Mask R-CNN and OBIA Fusion Improves the Segmentation of Scattered Vegetation in Very High-Resolution Optical Sensors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Dataset
- A 0.5 × 0.5 m spatial resolution RGB image obtained from Google Earth [52].
- A 0.1 × 0.1 m spatial resolution image acquired using an RGB camera sensor of 50 megapixels (Hasselblad H4D) equipped with a 50 mm lens and charge-coupled device (CCD) sensor of 8176 pixels × 6132 pixels mounted on a helicopter with a flight height of 550 m.
- A 0.03 × 0.03 m spatial resolution image acquired using a 4K pixels resolution RGB camera sensor on a professional UAV Phantom 4 UAV (DJI, Shenzhen, China) and with a flight height of 40 m.
2.3. OBIA
2.4. Mask R-CNN
2.5. Segmentation Accuracy Assessment
3. Experiments
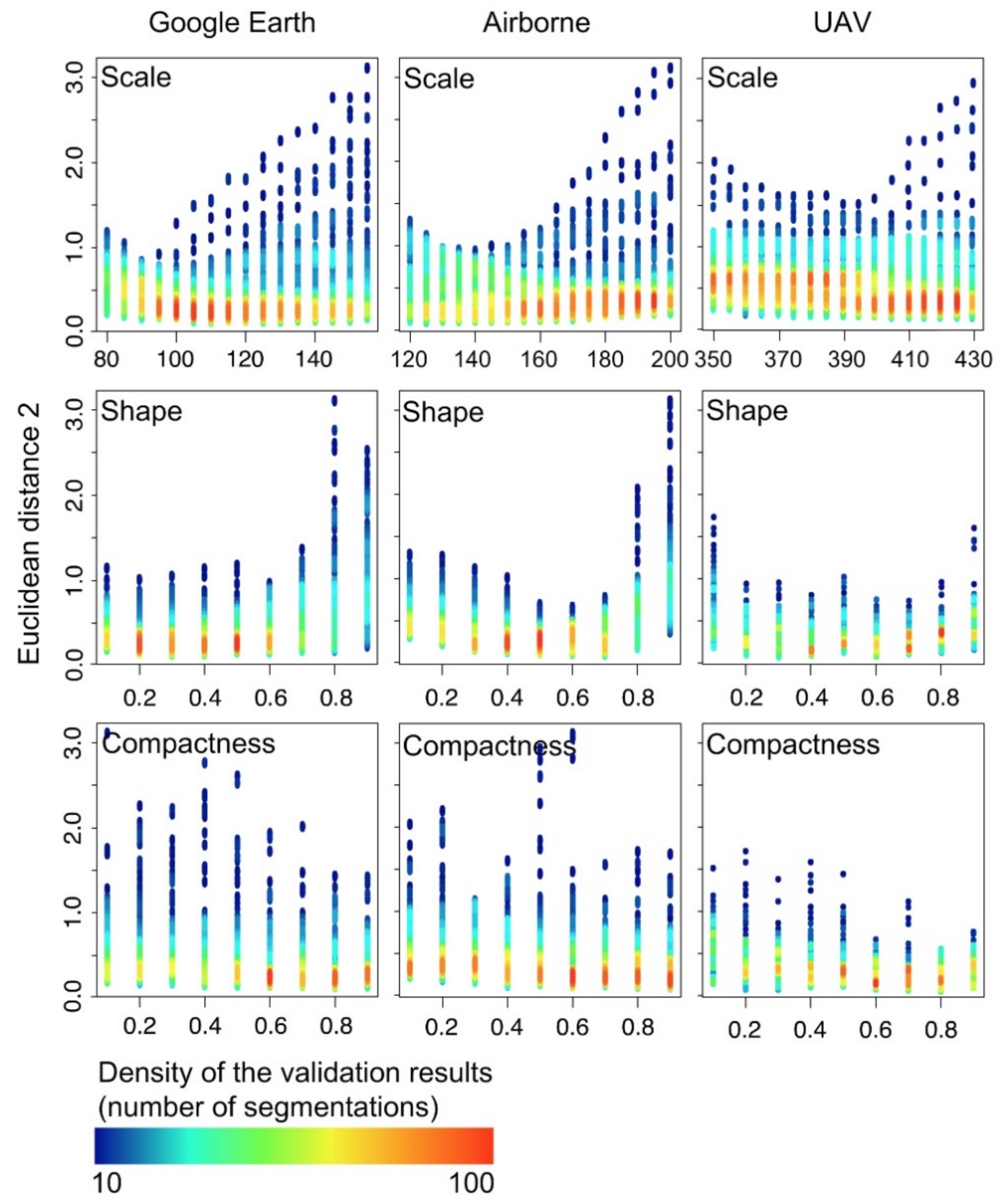
3.1. OBIA Experiments
- (i)
- A ruleset called Segmentation Parameters Range (SPR) in eCognition v8.9 (Definiens, Munich, Germany) with the “multi-resolution” algorithm that segmented the images of Plot 1 by systematically increasing the Scale parameter in steps of 5 and the Shape and Compactness parameters in steps of 0.1. The Scale parameter ranged from 80 to 430, and the Shape and the Compactness from 0.1 to 0.9. We generated a total of 9234 results with possible segmentations of Z. lotus shrubs. The Scale parameter ranges were evaluated considering the minimum cover size (12 m2) and maximum cover size (311 m2) of the shrubs measured in the plot and the pixel size.
- (ii)
- We also performed the semi-automatic method Estimation of Scale Parameter v.2 (ESP2; [70]) to select the best scale parameter. This tool performs semi-automatic segmentation of multiband images within a range of increasing Scale values (Levels), while the user previously defines the values of the Compactness and Shape parameters. Three options available in the ESP2 tool were tested: a) the hierarchical analysis Top-down (HT), starting from the highest level and segmenting these objects for lower levels; b) the hierarchical analysis Bottom-up (HB), which starts from the lower level and combines objects to get larger levels; and c) analysis without hierarchy (NH), where each scale parameter is generated independently, based only on the level of the pixel [64].
