Computer-Aided Diagnosis of Alzheimer’s Disease through Weak Supervision Deep Learning Framework with Attention Mechanism
Abstract
1. Introduction
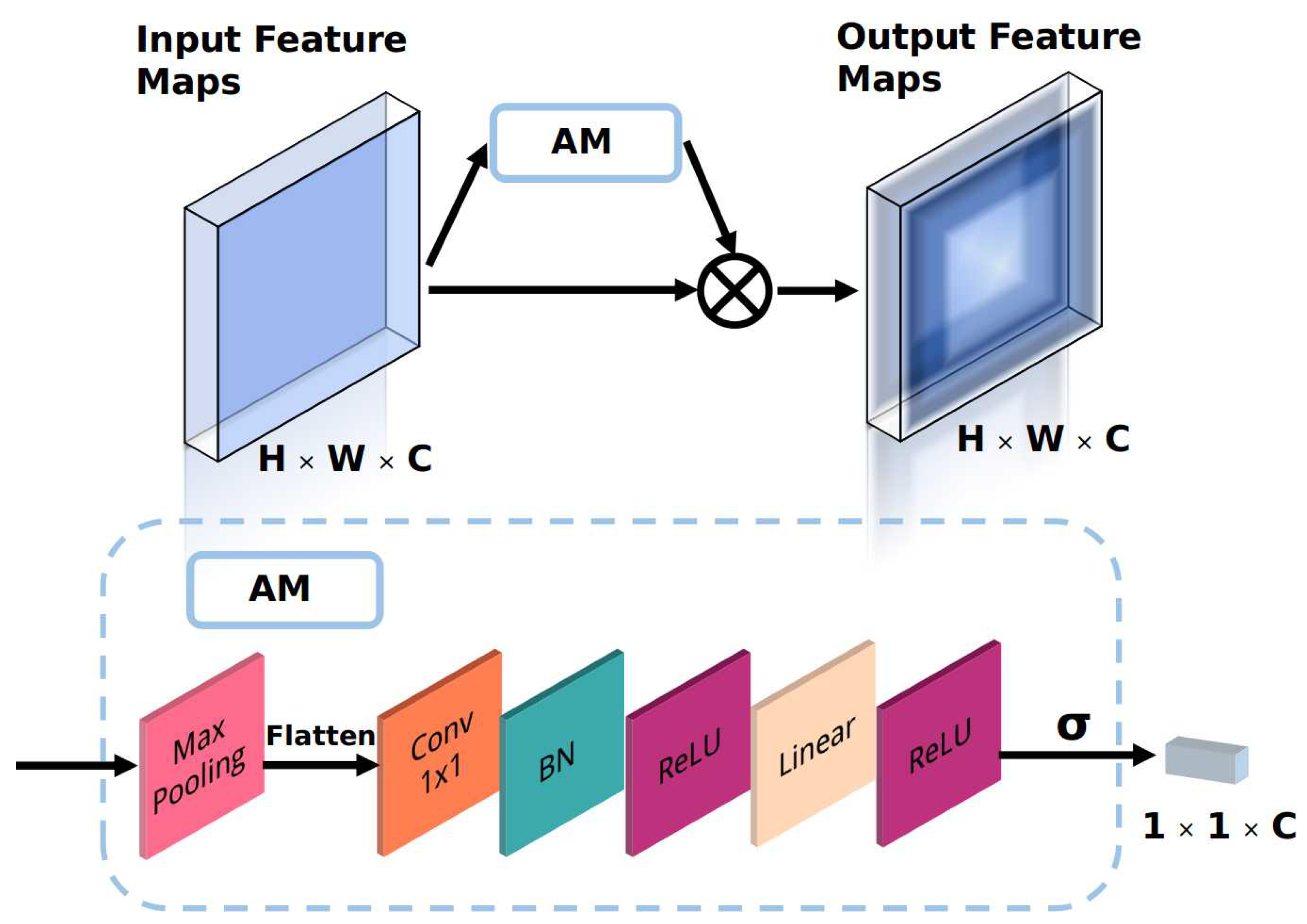
- An attention module (AM) is proposed to improve the discriminative ability of the backbone network with a low computational cost. The AM is an automatic weighting module that adjusts the weights of the channels in the feature maps so that the backbone network selectively focuses on the significant parts of the input.
