Lateral Flow Serodiagnosis in the Double-Antigen Sandwich Format: Theoretical Consideration and Confirmation of Advantages
Abstract
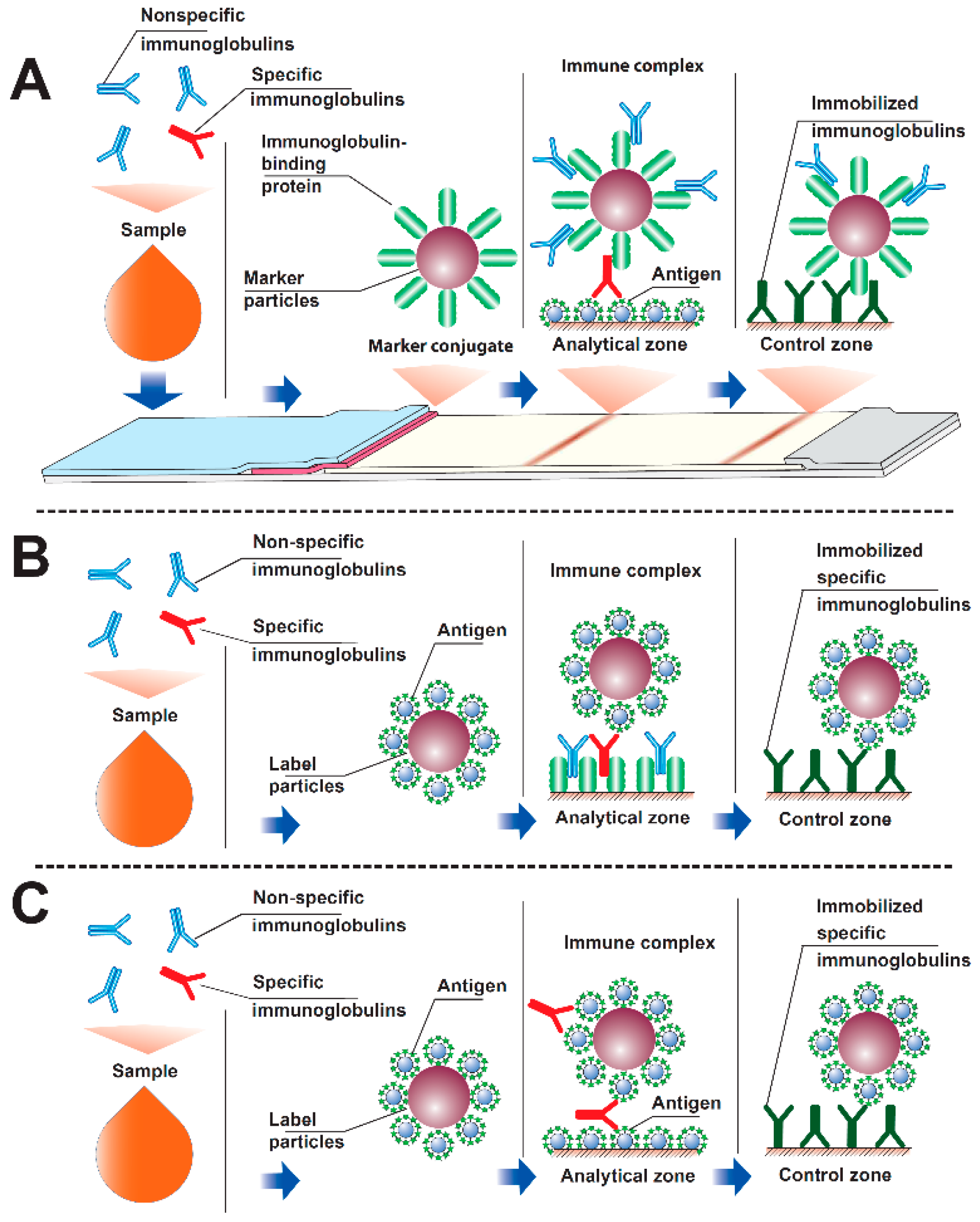
1. Introduction
2. Materials and Methods
2.1. Reactants and Membranes
2.2. Gold Nanoparticles Preparation
2.3. Transmission Electron Microscopy
2.4. Immobilization of Rv0934 Antigen on Gold Nanoparticles
2.5. Application of Reagents to Immunochromatographic Membranes
2.6. Manufacturing of Immunochromatographic Test Systems
2.7. Immunochromatographic Analysis
2.8. Determination of Kinetic and Equilibrium Parameters of Interactions between Immunoreagents
2.9. Determination of the Concentration of Binding Sites on the Gold Nanoparticles Conjugate
2.10. Numerical Simulation of an Immunochromatographic Assay
3. Results and Discussion (Theoretical)
3.1. Modeling Interactions in an Immunochromatographic System
3.2. Formation of PAb and P2Ab Complexes
3.3. Analytical Model of the Reactions (1) and (2) in Equilibrium Conditions
4. Results and Discussion (Experimental)
4.1. Overview
4.2. Characterization of the Preparation of Gold Nanoparticles
4.3. Determination of the Constants of Immune Interaction
4.4. Determination of the Concentration of the Labeled Antigen
4.5. Experimental Verification of Theoretical Relationships
5. Conclusions
- Use the concentration of the labeled antigen with approximately twice the concentration of active antibodies in the sample. As the concentration of antibodies against the antigen used is initially unknown, it is necessary to experimentally titrate the amount of labeled antigen to achieve the target sensitivity.