3.2. Mask R-CNN Experiments
- Data augmentation aims to artificially increase the size of the dataset by slightly modifying the original images. We applied the filters of vertical and horizontal flip; Scale decrease and increase in the horizontal and vertical axis between 0.8 to 1.2; Rotation of 0 to 365 degrees; Shearing factor between −8 to 8; Contrast normalization with values of 0.75 and 1.5 per channel; Emboss with alpha 0, 0.1; Strength with 0 to 2.0; Multiply 0.5 and 1.5, per channel to change the brightness of the image (50–150% of the original value).
- Transfer-learning consists in using knowledge learnt from one problem to another related one [73], and we used it to improve the neural network. Since the first layers of a neural network extract low-level characteristics, such as colour and edges, they do not change significantly and can be used for other visual recognition works. As our new dataset was small, we applied fine adjustment to the last part of the network by updating the penultimate weights, so that the model was not overfitting, as mainly occurs between the first layers of the network. We specifically used transfer-learning on ResNet 101 [69] and used Region-based CNN with the pre-trained weights of the same architectures on COCO dataset (around 1.28 million images over 1000 generic object classes) [74].
- (A)
- 40 epochs with transfer-learning in heads,
- (B)
- 80 epochs with 4 fist layers transfer-learning,
- (C)
- 160 epochs with all layers transfer-learning.
- (1.1)
- Trained with UAV images.
- (1.2)
- Trained with UAV images and data-augmentation.
- (2.1)
- Trained with airborne images.
- (2.2)
- Trained with airborne images and with data-augmentation.
- (3.1)
- Trained with Google Earth images.
- (3.2)
- Trained with Google Earth images and data-augmentation.
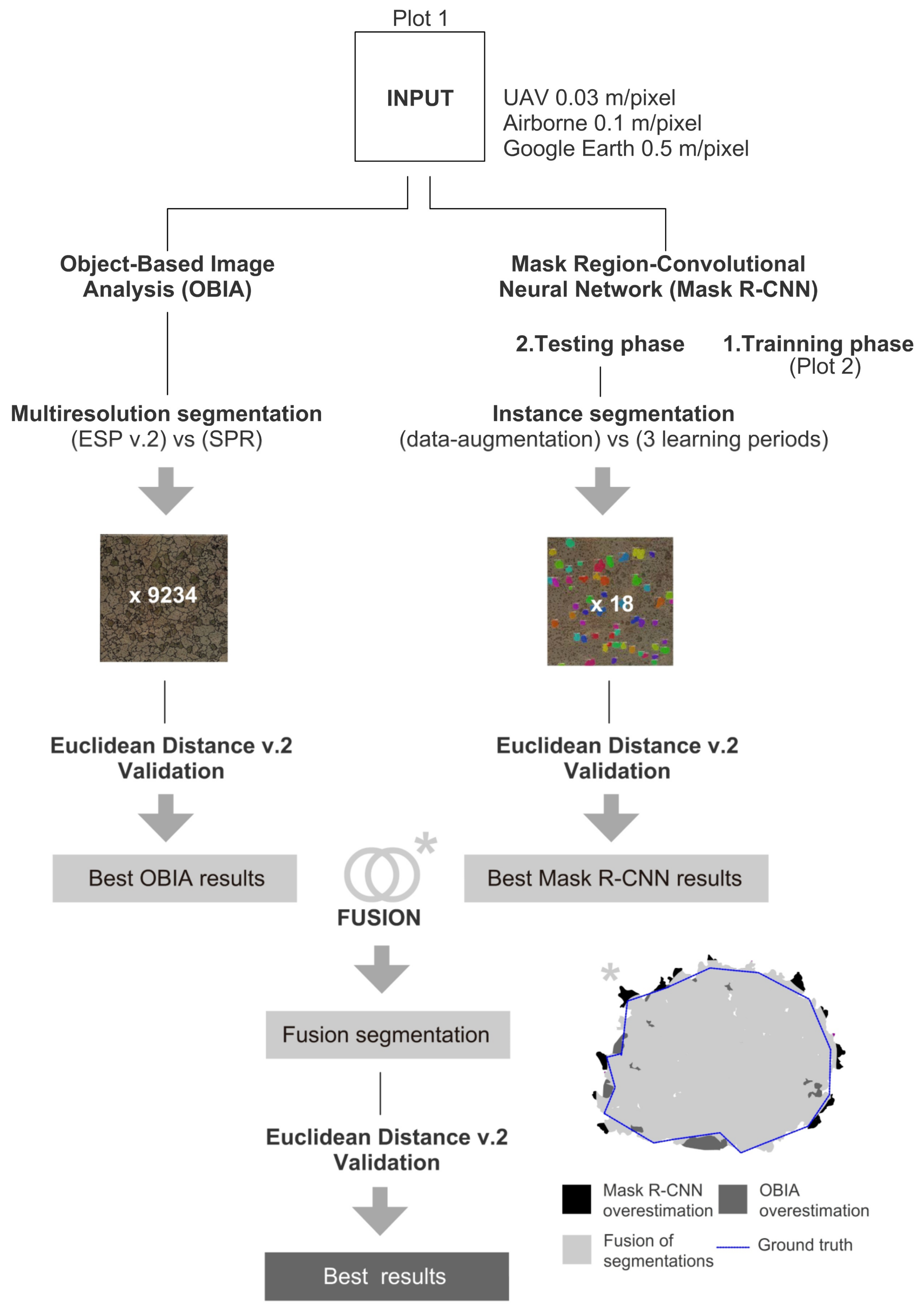
3.3. Fusion of OBIA and Mask R-CNN
4. Results and Discussion
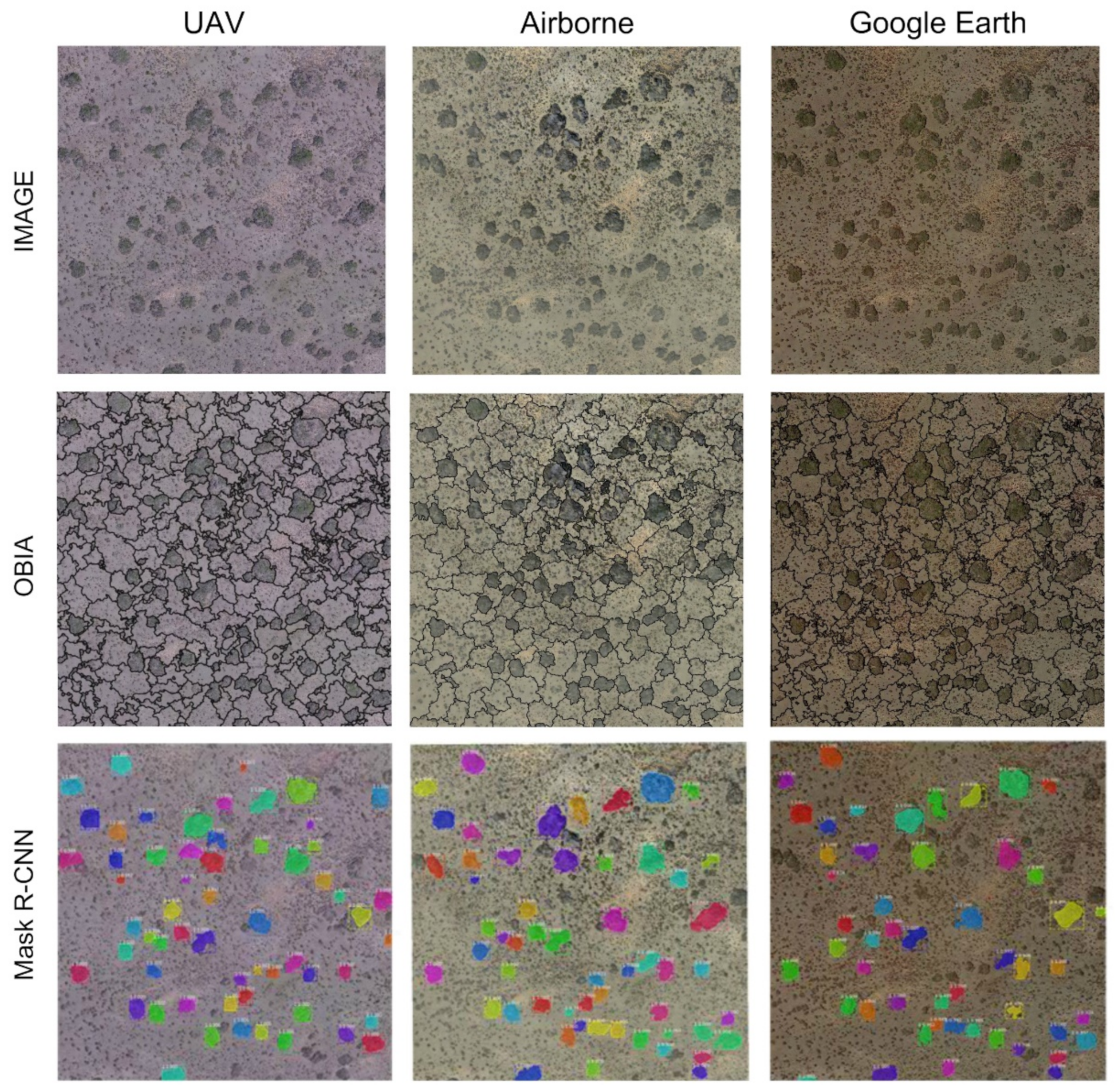
4.1. OBIA Segmentation
4.2. Mask R-CNN Segmentation
4.2.1. Detection of Scattered Shrubs
4.2.2. Segmentation Accuracy for Detected Shrubs
4.3. Fusion of OBIA and Mask R-CNN
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Description |
| CCD | Charge-Coupled Device |
| ED2 | Euclidean Distance v.2 |
| ESP2 | Estimation Scale Parameter v.2 |
| ETRS | European Terrestrial Reference System |
| HB | Bottom-up Hierarchy |
| HT | Top-down Hierarchy |
| JSON | JavaScript Object Notation |
| NH | Non-Hierarchical |
| NSR | Number-of-Segmentation Ratio |
| OBIA | Object-based Image Analysis |
| R-CNN | Region—Convolutional Neural Networks |
| RGB | Red Green Blue |
| SPR | Segmentation Parameters Range |
| UAV | Unmanned aerial vehicle |
| UTM | Universal Transverse Mercator |
| VGG | Visual Geometry Group |
References
- Koutroulis, A.G. Dryland changes under different levels of global warming. Sci. Total Environ. 2019, 655, 482–511. [Google Scholar] [CrossRef] [PubMed]
- Puigdefábregas, J. The role of vegetation patterns in structuring runoff and sediment fluxes in drylands. Earth Surf. Process. Landf. 2005, 30, 133–147. [Google Scholar] [CrossRef]
- Ravi, S.; Breshears, D.D.; Huxman, T.E.; D’Odorico, P. Land degradation in drylands: Interactions among hydrologic–aeolian erosion and vegetation dynamics. Geomorphology 2010, 116, 236–245. [Google Scholar] [CrossRef]
- Gao, Z.; Sun, B.; Li, Z.; Del Barrio, G.; Li, X. Desertification monitoring and assessment: A new remote sensing method. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 June 2016. [Google Scholar]
- Guirado, E.; Blanco-Sacristán, J.; Rigol-Sánchez, J.; Alcaraz-Segura, D.; Cabello, J. A Multi-Temporal Object-Based Image Analysis to Detect Long-Lived Shrub Cover Changes in Drylands. Remote Sens. 2019, 11, 2649. [Google Scholar] [CrossRef]