- The task networks perform two tasks (image classification and image reconstruction) in parallel. The task networks utilize the feature vector generated by the backbone network and use fully-connected (FC) layers and a decoder for label prediction and image reconstruction.
- A multi-task learning (MTL) framework is proposed for conducting image recognition and reconstruction in parallel, with low computational requirements and good performance (with the best F1-score of 99.61% and a sensitivity of 99.69%) using only 20% of the labels from the datasets for fine-tuning.
2. Related Works
2.1. Multi-Task Learning
2.2. Weakly Supervised Learning
2.3. Image Classification
2.4. Image Reconstruction
3. Materials and Methods
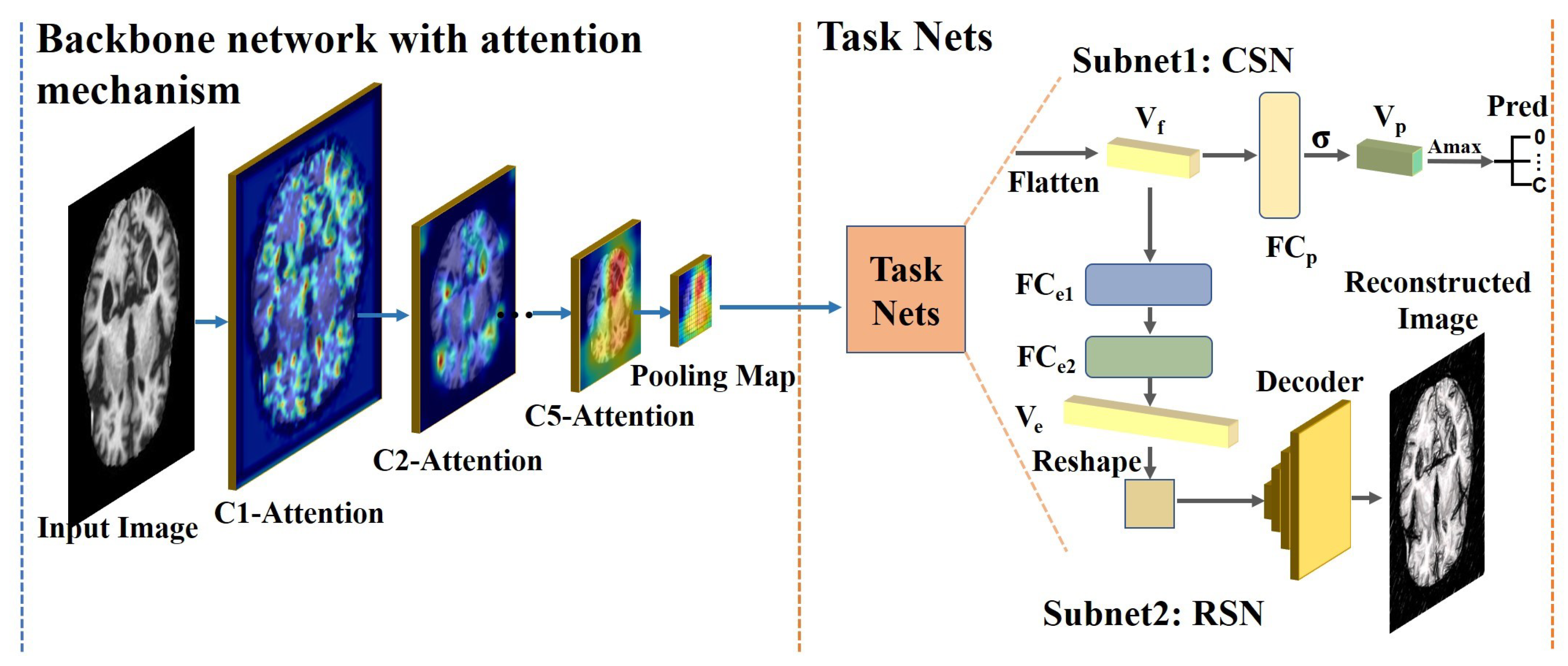
3.1. The Pipeline of the Proposed Framework
3.2. Backbone Network with the Proposed Attention Module
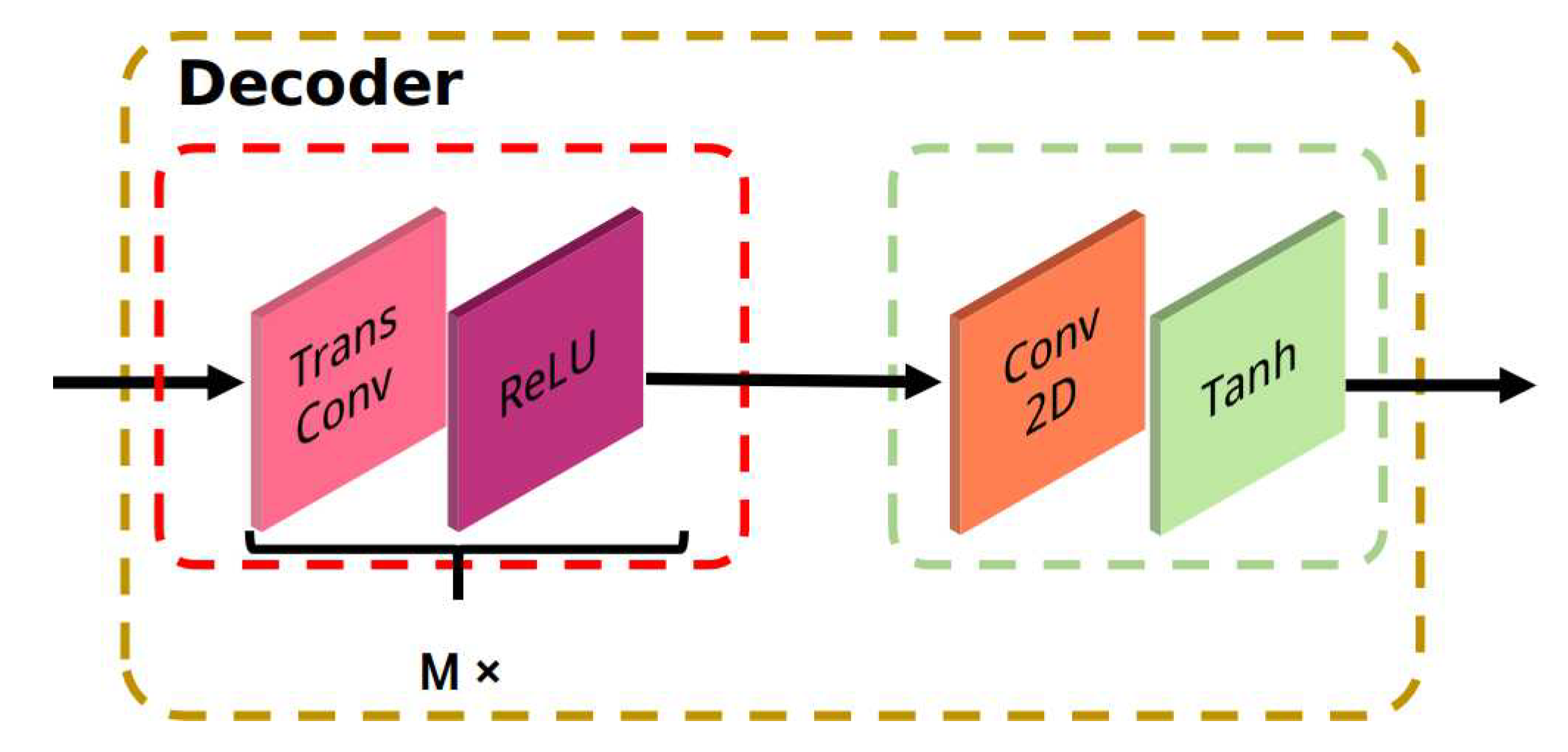
3.3. Task Sub-Networks
3.4. Loss Function
4. Results
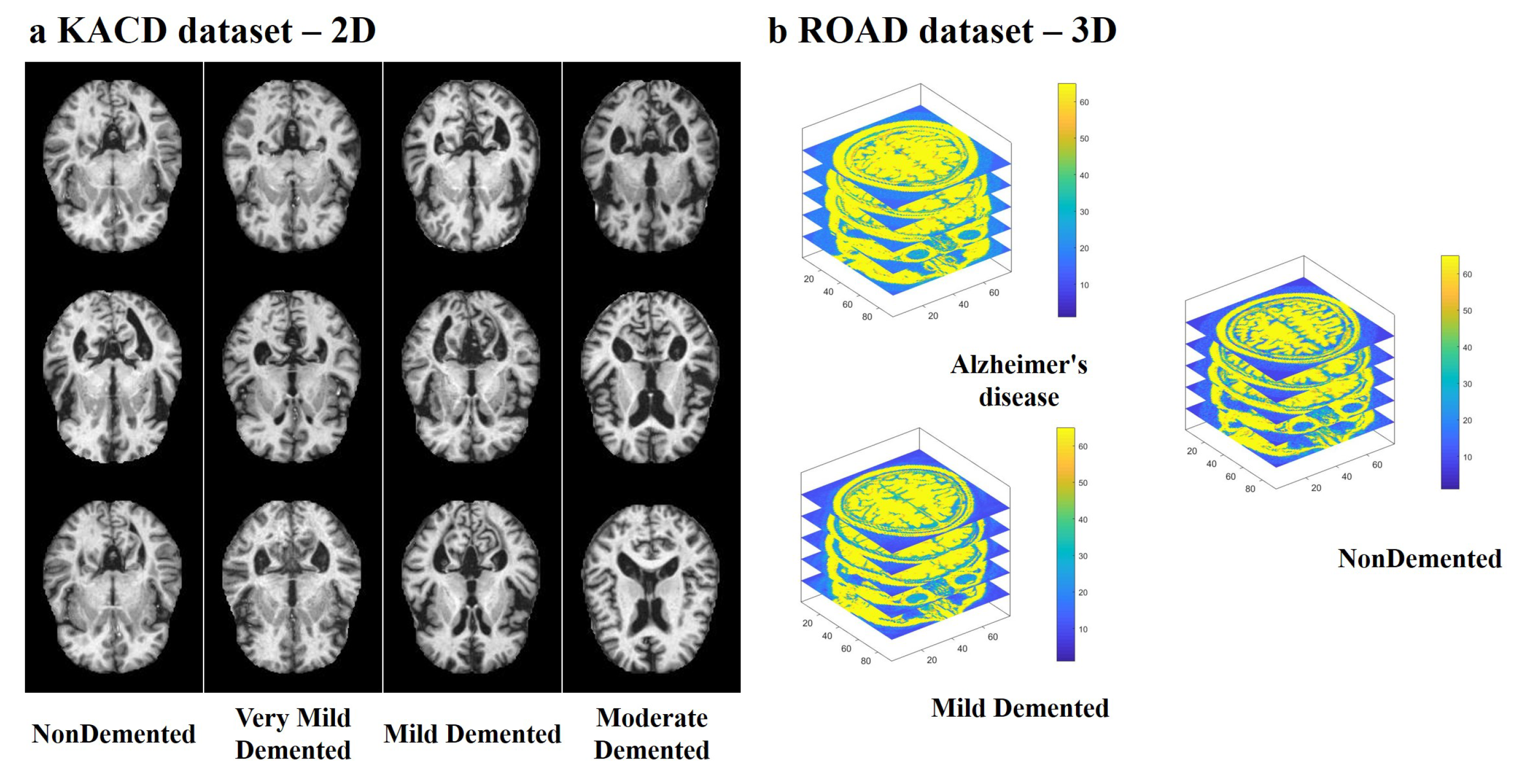
4.1. Multi-Modal Brain Imaging Dataset
4.2. Evaluation Metrics
4.3. Experimental Results
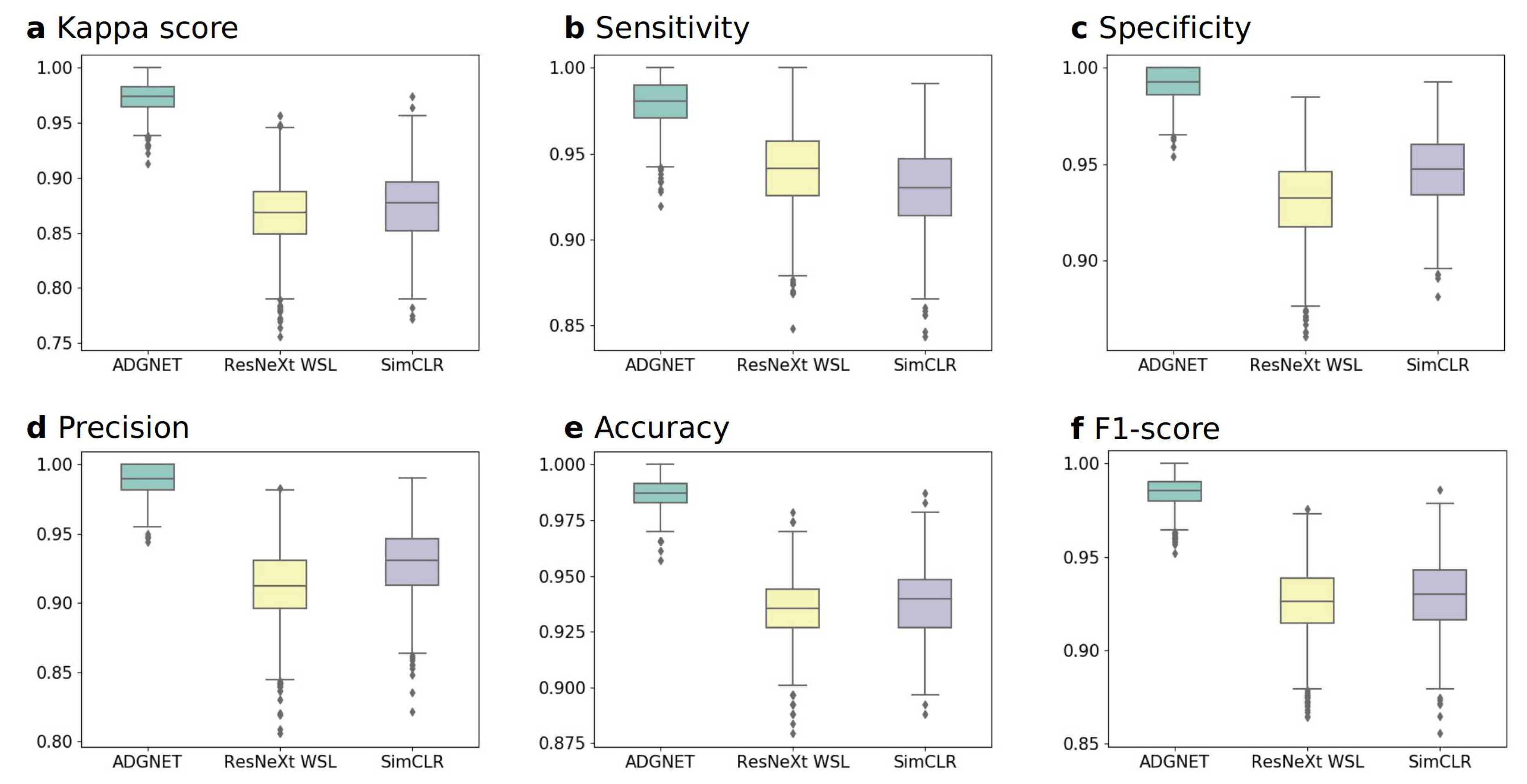
4.3.1. Experiment A: Performance on the KACD Dataset (Comparison between the Proposed ADGNET, ResNeXt WSL and SimCLR)
4.3.2. Experiment B: Performance on the ROAD Dataset (Comparison between the Proposed ADGNET, ResNeXt WSL and SimCLR)