- Use the highest possible concentration of the immobilized antigen in the analytical zone, but not exceeding the sorption capacity of the working membrane.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parija, S.C. Textbook of Microbiology & Immunology-E-Book; Reed Elsevier India Pvt. Ltd.: Maharashtra, India, 2014; p. 684. [Google Scholar]
- Tille, P. Bailey & Scott’s Diagnostic Microbiology-E-Book; Elsevier Health Sciences: New York, NY, USA, 2013; p. 1056. [Google Scholar]
- Hamilton, R.G.; Hemmer, W.; Nopp, A.; Kleine-Tebbe, J. Advances in IgE testing for diagnosis of allergic disease. J. Allergy Clin. Immunol. Pract. 2020, 8, 2495–2504. [Google Scholar] [CrossRef]
- Mohiuddin, M. Tuberculosis–burden and serodiagnosis. IMC J. Med. Sci. 2020, 14, 59–69. [Google Scholar] [CrossRef]
- Jenabian, M.A.; Costiniuk, C.T.; Talla, P.; Robin, L.; Tonen Wolyec, S.; Mboumba Bouassa, R.S.; Bélec, L. Potential for False-Positive Results with Serological Assays for HIV in Central Africa: Implications for the HIV Serodiagnosis Algorithm According to the 2015 Consolidated WHO Recommendations for Resource-Constrained Countries. AIDS Res. Hum. Retrovir. 2017, 33, 1077–1079. [Google Scholar] [CrossRef]
- Feng, X.; Xiu, B.; Chen, K.; Yang, X.; Zhang, H.; Yue, J.; Tan, Y.; Li, H.; Nicholson, R.A.; Tam, A.W.; et al. Enhanced serodiagnostic utility of novel Mycobacterium tuberculosis polyproteins. J. Infect. 2013, 66, 366–375. [Google Scholar] [CrossRef]
- Mani, V.; Paleja, B.; Larbi, K.; Kumar, P.; Tay, J.A.; Siew, J.Y.; Inci, F.; Wang, S.; Chee, C.; Wang, Y.T.; et al. Microchip-based ultrafast serodiagnostic assay for tuberculosis. Sci. Rep. 2016, 6, 35845. [Google Scholar] [CrossRef]
- Mezzasoma, L.; Bacarese-Hamilton, T.; Di Cristina, M.; Rossi, R.; Bistoni, F.; Crisanti, A. Antigen microarrays for serodiagnosis of infectious diseases. Clin. Chem. 2002, 48, 121–130. [Google Scholar] [CrossRef]
- Cheng, M.P.; Yansouni, C.P.; Basta, N.E.; Desjardins, M.; Kanjilal, S.; Paquette, K.; Caya, C.; Semret, M.; Quach, C.; Libman, M.; et al. Serodiagnostics for severe acute respiratory syndrome-related Coronavirus 2: A narrative review. Ann. Intern. Med. 2020, 173, 450–460. [Google Scholar] [CrossRef]
- Pecora, N.D.; Zand, M.S. Measuring the serologic response to severe acute respiratory syndrome coronavirus 2: Methods and meaning. Clin. Lab. Med. 2020, 40, 603–614. [Google Scholar] [CrossRef]
- Bastos, M.L.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Lehman, D.C. Immunodiagnosis of Infectious Diseases. In Textbook of Diagnostic Microbiology, 4th ed.; Mahon, C., Lehman, D., Eds.; Saunders: Yonkers, NY, USA, 2014; p. 200. [Google Scholar]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care testing for infectious diseases: Past, present, and future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Hanafiah, K.M.; Garcia, M.; Anderson, D. Point-of-care testing and the control of infectious diseases. Biomark. Med. 2013, 7, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Katoba, J.; Kuupiel, D.; Mashamba-Thompson, T.P. Toward improving accessibility of point-of-care diagnostic services for maternal and child health in low- and middle-income countries. Point Care 2019, 18, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Riccò, M.; Ferraro, P.; Gualerzi, G.; Ranzieri, S.; Henry, B.M.; Said, Y.B.; Pyatigorskaya, N.; Nevolina, E.; Wu, J.; Bragazzi, N.L.; et al. Point-of-Care diagnostic tests for detecting SARS-CoV-2 antibodies: A systematic review and meta-analysis of real-world data. J. Clin. Med. 2020, 9, 1515. [Google Scholar] [CrossRef] [PubMed]