- Guirado, E.; Alcaraz-Segura, D.; Rigol-Sánchez, J.P.; Gisbert, J.; Martínez-Moreno, F.J.; Galindo-Zaldívar, J.; González-Castillo, L.; Cabello, J. Remote-sensing-derived fractures and shrub patterns to identify groundwater dependence. Ecohydrology 2018, 11, e1933. [Google Scholar] [CrossRef]
- Guirado, E.; Tabik, S.; Alcaraz-Segura, D.; Cabello, J.; Herrera, F. Deep-learning Versus OBIA for Scattered Shrub Detection with Google Earth Imagery: Ziziphus lotus as Case Study. Remote Sens. 2017, 9, 1220. [Google Scholar] [CrossRef]
- Kéfi, S.; Guttal, V.; Brock, W.A.; Carpenter, S.R.; Ellison, A.M.; Livina, V.N.; Seekell, D.A.; Scheffer, M.; van Nes, E.H.; Dakos, V. Early warning signals of ecological transitions: Methods for spatial patterns. PLoS ONE 2014, 9, e92097. [Google Scholar] [CrossRef]
- Cunliffe, A.M.; Brazier, R.E.; Anderson, K. Ultra-fine grain landscape-scale quantification of dryland vegetation structure with drone-acquired structure-from-motion photogrammetry. Remote Sens. Environ. 2016, 183, 129–143. [Google Scholar] [CrossRef]
- Brandt, M.; Hiernaux, P.; Rasmussen, K.; Mbow, C.; Kergoat, L.; Tagesson, T.; Ibrahim, Y.Z.; Wélé, A.; Tucker, C.J.; Fensholt, R. Assessing woody vegetation trends in Sahelian drylands using MODIS based seasonal metrics. Remote Sens. Environ. 2016, 183, 215–225. [Google Scholar] [CrossRef]
- Berdugo, M.; Kéfi, S.; Soliveres, S.; Maestre, F.T. Author Correction: Plant spatial patterns identify alternative ecosystem multifunctionality states in global drylands. Nat. Ecol. Evol. 2018, 2, 574–576. [Google Scholar] [CrossRef]
- Mohammadi, A.; Costelloe, J.F.; Ryu, D. Application of time series of remotely sensed normalized difference water, vegetation and moisture indices in characterizing flood dynamics of large-scale arid zone floodplains. Remote Sens. Environ. 2017, 190, 70–82. [Google Scholar] [CrossRef]
- Tian, F.; Brandt, M.; Liu, Y.Y.; Verger, A.; Tagesson, T.; Diouf, A.A.; Rasmussen, K.; Mbow, C.; Wang, Y.; Fensholt, R. Remote sensing of vegetation dynamics in drylands: Evaluating vegetation optical depth (VOD) using AVHRR NDVI and in situ green biomass data over West African Sahel. Remote Sens. Environ. 2016, 177, 265–276. [Google Scholar] [CrossRef]
- Taddeo, S.; Dronova, I.; Depsky, N. Spectral vegetation indices of wetland greenness: Responses to vegetation structure, composition, and spatial distribution. Remote Sens. Environ. 2019, 234, 111467. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Zhang, J.; Jia, L. A comparison of pixel-based and object-based land cover classification methods in an arid/semi-arid environment of Northwestern China. In Proceedings of the 2014 Third International Workshop on Earth Observation and Remote Sensing Applications (EORSA), Changsha, China, 11–14 June 2014. [Google Scholar]
- Amitrano, D.; Guida, R.; Iervolino, P. High Level Semantic Land Cover Classification of Multitemporal Sar Images Using Synergic Pixel-Based and Object-Based Methods. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019. [Google Scholar]
- Li, S.; Yan, M.; Xu, J. Garbage object recognition and classification based on Mask Scoring RCNN. In Proceedings of the 2020 International Conference on Culture-oriented Science & Technology (ICCST), Beijing, China, 28–31 October 2020. [Google Scholar]
- Zhang, Q.; Chang, X.; Bian, S.B. Vehicle-Damage-Detection Segmentation Algorithm Based on Improved Mask RCNN. IEEE Access 2020, 8, 6997–7004. [Google Scholar] [CrossRef]
- Ghassemian, H. A review of remote sensing image fusion methods. Inf. Fusion 2016, 32, 75–89. [Google Scholar] [CrossRef]
- Belgiu, M.; Stein, A. Spatiotemporal Image Fusion in Remote Sensing. Remote Sens. 2019, 11, 818. [Google Scholar] [CrossRef]
