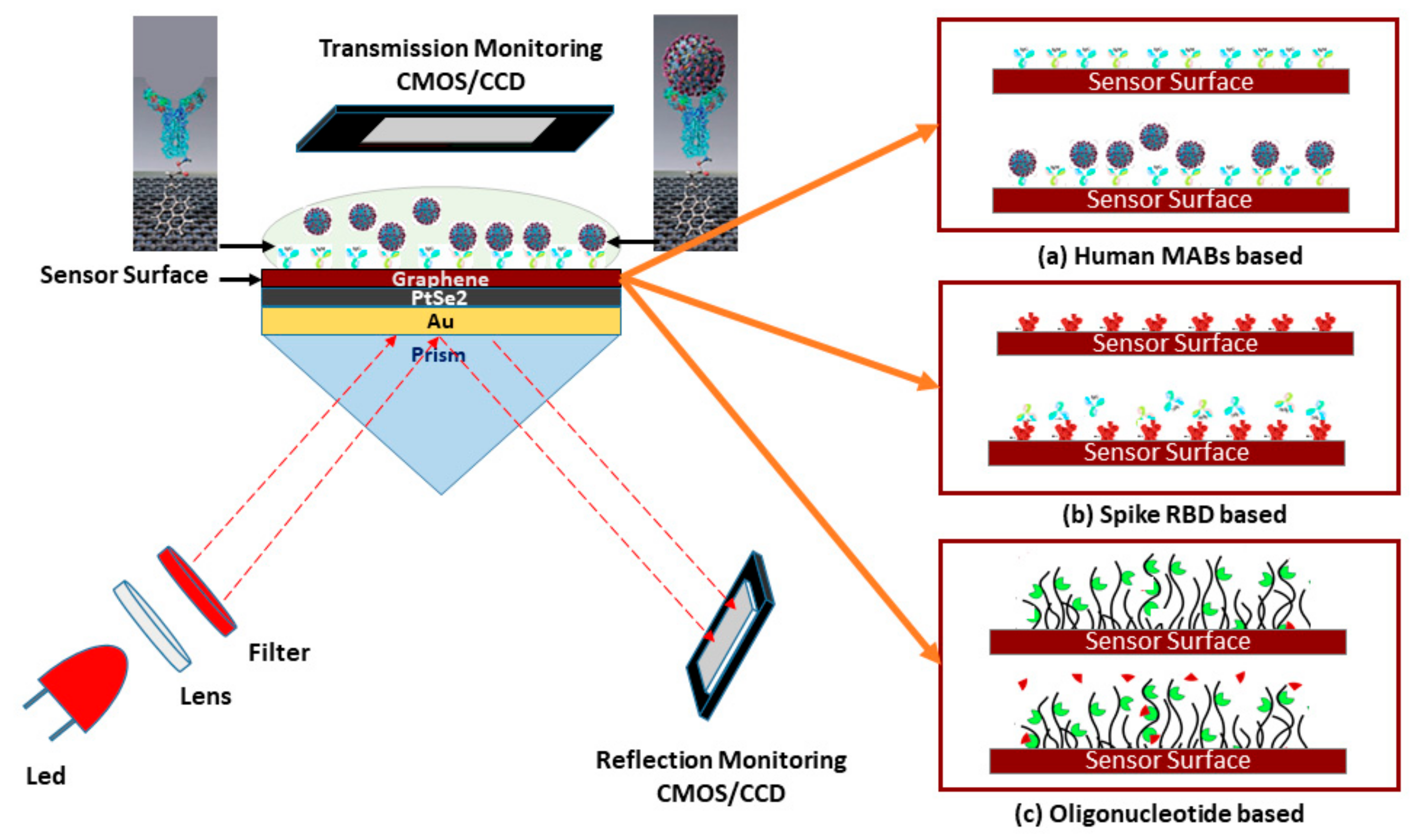
Schematic diagram of the five-layered (Bk7/Au/PtSe2/Graphene/PBS) SPR biosensor for diagnosis of SARS-CoV-2 cultured virus; three operating modes are represented to detect the SARS-CoV-2 virus: (a) rapid recognition of whole virus spike RBD with immobilized human mAbs (mAbs as ligand and spike RBD as analyte), (b) rapid recognition of mAbs with immobilized virus spike RBD (spike RBD as ligand and mAbs as analyte), and (c) or recognition of the virus RNA sequence with immobilized probe sequence onto the graphene implicated sensor surface.
