2.1. Dataset
The cervical tissue biopsy image dataset used in this article came from the First Affiliated Hospital of Xinjiang Medical University. These data were reviewed by the Medical Ethics Committee and were desensitized. The patients’ permission was obtained. There were 468 RGB images in total, each with a resolution of 3456 × 4608, as shown in
Figure 1, of which 150 were normal, 85 were low-grade squamous intraepithelial lesion (LSIL), and 104 were high-grade squamous intraepithelial lesion (HSIL). There were 129 images of cancer. After processing via the image enhancement method proposed in this article, the enhanced, small-size cervical tissue biopsy image dataset had a total of 100,020 images, each with a resolution of 200 × 200 (RGB) pixels, as shown in
Figure 2, including 50,370 normal images, 11,914 LSIL images, 16,677 HSIL images, and 21,059 cancer images. Among them, 90% of the images were used as the training set, and 10% were used as the test set, as shown in
Figure 2.
In
Figure 1, it was observed that the epithelial cells showed an increase in the number of atypical immature cells from the top (Area 1) to bottom (Area 2). In addition, as the degree of lesions increased, the number of atypical immature cells in the cervical biopsy tissue images also increased. This number of cells increased sequentially and became increasingly cancerous, which was reflected in the phenomenon that the nucleus-to-cell ratio of the cells became larger, and the cytoplasm deepened and became thicker. We found that the difference between
Figure 1a,b was small, and the difference between
Figure 1c,d was small. The cell morphology in the four pictures was varied and contained very rich information, and the similarity was very high. From this perspective, it is very difficult to describe the cervical pathological tissue image comprehensively through the use of traditional image features, which leads to an unsatisfactory final classification effect, especially in the early stage of disease.
2.1.1. New Classification Standards
The naming scheme of the WHO (2014) classification of female reproductive system tumors was used for cervical squamous cell precancerous lesions (
Table 1), where LSIL is defined as a kind of clinical and pathological change caused by HPV infection. Squamous intraepithelial lesions have a low risk of canceration currently or in the future. Synonyms for LSIL include cervical intraepithelial neoplasia Grade I (CIN1), mild atypical hyperplasia, flat condyloma, and keratocytosis. HSIL is defined as follows: If left untreated, this squamous intraepithelial lesion has a significant risk of progressing to invasive cancer. Synonyms for HSIL include cervical intraepithelial neoplasia grade II (CIN2), cervical intraepithelial neoplasia grade III (CIN3), moderate atypical hyperplasia, severe atypical hyperplasia, and squamous epithelial carcinoma in situ [
2].
2.1.2. Introduction to Image Features
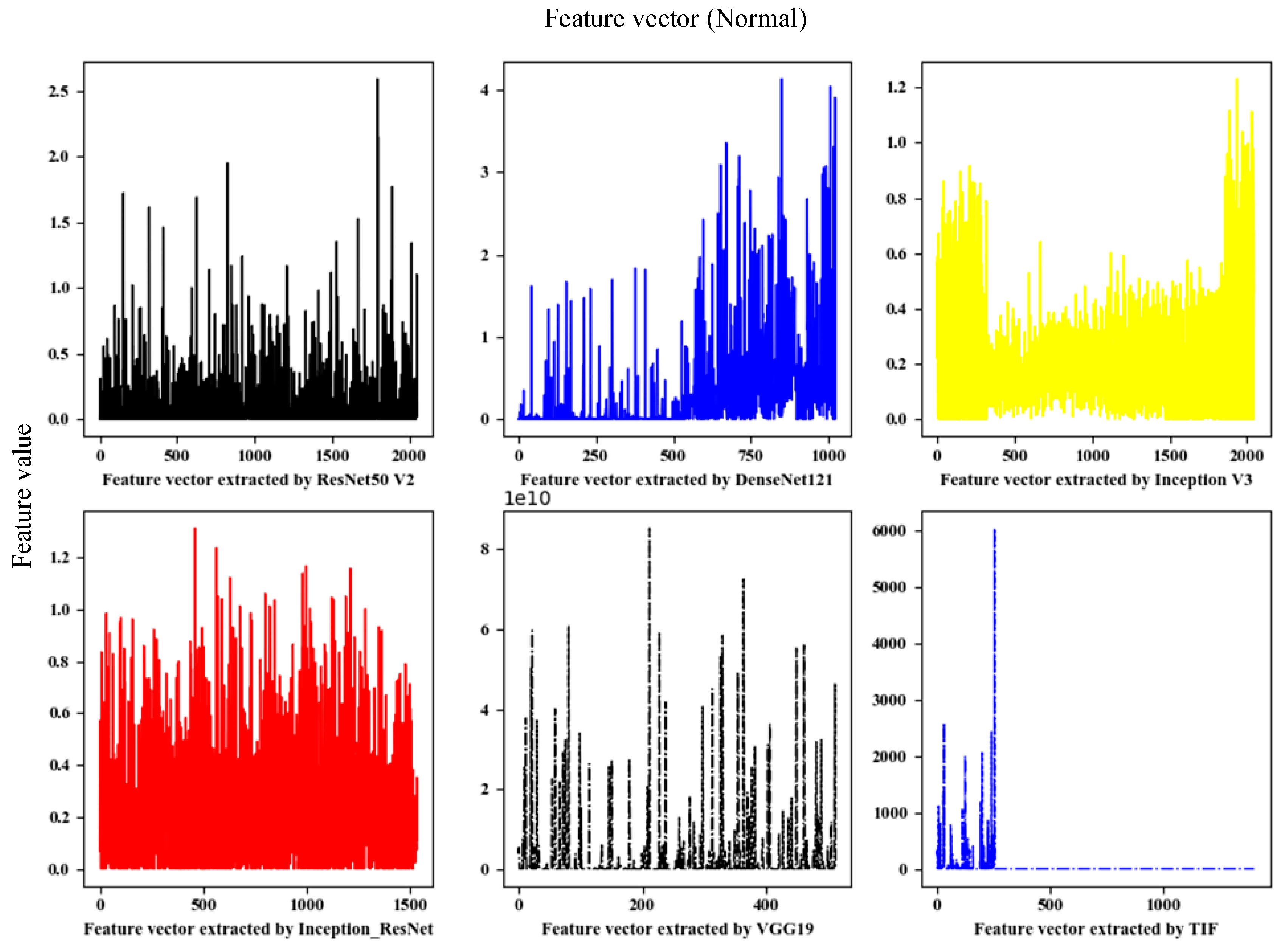
This paper used the fine-tuned deep network model to extract deep convolution features. A total of five depth models were trained. The dimensions of the extracted deep convolution features are shown in
Table 2, and the feature visualization is shown in
Figure 3.
Traditional image features (TIF): This paper mainly used a local binary pattern (LBP) and a histogram of oriented gradient (HOG) to extract features separately and then serially merged them into TIF vectors.
This paper used an LBP [
30,
31] algorithm to extract image texture features. LBP is a parameterless texture descriptor. LBP has the advantages of being simple and effective and having a strong recognition ability and low computational complexity. The gray value extracted by LBP was used to draw the gray statistics histogram, and the specific method is shown in Formula (1). A neighborhood in standard LBP is defined by a radius because square neighborhoods do not cover the entire image. The gray values of each circular neighborhood were obtained by comparing the gray values of the pixels on the circular border with the center pixel and then clockwise encoding at 90 degrees to obtain a re-encoded grayscale image in turn. The specific process is shown in