- Moreno-Martínez, Á.; Izquierdo-Verdiguier, E.; Maneta, M.P.; Camps-Valls, G.; Robinson, N.; Muñoz-Marí, J.; Sedano, F.; Clinton, N.; Running, S.W. Multispectral high resolution sensor fusion for smoothing and gap-filling in the cloud. Remote Sens. Environ. 2020, 247, 111901. [Google Scholar] [CrossRef]
- Alphan, H.; Çelik, N. Monitoring changes in landscape pattern: Use of Ikonos and Quickbird images. Environ. Monit. Assess. 2016, 188, 81. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Granger, J.E.; Mohammadimanesh, F.; Warren, S.; Puestow, T.; Salehi, B.; Brisco, B. Smart solutions for smart cities: Urban wetland mapping using very-high resolution satellite imagery and airborne LiDAR data in the City of St. John’s, NL, Canada. J. Environ. Manag. 2020, 111676, In press. [Google Scholar] [CrossRef]
- Mahdavi Saeidi, A.; Babaie Kafaky, S.; Mataji, A. Detecting the development stages of natural forests in northern Iran with different algorithms and high-resolution data from GeoEye-1. Environ. Monit. Assess. 2020, 192, 653. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, D.; Bennie, J.; Anderson, K. Monitoring spring phenology of individual tree crowns using drone—Acquired NDVI data. Remote Sens. Ecol. Conserv. 2020. [Google Scholar] [CrossRef]
- Hu, Q.; Wu, W.; Xia, T.; Yu, Q.; Yang, P.; Li, Z.; Song, Q. Exploring the Use of Google Earth Imagery and Object-Based Methods in Land Use/Cover Mapping. Remote Sens. 2013, 5, 6026–6042. [Google Scholar] [CrossRef]
- Venkatappa, M.; Sasaki, N.; Shrestha, R.P.; Tripathi, N.K.; Ma, H.O. Determination of Vegetation Thresholds for Assessing Land Use and Land Use Changes in Cambodia using the Google Earth Engine Cloud-Computing Platform. Remote Sens. 2019, 11, 1514. [Google Scholar] [CrossRef]
- Sowmya, D.R.; Deepa Shenoy, P.; Venugopal, K.R. Feature-based Land Use/Land Cover Classification of Google Earth Imagery. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019. [Google Scholar]
- Li, W.; Buitenwerf, R.; Munk, M.; Bøcher, P.K.; Svenning, J.-C. Deep-learning based high-resolution mapping shows woody vegetation densification in greater Maasai Mara ecosystem. Remote Sens. Environ. 2020, 247, 111953. [Google Scholar] [CrossRef]
- Uyeda, K.A.; Stow, D.A.; Richart, C.H. Assessment of volunteered geographic information for vegetation mapping. Environ. Monit. Assess. 2020, 192, 1–14. [Google Scholar] [CrossRef]
- Ancin--Murguzur, F.J.; Munoz, L.; Monz, C.; Hausner, V.H. Drones as a tool to monitor human impacts and vegetation changes in parks and protected areas. Remote Sens. Ecol. Conserv. 2020, 6, 105–113. [Google Scholar] [CrossRef]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-based Detailed Vegetation Classification with Airborne High Spatial Resolution Remote Sensing Imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Herrick, J.E.; Rango, A.; Winters, C. Acquisition, Orthorectification, and Object-based Classification of Unmanned Aerial Vehicle (UAV) Imagery for Rangeland Monitoring. Photogramm. Eng. Remote Sens. 2010, 76, 661–672. [Google Scholar] [CrossRef]
- Whiteside, T.G.; Boggs, G.S.; Maier, S.W. Comparing object-based and pixel-based classifications for mapping savannas. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 884–893. [Google Scholar] [CrossRef]
- Hossain, M.D.; Chen, D. Segmentation for Object-Based Image Analysis (OBIA): A review of algorithms and challenges from remote sensing perspective. ISPRS J. Photogramm. Remote Sens. 2019, 150, 115–134. [Google Scholar] [CrossRef]
- Arvor, D.; Durieux, L.; Andrés, S.; Laporte, M.-A. Advances in Geographic Object-Based Image Analysis with ontologies: A review of main contributions and limitations from a remote sensing perspective. ISPRS J. Photogramm. Remote Sens. 2013, 82, 125–137. [Google Scholar] [CrossRef]