Figure 2.
Schematic diagram of the five-layered (Bk7/Au/PtSe2/Graphene/PBS) SPR biosensor for diagnosis of SARS-CoV-2 cultured virus; three operating modes are represented to detect the SARS-CoV-2 virus: (a) rapid recognition of whole virus spike RBD with immobilized human mAbs (mAbs as ligand and spike RBD as analyte), (b) rapid recognition of mAbs with immobilized virus spike RBD (spike RBD as ligand and mAbs as analyte), and (c) or recognition of the virus RNA sequence with immobilized probe sequence onto the graphene implicated sensor surface.
Figure 3.
Schematic diagram of the possible multi-layer film fabrication process of the proposed SPR sensor for detection of the SARS-Cov-2 virus;—in step 1, the sensor Bk7/Au/PtSe2/graphene layers are deposited sequentially, the synthesis technique of PtSe2 and graphene film are also depicted. In step 2, the ligands;—(a) mAbs, (b) spike RBD, and (c) probe oligo are immobilized with the graphene layer through PBSE.
Figure 3.
Schematic diagram of the possible multi-layer film fabrication process of the proposed SPR sensor for detection of the SARS-Cov-2 virus;—in step 1, the sensor Bk7/Au/PtSe2/graphene layers are deposited sequentially, the synthesis technique of PtSe2 and graphene film are also depicted. In step 2, the ligands;—(a) mAbs, (b) spike RBD, and (c) probe oligo are immobilized with the graphene layer through PBSE.
Figure 4.
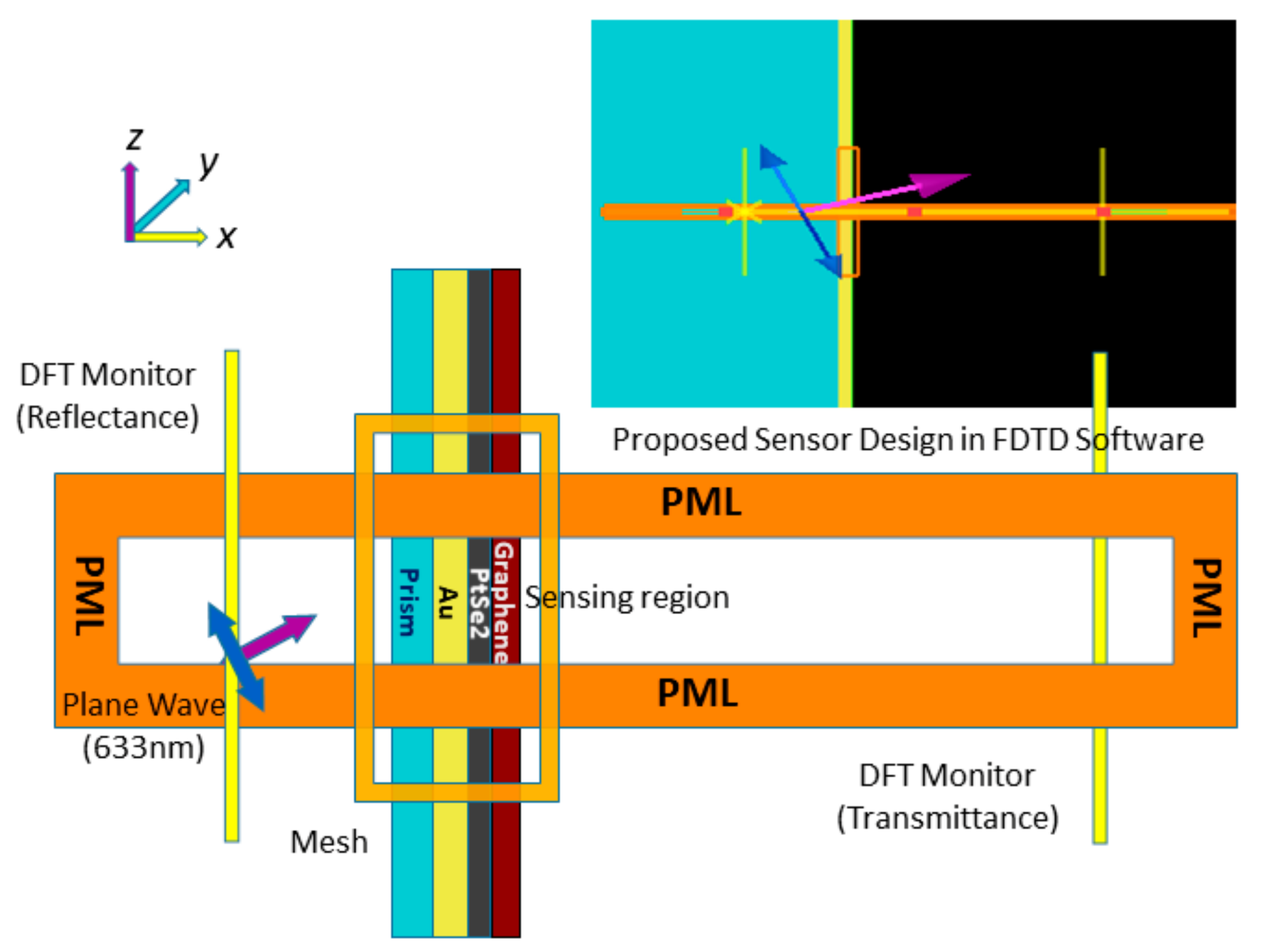
FDTD simulation schematic for the proposed SPR sensor.