4.3.3. Training Details
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| ADGNET | Alzheimer’s Disease Grade Network |
| WSL | Weakly Supervised Learning |
| DL | Deep Learning |
| WHO | World Health Organization |
| MCI | Mild Cognitive Impairment |
| CT | Computed Tomography |
| PET | Positron Emission Tomography |
| MRI | Magnetic Resonance Imaging |
| CAD | Computer-Aided Diagnosis |
| CNN | Convolution Neural Network |
| SOTA | State-of-the-Art |
| AM | Attention Module |
| MTL | Multi-Task Learning |
| SIMO | Single-Input-Multi-Output |
| CSN | Classification Sub-Network |
| RSN | Reconstruction Sub-Network |
| GAP | Global Average Pooling |
| FC | Fully Connected |
| CAF | Channel Attention Factors |
| EWMO | Element-Wise Multiplication Operation |
| BN | Batch Norm |
| KACD | Kaggle Alzheimer’s Classification Dataset |
| ROAD | Recognition of Alzheimer’s Disease Dataset |
| TTSF | Train-Test-Split Function |
| TVS | Training-Val Set |
| TS | Testing Set |
| Sen | Sensitivity |
| Spe | Specificity |
| Pr | Precision |
| Acc | Accuracy |
References
- Alzheimer’s Disease International. World Alzheimer Report 2019: Attitudes to Dementia. Available online: https://www.alz.co.uk/research/WorldAlzheimerReport2019.pdf (accessed on 20 November 2020).