- Reali, S.; Najib, E.Y.; Balázs, K.E.T.; Tan, A.C.H.; Váradi, L.; Hibbs, D.E.; Groundwater, P.W. Novel diagnostics for point-of-care bacterial detection and identification. RSC Adv. 2019, 9, 21486–21497. [Google Scholar] [CrossRef]
- Heidt, B.; Siqueira, W.F. Point of care diagnostics in resource-limited settings: A review of the present and future of PoC in its most needed environment. Biosensors 2020, 10, 133. [Google Scholar] [CrossRef]
- Quesada-González, D.; Merkoçi, A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015, 73, 47–63. [Google Scholar] [CrossRef]
- Huang, X.; Aguilar, Z.P.; Xu, H.; Lai, W.; Xiong, Y. Membrane-based lateral flow immunochromatographic strip with nanoparticles as reporters for detection: A review. Biosens. Bioelectron. 2016, 75, 166–180. [Google Scholar] [CrossRef]
- Goryacheva, I.Y.; Lenain, P.; De Saeger, S. Nanosized labels for rapid immunotests. Trends Analyt. Chem. 2013, 46, 30–43. [Google Scholar] [CrossRef]
- Van Amerongen, A.; Veen, J.; Arends, H.A.; Koets, M. Lateral Flow Immunoassays. In Handbook of Immunoassay Technologies, 1st ed.; Vashist, S.K., Luong, J.H.T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 157–182. [Google Scholar]
- Soh, J.H.; Chan, H.-M.; Ying, J.Y. Strategies for developing sensitive and specific nanoparticle-based lateral flow assays as point-of-care diagnostic device. Nano Today 2020, 30, 100831. [Google Scholar] [CrossRef]
- Abdoel, T.; Dias, I.T.; Cardoso, R.; Smits, H.L. Simple and rapid field tests for brucellosis in livestock. Vet. Microbiol. 2008, 130, 312–319. [Google Scholar] [CrossRef]
- Sotnikov, D.V.; Byzova, N.A.; Zherdev, A.V.; Eskendirova, S.Z.; Baltin, K.K.; Mukanov, K.K.; Ramankulov, E.M.; Sadykhov, E.G.; Dzantiev, B.B. Express immunochromatographic detection of antibodies against Brucella abortus in cattle sera based on quantitative photometric registration and modulated cut-off level. J. Immunoass. Immunochem. 2015, 36, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Tsukimura, T.; Togawa, T.; Ohashi, T.; Kobayashi, M.; Takayama, K.; Kobayashi, Y.; Abiko, H.; Satou, M.; Nakahata, T.; et al. Rapid immunochromatographic detection of serum Anti-α-Galactosidase A antibodies in fabry patients after enzyme replacement therapy. PLoS ONE 2015, 10, e0128351. [Google Scholar] [CrossRef] [PubMed]
- Janwan, P.; Intapan, P.M.; Yamasaki, H.; Rodpai, R.; Laummaunwai, P.; Thanchomnang, T.; Sanpool, O.; Kobayashi, K.; Takayama, K.; Kobayashi, Y.; et al. Development and usefulness of an immunochromatographic device to detect antibodies for rapid diagnosis of human gnathostomiasis. Parasit. Vectors 2016, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Struyf, F.; Lemmens, A.; Valadas, E.; Verhaegen, J.; Van Ranst, M. Usefulness of immunochromatographic detection of antibodies to mycobacterium tuberculosis as an adjunct to auramine staining for rapid diagnosis of tuberculosis in a low-prevalence setting. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 740–742. [Google Scholar] [CrossRef]