- Johnson, B.A.; Ma, L. Image Segmentation and Object-Based Image Analysis for Environmental Monitoring: Recent Areas of Interest, Researchers’ Views on the Future Priorities. Remote Sens. 2020, 12, 1772. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef]
- Singh, R.; Rani, R. Semantic Segmentation using Deep Convolutional Neural Network: A Review. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Aguilar, M.A.; Aguilar, F.J.; García Lorca, A.; Guirado, E.; Betlej, M.; Cichon, P.; Nemmaoui, A.; Vallario, A.; Parente, C. Assessment of multiresolution segmentation for extracting greenhouses from worldview-2 imagery. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B7, 145–152. [Google Scholar] [CrossRef]
- Brandt, M.; Tucker, C.J.; Kariryaa, A.; Rasmussen, K.; Abel, C.; Small, J.; Chave, J.; Rasmussen, L.V.; Hiernaux, P.; Diouf, A.A.; et al. An unexpectedly large count of trees in the West African Sahara and Sahel. Nature 2020, 587, 78–82. [Google Scholar] [CrossRef]
- Guirado, E.; Tabik, S.; Rivas, M.L.; Alcaraz-Segura, D.; Herrera, F. Whale counting in satellite and aerial images with deep learning. Sci. Rep. 2019, 9, 14259. [Google Scholar] [CrossRef]
- Guirado, E.; Alcaraz-Segura, D.; Cabello, J.; Puertas-Ruíz, S.; Herrera, F.; Tabik, S. Tree Cover Estimation in Global Drylands from Space Using Deep Learning. Remote Sens. 2020, 12, 343. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, R.; Wang, S.; Wang, F. Feature Selection Method Based on High-Resolution Remote Sensing Images and the Effect of Sensitive Features on Classification Accuracy. Sensors 2018, 18, 2013. [Google Scholar] [CrossRef]
- Foody, G.; Pal, M.; Rocchini, D.; Garzon-Lopez, C.; Bastin, L. The Sensitivity of Mapping Methods to Reference Data Quality: Training Supervised Image Classifications with Imperfect Reference Data. ISPRS Int. J. Geo-Inf. 2016, 5, 199. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Torres-García, M.; Salinas-Bonillo, M.J.; Gázquez-Sánchez, F.; Fernández-Cortés, A.; Querejeta, J.L.; Cabello, J. Squandering Water in Drylands: The Water Use Strategy of the Phreatophyte Ziziphus lotus (L.) Lam in a Groundwater Dependent Ecosystem. Am. J. Bot. 2021, 108, 2, in press. [Google Scholar]
- Tirado, R.; Pugnaire, F.I. Shrub spatial aggregation and consequences for reproductive success. Oecologia 2003, 136, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Tengberg, A.; Chen, D. A comparative analysis of nebkhas in central Tunisia and northern Burkina Faso. Geomorphology 1998, 22, 181–192. [Google Scholar] [CrossRef]
- Fisher, G.B.; Burch Fisher, G.; Amos, C.B.; Bookhagen, B.; Burbank, D.W.; Godard, V. Channel widths, landslides, faults, and beyond: The new world order of high-spatial resolution Google Earth imagery in the study of earth surface processes. Google Earth Virtual Vis. Geosci. Educ. Res. 2012, 492, 1–22. [Google Scholar] [CrossRef][Green Version]
- Li, M.; Ma, L.; Blaschke, T.; Cheng, L.; Tiede, D. A systematic comparison of different object-based classification techniques using high spatial resolution imagery in agricultural environments. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 87–98. [Google Scholar] [CrossRef]
- Yu, W.; Zhou, W.; Qian, Y.; Yan, J. A new approach for land cover classification and change analysis: Integrating backdating and an object-based method. Remote Sens. Environ. 2016, 177, 37–47. [Google Scholar] [CrossRef]
- Yan, J.; Lin, L.; Zhou, W.; Ma, K.; Pickett, S.T.A. A novel approach for quantifying particulate matter distribution on leaf surface by combining SEM and object-based image analysis. Remote Sens. Environ. 2016, 173, 156–161. [Google Scholar] [CrossRef]