- World Health Organization. World Health Organization (2018) The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 24 May 2018).
- Korczyn, A.D. Why have we failed to cure Alzheimer’s disease? J. Alzheimers Dis. 2012, 29, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Sanford, A.M. Mild cognitive impairment. Clin. Geriatr. Med. 2017, 33, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C.; Stevens, J.C.; Ganguli, M.; Tangalos, E.G.; Cummings, J.; DeKosky, S.T. Practice parameter: Early detection of dementia: Mild cognitive impairment (an evidence-based review): Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology 2001, 56, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aztiria, A.; Basarab, A. On the early diagnosis of Alzheimer’s Disease from multimodal signals: A survey. Artif. Intell. Med. 2016, 71, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Trombella, S.; Assal, F.; Zekry, D.; Gold, G.; Giannakopoulos, P.; Garibotto, V.; Démonet, J.F.; Frisoni, G.B. Brain imaging of Alzheimer’disease: State of the art and perspectives for clinicians. Rev. Medicale Suisse 2016, 12, 795–798. [Google Scholar]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629. [Google Scholar] [CrossRef]
- Beaulieu, J.; Dutilleul, P. Applications of computed tomography (CT) scanning technology in forest research: A timely update and review. Can. J. For. Res. 2019, 49, 1173–1188. [Google Scholar] [CrossRef]
- Zhang, B.; Gu, G.j.; Jiang, H.; Guo, Y.; Shen, X.; Li, B.; Zhang, W. The value of whole-brain CT perfusion imaging and CT angiography using a 320-slice CT scanner in the diagnosis of MCI and AD patients. Eur. Radiol. 2017, 27, 4756–4766. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Wiste, H.J.; Schwarz, C.G.; Lowe, V.J.; Senjem, M.L.; Vemuri, P.; Weigand, S.D.; Therneau, T.M.; Knopman, D.S.; Gunter, J.L. Longitudinal tau PET in ageing and Alzheimer’s disease. Brain 2018, 141, 1517–1528. [Google Scholar] [CrossRef]
- Domingues, I.; Pereira, G.; Martins, P.; Duarte, H.; Santos, J.; Abreu, P.H. Using deep learning techniques in medical imaging: A systematic review of applications on CT and PET. Artif. Intell. Rev. 2020, 53, 4093–4160. [Google Scholar] [CrossRef]
- Debette, S.; Schilling, S.; Duperron, M.G.; Larsson, S.C.; Markus, H.S. Clinical significance of magnetic resonance imaging markers of vascular brain injury: A systematic review and meta-analysis. JAMA Neurol. 2019, 76, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Battineni, G.; Chintalapudi, N.; Amenta, F.; Traini, E. A Comprehensive Machine-Learning Model Applied to Magnetic Resonance Imaging (MRI) to Predict Alzheimer’s Disease (AD) in Older Subjects. J. Clin. Med. 2020, 9, 2146. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Mansour, R.F. Deep-learning-based automatic computer-aided diagnosis system for diabetic retinopathy. Biomed. Eng. Lett. 2018, 8, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, Y.D.; Yan, X.; Liu, H.; Zhou, M.; Hu, B.; Yang, G. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Cao, C.; Wang, Z.; Xu, G.; Qiao, J. Anatomical Landmarks and DAG Network Learning for Alzheimer’s Disease Diagnosis. IEEE Access 2020, 8, 206063–206073. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Mahajan, D.; Girshick, R.; Ramanathan, V.; He, K.; Paluri, M.; Li, Y.; Bharambe, A.; van der Maaten, L. Exploring the limits of weakly supervised pretraining. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 181–196. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. arXiv 2020, arXiv:2002.05709. [Google Scholar]