Figure 4.
FDTD simulation schematic for the proposed SPR sensor.
Figure 5.
Schematic diagram of: (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm)/PBS (RI = 1.3348) layered model with variation in incident light wavelength .
Figure 5.
Schematic diagram of: (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm)/PBS (RI = 1.3348) layered model with variation in incident light wavelength .
Figure 6.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of the Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (1.7 nm) sensor with other conventional sensors with PBS analyte (RI = 1.3348).
Figure 6.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of the Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (1.7 nm) sensor with other conventional sensors with PBS analyte (RI = 1.3348).
Figure 7.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (0.34 nm × L) coated sensor reflectance and transmittance spectra with the thickness of the graphene layers (L = 0, 1, …, 5) before adsorption of the analyte.
Figure 7.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (0.34 nm × L) coated sensor reflectance and transmittance spectra with the thickness of the graphene layers (L = 0, 1, …, 5) before adsorption of the analyte.
Figure 8.
The Schematic of: (a) SPR, and (b) SPR sensorgram absorption curve.
Figure 8.
The Schematic of: (a) SPR, and (b) SPR sensorgram absorption curve.
Figure 9.
The schematic of (a) the angle of incidence, and (b) the electric field intensity as a function of normal distance from the interface for (i) three-layer, (ii) four-layer, and (iii) five-layer SPR sensor configuration.
Figure 9.
The schematic of (a) the angle of incidence, and (b) the electric field intensity as a function of normal distance from the interface for (i) three-layer, (ii) four-layer, and (iii) five-layer SPR sensor configuration.
Figure 10.
Schematic diagram of: (a) SPR and (b) SPRF characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm) substrates with immobilized IgG (H014 or S309) as ligand and different concentration levels of SARS-CoV-2 spike RBDs (concentration of 1.953125 nM to 62.5 nM) as the analyte.
Figure 10.
Schematic diagram of: (a) SPR and (b) SPRF characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm) substrates with immobilized IgG (H014 or S309) as ligand and different concentration levels of SARS-CoV-2 spike RBDs (concentration of 1.953125 nM to 62.5 nM) as the analyte.
Figure 11.