- Colkesen, I.; Kavzoglu, T. Selection of Optimal Object Features in Object-Based Image Analysis Using Filter-Based Algorithms. J. Indian Soc. Remote Sens. 2018, 46, 1233–1242. [Google Scholar] [CrossRef]
- Lefèvre, S.; Sheeren, D.; Tasar, O. A Generic Framework for Combining Multiple Segmentations in Geographic Object-Based Image Analysis. ISPRS Int. J. Geo-Inf. 2019, 8, 70. [Google Scholar] [CrossRef]
- Hurskainen, P.; Adhikari, H.; Siljander, M.; Pellikka, P.K.E.; Hemp, A. Auxiliary datasets improve accuracy of object-based land use/land cover classification in heterogeneous savanna landscapes. Remote Sens. Environ. 2019, 233, 111354. [Google Scholar] [CrossRef]
- Gonçalves, J.; Pôças, I.; Marcos, B.; Mücher, C.A.; Honrado, J.P. SegOptim—A new R package for optimizing object-based image analyses of high-spatial resolution remotely-sensed data. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 218–230. [Google Scholar] [CrossRef]
- Ma, L.; Cheng, L.; Li, M.; Liu, Y.; Ma, X. Training set size, scale, and features in Geographic Object-Based Image Analysis of very high resolution unmanned aerial vehicle imagery. ISPRS J. Photogramm. Remote Sens. 2015, 102, 14–27. [Google Scholar] [CrossRef]
- Zhang, X.; Du, S.; Ming, D. Segmentation Scale Selection in Geographic Object-Based Image Analysis. High Spat. Resolut. Remote Sens. 2018, 201–228. [Google Scholar]
- Yang, L.; Mansaray, L.; Huang, J.; Wang, L. Optimal Segmentation Scale Parameter, Feature Subset and Classification Algorithm for Geographic Object-Based Crop Recognition Using Multisource Satellite Imagery. Remote Sens. 2019, 11, 514. [Google Scholar] [CrossRef]
- Mao, C.; Meng, W.; Shi, C.; Wu, C.; Zhang, J. A Crop Disease Image Recognition Algorithm Based on Feature Extraction and Image Segmentation. Traitement Signal 2020, 37, 341–346. [Google Scholar] [CrossRef]
- Drăguţ, L.; Csillik, O.; Eisank, C.; Tiede, D. Automated parameterisation for multi-scale image segmentation on multiple layers. ISPRS J. Photogramm. Remote Sens. 2014, 88, 119–127. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Peña, J.M. An automatic object-based method for optimal thresholding in UAV images: Application for vegetation detection in herbaceous crops. Comput. Electron. Agric. 2015, 114, 43–52. [Google Scholar] [CrossRef]
- Josselin, D.; Louvet, R. Impact of the Scale on Several Metrics Used in Geographical Object-Based Image Analysis: Does GEOBIA Mitigate the Modifiable Areal Unit Problem (MAUP)? ISPRS Int. J. Geo-Inf. 2019, 8, 156. [Google Scholar] [CrossRef]
- Blaschke, T.; Lang, S.; Hay, G. Object-Based Image Analysis: Spatial Concepts for Knowledge-Driven Remote Sensing Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008; ISBN 9783540770589. [Google Scholar]
- Watanabe, T.; Wolf, D.F. Instance Segmentation as Image Segmentation Annotation. In Proceedings of the 2019 IEEE Intelligent Vehicles Symposium (IV), Paris, France, 9–12 June 2019. [Google Scholar]
- Demir, A.; Yilmaz, F.; Kose, O. Early detection of skin cancer using deep learning architectures: Resnet-101 and inception-v3. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Izmir, Turkey, 3–5 October 2019. [Google Scholar]
- Liu, Y.; Bian, L.; Meng, Y.; Wang, H.; Zhang, S.; Yang, Y.; Shao, X.; Wang, B. Discrepancy measures for selecting optimal combination of parameter values in object-based image analysis. ISPRS J. Photogramm. Remote Sens. 2012, 68, 144–156. [Google Scholar] [CrossRef]
- Nussbaum, S.; Menz, G. eCognition Image Analysis Software. In Object-Based Image Analysis and Treaty Verification; Springer: Berlin/Heidelberg, Germany, 2008; pp. 29–39. [Google Scholar]
- Dutta, A.; Gupta, A.; Zissermann, A. VGG Image Annotator (VIA). Available online: http://www.robots.ox.ac.uk/~vgg/software/via (accessed on 11 December 2020).