- Zhang, Y.; Yang, Q. An overview of multi-task learning. Natl. Sci. Rev. 2018, 5, 30–43. [Google Scholar] [CrossRef]
- Caruana, R. Multitask learning. Mach. Learn. 1997, 28, 41–75. [Google Scholar] [CrossRef]
- Misra, I.; Shrivastava, A.; Gupta, A.; Hebert, M. Cross-stitch networks for multi-task learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3994–4003. [Google Scholar]
- Lu, Y.; Kumar, A.; Zhai, S.; Cheng, Y.; Javidi, T.; Feris, R. Fully-adaptive feature sharing in multi-task networks with applications in person attribute classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5334–5343. [Google Scholar]
- Zhang, Z.; Luo, P.; Loy, C.C.; Tang, X. Facial landmark detection by deep multi-task learning. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 94–108. [Google Scholar]
- Li, Y.F.; Guo, L.Z.; Zhou, Z.H. Towards safe weakly supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018, 5, 44–53. [Google Scholar] [CrossRef]
- Hu, Y.; Modat, M.; Gibson, E.; Li, W.; Ghavami, N.; Bonmati, E.; Wang, G.; Bandula, S.; Moore, C.M.; Emberton, M. Weakly-supervised convolutional neural networks for multimodal image registration. Med. Image Anal. 2018, 49, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, W.; Xie, S.M.; Azzari, G.; Lobell, D.B. Weakly supervised deep learning for segmentation of remote sensing imagery. Remote Sens. 2020, 12, 207. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Wang, X.; Wang, W.; Li, J. Development of convolutional neural network and its application in image classification: A survey. Opt. Eng. 2019, 58, 040901. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Zhu, B.; Liu, J.Z.; Cauley, S.F.; Rosen, B.R.; Rosen, M.S. Image reconstruction by domain-transform manifold learning. Nature 2018, 555, 487–492. [Google Scholar] [CrossRef]
- Xu, W.; Keshmiri, S.; Wang, G. Adversarially approximated autoencoder for image generation and manipulation. IEEE Trans. Multimed. 2019, 21, 2387–2396. [Google Scholar] [CrossRef]
- Goyal, P.; Kaiming, H. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 39, 2999–3007. [Google Scholar]
- Li, X.; Wang, W.; Wu, L.; Chen, S.; Hu, X.; Li, J.; Tang, J.; Yang, J. Generalized Focal Loss: Learning Qualified and Distributed Bounding Boxes for Dense Object Detection. arXiv 2020, arXiv:2006.04388. [Google Scholar]
- Dubey, S. Alzheimer’s Dataset (4 Class of Images). Available online: https://www.kaggle.com/tourist55/alzheimers-dataset-4-class-of-images (accessed on 29 November 2020).
- CCF BDCI. Recognition of Alzheimer’s Disease Dataset. Available online: https://www.datafountain.cn/competitions/369 (accessed on 29 November 2020).


| KACD | Train-Val | Test | Total | ROAD | Train-Val | Test | Total |
|---|---|---|---|---|---|---|---|
| NoneDemented | 2560 | 640 | 3200 | NoneDemented | 68 | 52 | 120 |
| Very Mild Demented | 1792 | 448 | 2240 | Very Mild Demented | - | - | - |
| Mild Demented | 717 | 179 | 896 | Mild Demented | 151 | 116 | 267 |