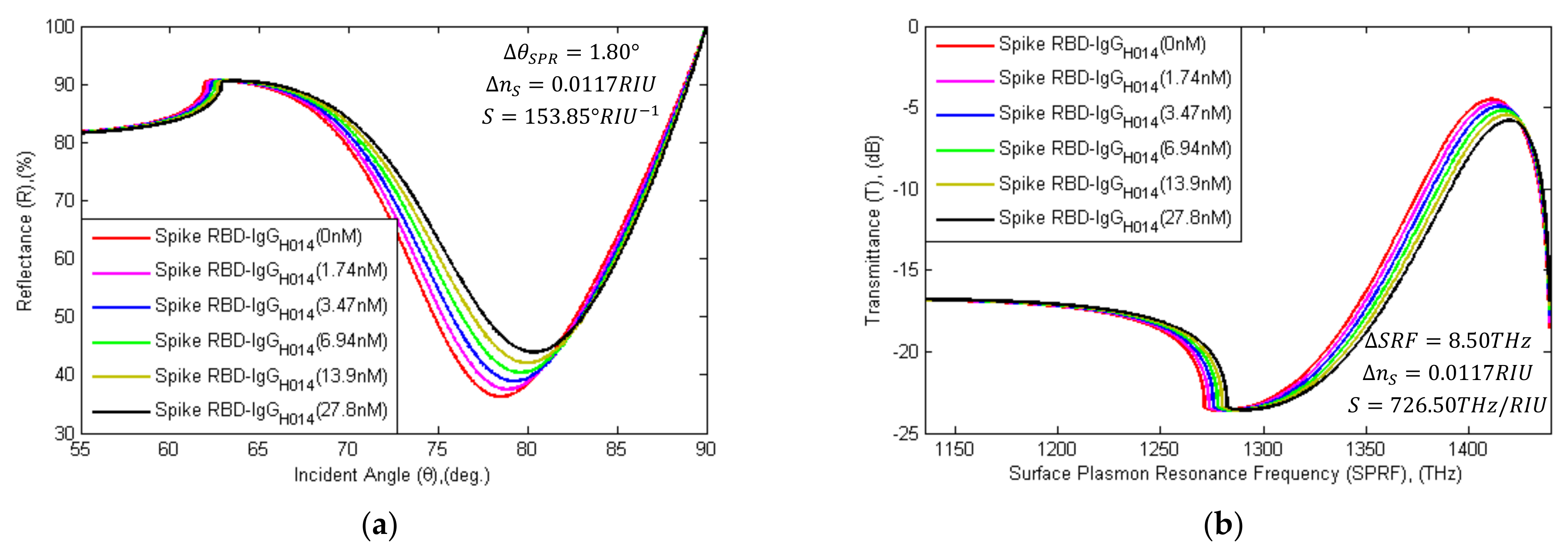
Schematic diagram of (a) SPR and (b) SPRF characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm) substrates with immobilized SARS-CoV-2 spike RBDs as the ligand and different concentration levels of IgG-H014 (concentration of 1.74 nM to 27.8 nM) as the analyte.
Figure 11.
Schematic diagram of (a) SPR and (b) SPRF characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene(1.7 nm) substrates with immobilized SARS-CoV-2 spike RBDs as the ligand and different concentration levels of IgG-H014 (concentration of 1.74 nM to 27.8 nM) as the analyte.
Figure 12.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (0.34 nm × L) substrates without probe (sh-Oligo), with sh-Oligo and different concentrated levels of oligonucleotide (wt or mr type) binding for recognition of SARS-Cov-2 virus RNA. angle and SPRF shift right due to binding with oligonucleotides.
Figure 12.
Schematic diagram of (a) SPR and (b) SPRF curve characteristics of Bk7/Au(50 nm)/PtSe2(2 nm)/Graphene (0.34 nm × L) substrates without probe (sh-Oligo), with sh-Oligo and different concentrated levels of oligonucleotide (wt or mr type) binding for recognition of SARS-Cov-2 virus RNA. angle and SPRF shift right due to binding with oligonucleotides.
Table 1.
RI of sensor materials at different wavelengths.
Table 1.
RI of sensor materials at different wavelengths.
| RI | | | | |
|---|
| BK7 [34] | 1.5195 | 1.5151 | 1.5112 | 1.5055 |
| Au [34,66] | 0.0990 + 2.9952i | 0.1377 + 3.6183i | 0.2063 + 4.5133i | 0.4417 + 6.7308i |
| PtSe2 [34,67] | 2.5917 + 1.1332i | 2.9029 + 0.8905i | 2.8229 + 0.5277i | 2.7986 + 0.2878i |
| Graphene [34,68] | 3 + 0.9658i | 3 + 1.1487i | 3 + 1.4160i | 3 + 2.0913i |
Table 2.
Data evaluation of , and sensitivity (S) attributors with change of incident wavelength in the proposed SPR sensor.
Table 2.
Data evaluation of , and sensitivity (S) attributors with change of incident wavelength in the proposed SPR sensor.
| Incident Light Wavelength, (λ) [nm] | | | | | | | | | Sensitivity (S) |
|---|
| |
|---|
| 532.0 nm | 74.56 | 80.70 | 13.689 | 1691.2 | 0.00 | 0.00 | 0.0000 | 0.00 | 50 | 100 |
| 632.8 nm | 31.22 | 76.25 | 3.7420 | 1399.6 | 43.34 | 4.45 | 9.9470 | 291.60 | 200 | 1000 |
| 780.0 nm | 15.04 | 70.05 | 1.6288 | 1098.6 | 59.52 | 10.65 | 12.0602 | 592.60 | 100 | 900 |
| 1152 nm | 19.12 | 65.70 | 2.1217 | 721.29 | 55.44 | 15.00 | 11.5673 | 969.91 | 100 | 570 |
Table 3.