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Caesar, H.; Uijlings, J.; Ferrari, V. COCO-Stuff: Thing and Stuff Classes in Context. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Wang, B.; Li, C.; Pavlu, V.; Aslam, J. A Pipeline for Optimizing F1-Measure in Multi-label Text Classification. In Proceedings of the 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, USA, 17–20 December 2018. [Google Scholar]
- Zhan, Q.; Molenaar, M.; Tempfli, K.; Shi, W. Quality assessment for geo--spatial objects derived from remotely sensed data. Int. J. Remote Sens. 2005, 26, 2953–2974. [Google Scholar] [CrossRef]
- Chen, G.; Weng, Q.; Hay, G.J.; He, Y. Geographic object-based image analysis (GEOBIA): Emerging trends and future opportunities. Gisci. Remote Sens. 2018, 55, 159–182. [Google Scholar] [CrossRef]
- Cook-Patton, S.C.; Leavitt, S.M.; Gibbs, D.; Harris, N.L.; Lister, K.; Anderson-Teixeira, K.J.; Briggs, R.D.; Chazdon, R.L.; Crowther, T.W.; Ellis, P.W.; et al. Mapping carbon accumulation potential from global natural forest regrowth. Nature 2020, 585, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Bastin, J.-F.; Berrahmouni, N.; Grainger, A.; Maniatis, D.; Mollicone, D.; Moore, R.; Patriarca, C.; Picard, N.; Sparrow, B.; Abraham, E.M.; et al. The extent of forest in dryland biomes. Science 2017, 356, 635–638. [Google Scholar] [CrossRef]
- Schepaschenko, D.; Fritz, S.; See, L.; Bayas, J.C.L.; Lesiv, M.; Kraxner, F.; Obersteiner, M. Comment on “The extent of forest in dryland biomes”. Science 2017, 358, eaao0166. [Google Scholar] [CrossRef]
- de la Cruz, M.; Quintana-Ascencio, P.F.; Cayuela, L.; Espinosa, C.I.; Escudero, A. Comment on “The extent of forest in dryland biomes”. Science 2017, 358, eaao0369. [Google Scholar] [CrossRef]
- Fernández, N.; Ferrier, S.; Navarro, L.M.; Pereira, H.M. Essential Biodiversity Variables: Integrating In-Situ Observations and Remote Sensing Through Modeling. Remote Sens. Plant Biodivers. 2020, 18, 485–501. [Google Scholar]
- Vihervaara, P.; Auvinen, A.-P.; Mononen, L.; Törmä, M.; Ahlroth, P.; Anttila, S.; Böttcher, K.; Forsius, M.; Heino, J.; Heliölä, J.; et al. How Essential Biodiversity Variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 2017, 10, 43–59. [Google Scholar] [CrossRef]
| Segmentation Parameters | Segmentation Quality | ||||||
|---|---|---|---|---|---|---|---|
| Image Source | Resolution (m/Pixel) | Segmentation Method | Scale | Shape | Compactness | ED2 | Average Time (s) |
| Google Earth | 0.5 | ESP2/HB | 100 | 0.6 | 0.9 | 0.25 | 365 |
| ESP2/HT | 105 | 0.7 | 0.5 | 0.26 | 414 | ||
| ESP2/NH | 105 | 0.5 | 0.1 | 0.28 | 2057 | ||
| SPR | 90 | 0.3 | 0.8 | 0.2 | 18 | ||
| Airborne | 0.1 | ESP2/HB | 170 | 0.5 | 0.9 | 0.14 | 416 |
| ESP2/HT | 160 | 0.5 | 0.9 | 0.15 | 650 | ||
| ESP2/NH | 160 | 0.5 | 0.5 | 0.14 | 3125 | ||
| SPR | 155 | 0.6 | 0.9 | 0.1 | 24 | ||
| UAV | 0.03 | ESP2/HB | 355 | 0.3 | 0.7 | 0.12 | 5537 |
| ESP2/HT | 370 | 0.5 | 0.7 | 0.11 | 8365 | ||
| ESP2/NH | 350 | 0.5 | 0.7 | 0.1 | 40,735 | ||
| SPR | 420 | 0.1 | 0.8 | 0.05 | 298 | ||
| Experiments/Image | TP | FP | FN | Precision | Recall | F1 | |
|---|---|---|---|---|---|---|---|
| 1.1.A | UAV | 55 | 5 | 10 | 0.92 | 0.85 | 0.88 |
| Airborne | 56 | 4 | 9 | 0.93 | 0.86 | 0.90 | |
| GE | 50 | 1 | 15 | 0.98 | 0.77 | 0.86 | |
| 1.1.B | UAV | 59 | 6 | 6 | 0.91 | 0.91 | 0.91 |
| Airborne | 60 | 7 | 5 | 0.90 | 0.92 | 0.91 | |
| GE | 55 | 2 | 10 | 0.96 | 0.85 | 0.90 | |
| 1.1.C | UAV | 55 | 1 | 10 | 0.98 | 0.85 | 0.91 |
| Airborne | 52 | 3 | 13 | 0.94 | 0.80 | 0.87 | |
| GE | 53 | 0 | 12 | 1 | 0.81 | 0.89 | |
| 1.2.A | UAV | 53 | 1 | 12 | 0.98 | 0.82 | 0.89 |
| Airborne | 54 | 1 | 11 | 0.98 | 0.83 | 0.90 | |
| GE | 42 | 3 | 23 | 0.93 | 0.65 | 0.76 | |
| 1.2.B | UAV | 55 | 1 | 10 | 0.98 | 0.85 | 0.91 |
| Airborne | 50 | 2 | 15 | 0.96 | 0.77 | 0.85 | |
| GE | 50 | 2 | 15 | 0.96 | 0.77 | 0.85 | |
| 1.2.C | UAV | 56 | 3 | 8 | 0.95 | 0.87 | 0.91 |