- Schrier, W.H.; Schoengold, R.J.; Baker, J.T.; Norell, J.L.; Jaseph, C.L.; Okin, Y.; Doe, J.Y.; Chandler, H. Development of FlexSure® HP—An immunochromatographic method to detect antibodies against Helicobacter pylori. Clin. Chem. 1998, 44, 293–298. [Google Scholar] [CrossRef]
- Sotnikov, D.V.; Berlina, A.N.; Zherdev, A.V.; Eskendirova, S.Z.; Mukanov, K.K.; Ramankulov, Y.M.; Eskendirova, S.Z.; Mukantayev, K.N.; Dzantiev, B.B. Comparison of three schemes of quantum dots-based immunochromatography for serodiagnosis of brucellosis in cattle. J. Eng. Stud. Res. 2019, 14, 3711–3718. [Google Scholar] [CrossRef][Green Version]
- Kemper, M.J.; Altrogge, H.; Ganschow, R.; Müller-Wiefel, D.E. Serum levels of immunoglobulins and IgG subclasses in steroid sensitive nephrotic syndrome. Pediatr. Nephrol. 2002, 17, 413–417. [Google Scholar] [CrossRef]
- Rajerison, M.; Dartevelle, S.; Ralafiarisoa, L.A.; Bitam, I.; Tuyet, D.T.N.; Andrianaivoarimanana, V.; Nato, F.; Rahalison, L. Development and evaluation of two simple, rapid immunochromatographic tests for the detection of Yersinia pestis antibodies in humans and reservoirs. PLoS Negl. Trop. Dis. 2009, 3, e421. [Google Scholar] [CrossRef]
- Karakus, C.; Salih, B.A. Comparison of the lateral flow immunoassays (LFIA) for the diagnosis of Helicobacter pylori infection. J. Immunol. Methods 2013, 396, 8–14. [Google Scholar] [CrossRef]
- Martínez-Sernández, V.; Muino, L.; Perteguer, M.J.; Garate, T.; Mezo, M.; Gonzalez-Warleta, M.; Muro, A.; Correia da Costa, J.M.; Romaris, F.; Ubeira, F.M. Development and evaluation of a new lateral flow immunoassay for serodiagnosis of human fasciolosis. PLoS Negl. Trop. Dis. 2011, 5, e1376. [Google Scholar] [CrossRef]
- Ben-Selma, W.; Harizi, H.; Boukadida, J. Immunochromatographic IgG/IgM test for rapid diagnosis of active tuberculosis. Clin. Vaccine Immunol. 2011, 18, 2090–2094. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Theoretical and experimental comparison of different formats of immunochromatographic serodiagnostics. Sensors 2017, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Zherdev, A.V.; Avdienko, V.G.; Dzantiev, B.B. Immunochromatographic assay for serodiagnosis of tuberculosis using an antigen-colloidal gold conjugate. Appl. Biochem. Microbiol. 2015, 51, 834–839. [Google Scholar] [CrossRef]
- Wu, H.S.; Chiu, S.C.; Tseng, T.C.; Lin, S.F.; Lin, J.H.; Hsu, Y.H.; Wang, M.C.; Lin, T.L.; Yang, W.Z.; Ferng, T.L.; et al. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg. Infect. Dis. 2004, 10, 304–310. [Google Scholar] [CrossRef]
- Sato, N.S.; Melo, C.S.D.; Zerbini, L.C.; Silveira, E.P.; Fagundes, L.J.; Ueda, M. Assessment of the rapid test based on an immunochromatography technique for detecting anti-Treponema pallidum antibodies. Rev. Inst. Med. Trop. Sao Paulo 2003, 45, 319–322. [Google Scholar] [CrossRef][Green Version]