| Moderate Demented | 52 | 12 | 64 | Alzheimer’s disease | 81 | 64 | 145 |
| Total | 5121 | 1279 | 6400 | Total | 300 | 232 | 532 |
| KACD Dataset | |||||
|---|---|---|---|---|---|
| ADGNET (Proposed) | ADGNET (No RSN) | ADGNET (No AM) | ResNeXt WSL | SimCLR | |
| Kappa (95%CI) | 0.9922 | 0.9781 | 0.9812 | 0.9672 | 0.9734 |
| (0.9844, 0.9984) | (0.9656, 0.9890) | (0.9703, 0.9906) | (0.9514, 0.9797) | (0.9609, 0.9844) | |
| Sen (95% CI) | 0.9969 | 0.9906 | 0.9922 | 0.9891 | 0.9875 |
| (0.9921, 1.0000) | (0.9824, 0.9970) | (0.9845, 0.9984) | (0.9804, 0.9955) | (0.9785, 0.9953) | |
| Spe (95% CI) | 0.9953 | 0.9875 | 0.9890 | 0.9781 | 0.9859 |
| (0.9890, 1.0000) | (0.9783, 0.9953) | (0.9799, 0.9955) | (0.9663, 0.9888) | (0.9769, 0.9937) | |
| Pr (95% CI) | 0.9953 | 0.9875 | 0.9891 | 0.9784 | 0.9860 |
| (0.9894, 1.0000) | (0.9780, 0.9953) | (0.9798, 0.9956) | (0.9670, 0.9889) | (0.9805, 0.9922) | |
| Acc (95% CI) | 0.9961 | 0.9891 | 0.9906 | 0.9836 | 0.9860 |
| (0.9922, 0.9992) | (0.9828, 0.9945) | (0.9851, 0.9953) | (0.9757, 0.9898) | (0.9805, 0.9922) | |
| F1-score (95% CI) | 0.9961 | 0.9891 | 0.9906 | 0.9837 | 0.9867 |
| (0.9922, 0.9992) | (0.9828, 0.9945) | (0.9849, 0.9956) | (0.9756, 0.9901) | (0.9806, 0.9922) | |
| ROAD Dataset | |||||
|---|---|---|---|---|---|
| ADGNET (Proposed) | ADGNET (No RSN) | ADGNET (No AM) | ResNeXt WSL | SimCLR | |
| Kappa (95% CI) | 0.9736 | 0.9210 | 0.9473 | 0.8687 | 0.8770 |
| (0.9387, 1.0000) | (0.8696, 0.9654) | (0.9032, 0.9825) | (0.7986, 0.9300) | (0.8143, 0.9308) | |
| Sen (95% CI) | 0.9800 | 0.9600 | 0.9700 | 0.9400 | 0.9300 |
| (0.9500, 1.0000) | (0.9175, 0.9906) | (0.9310, 1.0000) | (0.8889, 0.9804) | (0.8764, 0.9770) | |
| Spe (95% CI) | 0.9924 | 0.9621 | 0.9773 | 0.9318 | 0.9470 |
| (0.9754, 1.0000) | (0.9274, 0.9924) | (0.9503, 1.0000) | (0.8824, 0.9699) | (0.9091, 0.9835) | |
| Pr (95% CI) | 0.9899 | 0.9505 | 0.9700 | 0.9126 | 0.9300 |
| (0.9674, 1.0000) | (0.9053, 0.9897) | (0.9347, 1.0000) | (0.8509, 0.9619) | (0.8775, 0.9780) | |
| Acc (95% CI) | 0.9871 | 0.9612 | 0.9741 | 0.9353 | 0.9300 |
| (0.9698, 1.0000) | (0.9353, 0.9828) | (0.9526, 0.9914) | (0.9009, 0.9655) | (0.9095, 0.9655) | |
| F1-score (95% CI) | 0.9849 | 0.9552 | 0.9700 | 0.9261 | 0.9300 |
| (0.9655, 1.0000) | (0.9246, 0.9817) | (0.9436, 0.9903) | (0.8832, 0.9608) | (0.8912, 0.9630) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, S.; Gu, Y. Computer-Aided Diagnosis of Alzheimer’s Disease through Weak Supervision Deep Learning Framework with Attention Mechanism. Sensors 2021, 21, 220. https://doi.org/10.3390/s21010220
Liang S, Gu Y. Computer-Aided Diagnosis of Alzheimer’s Disease through Weak Supervision Deep Learning Framework with Attention Mechanism. Sensors. 2021; 21(1):220. https://doi.org/10.3390/s21010220
Chicago/Turabian StyleLiang, Shuang, and Yu Gu. 2021. "Computer-Aided Diagnosis of Alzheimer’s Disease through Weak Supervision Deep Learning Framework with Attention Mechanism" Sensors 21, no. 1: 220. https://doi.org/10.3390/s21010220
APA StyleLiang, S., & Gu, Y. (2021). Computer-Aided Diagnosis of Alzheimer’s Disease through Weak Supervision Deep Learning Framework with Attention Mechanism. Sensors, 21(1), 220. https://doi.org/10.3390/s21010220