| Airborne | 52 | 3 | 13 | 0.94 | 0.80 | 0.87 | |
| GE | 54 | 1 | 12 | 0.98 | 0.81 | 0.89 | |
| 2.1.A | UAV | 41 | 0 | 24 | 1 | 0.63 | 0.77 |
| Airborne | 38 | 0 | 27 | 1 | 0.58 | 0.74 | |
| GE | 34 | 1 | 31 | 0.97 | 0.52 | 0.68 | |
| 2.1.B | UAV | 47 | 0 | 18 | 1 | 0.72 | 0.84 |
| Airborne | 55 | 3 | 10 | 0.95 | 0.85 | 0.89 | |
| GE | 50 | 1 | 16 | 0.98 | 0.76 | 0.85 | |
| 2.1.C | UAV | 52 | 1 | 13 | 0.98 | 0.80 | 0.88 |
| Airborne | 58 | 3 | 7 | 0.95 | 0.88 | 0.91 | |
| GE | 54 | 1 | 12 | 0.98 | 0.82 | 0.89 | |
| 2.2.A | UAV | 31 | 0 | 34 | 1 | 0.48 | 0.65 |
| Airborne | 48 | 1 | 17 | 0.98 | 0.74 | 0.84 | |
| GE | 38 | 1 | 27 | 0.97 | 0.58 | 0.73 | |
| 2.2.B | UAV | 38 | 1 | 27 | 0.97 | 0.58 | 0.73 |
| Airborne | 46 | 1 | 19 | 0.98 | 0.71 | 0.82 | |
| GE | 47 | 3 | 18 | 0.94 | 0.72 | 0.82 | |
| 2.2.C | UAV | 46 | 1 | 19 | 0.98 | 0.70 | 0.82 |
| Airborne | 51 | 2 | 14 | 0.96 | 0.78 | 0.86 | |
| GE | 50 | 2 | 15 | 0.96 | 0.77 | 0.85 | |
| 3.1.A | UAV | 37 | 0 | 28 | 1 | 0.57 | 0.73 |
| Airborne | 43 | 0 | 22 | 1 | 0.66 | 0.80 | |
| GE | 41 | 1 | 24 | 0.98 | 0.63 | 0.77 | |
| 3.1.B | UAV | 48 | 1 | 17 | 0.98 | 0.74 | 0.84 |
| Airborne | 51 | 1 | 14 | 0.98 | 0.78 | 0.87 | |
| GE | 54 | 1 | 11 | 0.98 | 0.83 | 0.90 | |
| 3.1.C | UAV | 52 | 1 | 13 | 0.98 | 0.80 | 0.88 |
| Airborne | 52 | 1 | 13 | 0.98 | 0.80 | 0.88 | |
| GE | 54 | 2 | 11 | 0.96 | 0.83 | 0.89 | |
| 3.2.A | UAV | 54 | 1 | 11 | 0.98 | 0.83 | 0.90 |
| Airborne | 56 | 4 | 9 | 0.93 | 0.86 | 0.90 | |
| GE | 53 | 2 | 12 | 0.96 | 0.82 | 0.88 | |
| 3.2.B | UAV | 56 | 3 | 9 | 0.95 | 0.86 | 0.90 |
| Airborne | 54 | 5 | 11 | 0.92 | 0.83 | 0.87 | |
| GE | 53 | 3 | 12 | 0.95 | 0.82 | 0.88 | |
| 3.2.C | UAV | 54 | 3 | 11 | 0.95 | 0.83 | 0.89 |
| Airborne | 52 | 3 | 13 | 0.95 | 0.80 | 0.87 | |
| GE | 52 | 3 | 13 | 0.95 | 0.80 | 0.87 | |
| Best Experiment | Image Train | Image Test | PSE | NSR | ED2 |
|---|---|---|---|---|---|
| 1.1.C | UAV | UAV | 0.0532 | 0.1290 | 0.1396 |
| 1.2.C | UAV | UAV | 0.0512 | 0.0967 | 0.1095 |
| 2.1.C | Airborne | Airborne | 0.0408 | 0.0645 | 0.0763 |
| 2.2.C | Airborne | Airborne | 0.0589 | 0.0645 | 0.0873 |
| 3.1.B | GE | GE | 0.0414 | 0.0645 | 0.0767 |
| 3.2.B | GE | UAV | 0.0501 | 0.0645 | 0.0816 |
| Best Experiment | Best OBIA (ED2) | Best Mask R-CNN (ED2) | PSE | NSR | ED2 |
|---|---|---|---|---|---|
| 1.1.C | 0.05 | 0.13 | 0.02 | 0.03 | 0.0386 |
| 1.2.C | 0.05 | 0.10 | 0.02 | 0.03 | 0.0417 |
| 2.1.C | 0.10 | 0.07 | 0.02 | 0.03 | 0.0388 |
| 2.2.C | 0.10 | 0.08 | 0.05 | 0.06 | 0.0395 |
| 3.1.B | 0.20 | 0.07 | 0.00 | 0.06 | 0.0645 |
| 3.2.B | 0.20 | 0.08 | 0.00 | 0.06 | 0.0645 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guirado, E.; Blanco-Sacristán, J.; Rodríguez-Caballero, E.; Tabik, S.; Alcaraz-Segura, D.; Martínez-Valderrama, J.; Cabello, J. Mask R-CNN and OBIA Fusion Improves the Segmentation of Scattered Vegetation in Very High-Resolution Optical Sensors. Sensors 2021, 21, 320. https://doi.org/10.3390/s21010320
Guirado E, Blanco-Sacristán J, Rodríguez-Caballero E, Tabik S, Alcaraz-Segura D, Martínez-Valderrama J, Cabello J. Mask R-CNN and OBIA Fusion Improves the Segmentation of Scattered Vegetation in Very High-Resolution Optical Sensors. Sensors. 2021; 21(1):320. https://doi.org/10.3390/s21010320
Chicago/Turabian StyleGuirado, Emilio, Javier Blanco-Sacristán, Emilio Rodríguez-Caballero, Siham Tabik, Domingo Alcaraz-Segura, Jaime Martínez-Valderrama, and Javier Cabello. 2021. "Mask R-CNN and OBIA Fusion Improves the Segmentation of Scattered Vegetation in Very High-Resolution Optical Sensors" Sensors 21, no. 1: 320. https://doi.org/10.3390/s21010320
APA StyleGuirado, E., Blanco-Sacristán, J., Rodríguez-Caballero, E., Tabik, S., Alcaraz-Segura, D., Martínez-Valderrama, J., & Cabello, J. (2021). Mask R-CNN and OBIA Fusion Improves the Segmentation of Scattered Vegetation in Very High-Resolution Optical Sensors. Sensors, 21(1), 320. https://doi.org/10.3390/s21010320