Evaluation of the proposed sensor model with other conventional models with regards to sensitivity (S), FWHM, detection accuracy (DA), the figure of merit (FOM), and quality factor (QF).
Table 3.
Evaluation of the proposed sensor model with other conventional models with regards to sensitivity (S), FWHM, detection accuracy (DA), the figure of merit (FOM), and quality factor (QF).
|
|
|
|
| | | |
|
| FW-HM
| DA
| FOM
| QF
|
|---|
| Bk7/Au(50 nm)/PBS | 0.54 | 72.60 | 0.054 | 104.19 | - | - | - | - | - | - | - | - | - |
| Bk7/Au(50 nm)/Graphene (5 layer)/PBS | 14.80 | 75.05 | 1.601 | 1391.5 | 14.26 | 2.46 | 1.547 | 1287.31 | 82.0 | 1.30 | 0.77 | 63.07 | 155.17 |
| Bk7/Au(50 nm)/PtSe2 (2 nm)/Graphene (5 layer)/PBS | 35.08 | 78.10 | 4.320 | 1409.3 | 34.54 | 5.50 | 4.266 | 1305.11 | 183.3 | 3.90 | 0.26 | 47.00 | 258.50 |
Table 4.
Comparison of change in , sensitivity, FWHM, DA, FOM, and QF with the variation of the graphene layer (L) in the proposed SPR biosensor.
Table 4.
Comparison of change in , sensitivity, FWHM, DA, FOM, and QF with the variation of the graphene layer (L) in the proposed SPR biosensor.
| Graphene layer thickness | |
|
|
| |
|
|
| S
| FW-HM
| DA
| FOM
| QF
|
|---|
| L = 0 × 0.34 nm | 15.20 | 75.45 | 1.6409 | 1394.2 | - | - | - | - | - | - | - | - | - |
| L = 1 × 0.34 nm | 19.05 | 76.00 | 2.1133 | 1397.6 | 3.85 | 0.55 | 0.4724 | 3.40 | 18.33 | 0.38 | 2.63 | 48.24 | 26.53 |
| L = 2 × 0.34 nm | 22.99 | 76.55 | 2.6124 | 1400.9 | 7.79 | 1.10 | 0.9715 | 6.70 | 36.67 | 0.78 | 1.28 | 47.01 | 51.72 |
| L = 3 × 0.34 nm | 27.00 | 77.10 | 3.1464 | 1404.0 | 11.8 | 1.65 | 1.5055 | 9.80 | 55.00 | 1.22 | 0.82 | 45.08 | 74.39 |
| L = 4 × 0.34 nm | 31.04 | 77.60 | 3.7156 | 1406.8 | 15.84 | 2.15 | 2.0747 | 12.60 | 71.67 | 1.73 | 0.58 | 41.43 | 89.07 |
| L = 5 × 0.34 nm | 35.08 | 78.10 | 4.3201 | 1409.3 | 19.88 | 2.65 | 2.6792 | 15.10 | 88.33 | 2.31 | 0.43 | 38.24 | 101.3 |
Table 5.
and characteristics for different concentrations of SARS-Cov-2 Spike RBD as an analyte.
Table 5.
and characteristics for different concentrations of SARS-Cov-2 Spike RBD as an analyte.
| | | | | | | | |
|---|
| | 0 nM | 36.25 | 78.50 | 4.5017 | 1411.3 | 0.00 | 0.00 | 0.0000 | 0.00 |
| | 1.953125 nM | 37.52 | 78.85 | 4.7035 | 1413.1 | 1.27 | 0.35 | 0.2018 | 1.80 |
| | 3.90625 nM | 38.91 | 79.25 | 4.9280 | 1414.9 | 2.66 | 0.75 | 0.4263 | 3.60 |
| RBD | 7.8125 nM | 40.42 | 79.60 | 5.1792 | 1416.6 | 4.17 | 1.10 | 0.6775 | 5.30 |
| | 15.625 nM | 42.09 | 79.95 | 5.4627 | 1418.2 | 5.84 | 1.45 | 0.9610 | 6.90 |
| | 31.25 nM | 43.94 | 80.30 | 5.7880 | 1419.8 | 7.69 | 1.80 | 1.2863 | 8.50 |
| | 62.5 nM | 46.06 | 80.70 | 6.1728 | 1421.3 | 9.81 | 2.20 | 1.6711 | 10.00 |
Table 6.
and characteristics for different concentrations of (H014) as an analyte.