- Vrublevskaya, V.V.; Afanasyev, V.N.; Grinevich, A.A.; Skarga, Y.Y.; Gladyshev, P.P.; Ibragimova, S.A.; Krylsky, D.V.; Dezhurov, S.V.; Morenkov, O.S. A sensitive and specific lateral flow assay for rapid detection of antibodies against glycoprotein B of Aujeszky’s disease virus. J. Virol. Methods 2017, 249, 175–180. [Google Scholar] [CrossRef]
- Berli, C.L.A.; Kler, P.A. A quantitative model for lateral flow assays. Microfluid. Nanofluid. 2016, 20, 104. [Google Scholar] [CrossRef]
- Gasperino, D.; Baughman, T.; Hsieh, H.V.; Bell, D.; Weigl, B.H. Improving lateral flow assay performance using computational modeling. Annu. Rev. Anal. Chem. 2018, 11, 219–244. [Google Scholar] [CrossRef]
- Schaumburg, F.; Kler, P.A.; Berli, C.L.A. Numerical prototyping of lateral flow biosensors. Sens. Act. B Chem. 2018, 259, 1099–1107. [Google Scholar] [CrossRef]
- Gasperino, D.J.; Leon, D.; Lutz, B.; Cate, D.M.; Nichols, K.P.; Bell, D.; Weigl, B.H. Threshold-based quantification in a multiline lateral flow assay via computationally designed capture efficiency. Anal. Chem. 2018, 90, 6643–6650. [Google Scholar] [CrossRef]
- Liu, Z.; Qu, Z.; Tang, R.; He, X.; Yang, H.; Bai, D.; Xu, F. An improved detection limit and working range of lateral flow assays based on a mathematical model. Analyst 2018, 143, 2775–2783. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Mathematical modeling of bioassays. Biochemistry (Moscow) 2017, 82, 1744–1766. [Google Scholar] [CrossRef] [PubMed]
- Hoops, S.; Sahle, S.; Gauges, R.; Lee, C.; Pahle, J.; Simus, N.; Singhal, M.; Xu, L.; Mendes, P.; Kummer, U. COPASI—A complex pathway simulator. Bioinformatics 2006, 22, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995, 360, 111–114. [Google Scholar] [CrossRef]
- Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Development and application of a label-free fluorescence method for determining the composition of gold nanoparticle-protein conjugates. Int. J. Mol. Sci. 2014, 16, 907–923. [Google Scholar] [CrossRef] [PubMed]







| Major Axis Length (nm) | Minor Axis Length (nm) | Axis Ratio | |
|---|---|---|---|
| Average diameter (nm) | 22.8 | 18.5 | 0.845 |
| Standard deviation (nm) | 4.1 | 2.9 | 0.095 |
| ka (1/Ms) | kd (1/s) | Ka (1/M) | Kd (M) |
|---|---|---|---|
| 1.08 × 105 | 2.54 × 10−3 | 4.27 × 107 | 2.34 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Lateral Flow Serodiagnosis in the Double-Antigen Sandwich Format: Theoretical Consideration and Confirmation of Advantages. Sensors 2021, 21, 39. https://doi.org/10.3390/s21010039
Sotnikov DV, Zherdev AV, Dzantiev BB. Lateral Flow Serodiagnosis in the Double-Antigen Sandwich Format: Theoretical Consideration and Confirmation of Advantages. Sensors. 2021; 21(1):39. https://doi.org/10.3390/s21010039
Chicago/Turabian StyleSotnikov, Dmitriy V., Anatoly V. Zherdev, and Boris B. Dzantiev. 2021. "Lateral Flow Serodiagnosis in the Double-Antigen Sandwich Format: Theoretical Consideration and Confirmation of Advantages" Sensors 21, no. 1: 39. https://doi.org/10.3390/s21010039
APA StyleSotnikov, D. V., Zherdev, A. V., & Dzantiev, B. B. (2021). Lateral Flow Serodiagnosis in the Double-Antigen Sandwich Format: Theoretical Consideration and Confirmation of Advantages. Sensors, 21(1), 39. https://doi.org/10.3390/s21010039