Table 6.
and characteristics for different concentrations of (H014) as an analyte.
| |
[deg.] |
[dB] | |
| |
| |
|---|
| H014 | 0 nM | 36.25 | 78.50 | 4.5017 | 1411.3 | 0.00 | 0.00 | 0.0000 | 0.00 |
| 1.74 nM | 37.52 | 78.85 | 4.7034 | 1413.1 | 1.27 | 0.35 | 0.2017 | 1.80 |
| 3.47 nM | 38.91 | 79.25 | 4.9277 | 1414.9 | 2.66 | 0.75 | 0.4260 | 3.60 |
| 6.94 nM | 40.42 | 79.60 | 5.1784 | 1416.6 | 4.17 | 1.10 | 0.6767 | 5.30 |
| 13.9 nM | 42.08 | 79.95 | 5.4608 | 1418.2 | 5.83 | 1.45 | 0.9591 | 6.90 |
| 27.8 nM | 43.92 | 80.30 | 5.7837 | 1419.8 | 7.67 | 1.80 | 1.2820 | 8.50 |
Table 7.
Primers and probe sequence orientation for recognition of SARS-CoV-2 virus RNA.
Table 7.
Primers and probe sequence orientation for recognition of SARS-CoV-2 virus RNA.
| Name | Target Genes | Sequence 5′→3′ | Nucleotide position | Target Genes | Sequence 5′→3′ | Nucleotide Position |
|---|
| Forward Primer: | ORF 1ab | CCCTGTGGGTTTTACACTTAA | 13342-13362 | N gene | GGGGAACTTCTCCTGCTAGAAT | 28881-28902 |
| Reverse Primer: | ACGATTGTGCATCAGCTGA | 13442-13460 | CAG ACATTTTGCTCTCAAGCTG | 28958-28979 |
| Immobilized Mutant Probe (sh): | FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ | 13377-13404 | FAM-TTGCTGCTGCTTGACAGATT-TAMRA | 28934-28953 |
| Matching type (mr) Sequence: | TCCAACCTTTCCACATACCGCAGCGGA | - | AACTCTGTCAAGCAGCAGCAA | - |
| Mismatching type (wt) Sequence: | TCCAAGCAAACCCAATACCGCAGCGGA | - | AACTCTACTAATTAGCAGCAA | - |
Table 8.
for different concentrations of the target oligonucleotides as an analyte.
Table 8.
for different concentrations of the target oligonucleotides as an analyte.
| | | | | | |
| |
|---|
| 0 nM (no-sh-Oligo) | 36.25 | 78.50 | 4.5017 | 1411.3 | - | - | - | - |
| 150 nM (sh-Oligo) | 37.92 | 78.95 | 4.7670 | 1413.7 | 0.00 | 0.00 | 0.0000 | 0.00 |
| 165 nM (wt-Oligo) | 39.82 | 79.45 | 5.0778 | 1416.0 | 1.90 | 0.50 | 0.3108 | 2.30 |
| 180 nM (mr-Oligo) | 41.97 | 79.95 | 5.4429 | 1418.1 | 4.05 | 1.00 | 0.6759 | 4.40 |
| 210 nM (mr-Oligo) | 44.47 | 80.40 | 5.8827 | 1420.2 | 6.55 | 1.45 | 1.1157 | 6.50 |
| 240 nM (mr-Oligo) | 47.34 | 80.90 | 6.4130 | 1422.1 | 9.42 | 1.95 | 1.6460 | 8.40 |
| 270 nM (mr-Oligo) | 50.59 | 81.30 | 7.0506 | 1423.7 | 12.67 | 2.35 | 2.2836 | 10.00 |
| 300 nM (mr-Oligo) | 54.21 | 81.70 | 7.8111 | 1425.1 | 16.29 | 2.75 | 3.0441 | 11.40 |
Table 9.
COVID-19 test results based on the detection of attributors’ characteristics.
Table 9.
COVID-19 test results based on the detection of attributors’ characteristics.
| Pathology No. | COVID-19 Test Result Making Condition | Result |
|---|
| 01 | & | OR | & | COVID-19 Positive |
| 02 | & | & | COVID-19 Negative |
| 03 | & | & | Try again |
| 04 | & | & | Try again |