Figure 4. The form of the LBP descriptor is shown in Formula (2).
where
represents the frequency of gray value k after encoding,
n represents the number of pixels in the numbered image,
represents the sum of pixels of the gray value k, and M represents the number of gray values in the encoded image.
where R represents the radius of the circular neighborhood. The minimum unit is the Euclidean distance D between the four neighboring pixels of the image, and the distance is 1. Calculated through the defined R formula, the D value of the center pixel four-neighborhood is 1, the D value of the eight-neighborhood is 2, the D value of the 16-neighborhood is 3, and so on.
n represents the number of pixels in the circular area, and
and
represent the gray values of the central pixel and the i-th pixel in the circular neighborhood, respectively. When R = 1, the boundary point is the eight-neighborhood of the center pixel, and when R = 2,
p = 16, the center pixel and the eight-neighborhood are considered as a whole to form a new center pixel. The boundary point is the 16-neighborhood of the new central pixel, and so on.
HOG [
32,
33] features have a strong image structure and contour description capabilities as well as a strong recognition effect on the description of local areas. HOG features are also suitable for describing texture features. Texture features have local irregularity and macro regularity. Using appropriate HOG cell units to divide the image and extracting HOG features can obtain the changing pattern of the overall texture features of the image. Choosing HOG cell units that are too small results in local features that are too fine and macro features that are unclear and computationally complex. If the selected HOG cell unit is too large, the local feature description is incomplete, which is not conducive to generalizing the macro features. The cell unit size used in this paper was 10 × 10.
2.2. Image Processing
Random cropping based on grayscale matching: Random cropping was performed for each cervical image. The cropped size was 200 × 200 × 3, but there were images without cell nuclei. Obviously, such an image was useless at the training of the depth model. The sum of absolute differences (SAD) [
34] used grayscale matching to remove such cropped subimages, as expressed in Formula (3) and manually cropped the verification set to obtain the optimal threshold
. The random cropping function is defined as
, and its core formula is shown in (4) and (5).
where
represents the length of the row of the image matrix;
is the length of the column,
,
represent the variables of the number of cropping times;
,
represent the lengths of the row and column of the original image matrix, respectively;
,
represent the value range of randomly cropped row and column position variables;
represent randomly generated row and column values;
represents the cropped image matrix;
represents the template cropped image that meets the requirements; and
are used to match the row and column variables of the cropped image.
Random translation: The random translation function is defined as
. When implementing this function, a movement matrix M is constructed first. The specific formula is shown in (6).
Random rotation: The random rotation function is defined as
. When implementing this function, a rotation matrix M is constructed first. The specific formula is shown in (7).
Random zoom: The random zoom function is defined as
. Zooming is performed by dividing the image and selection points. The specific formula is shown in (8).
Random brightness adjustment: The random brightness adjustment function is defined as
. When implementing this function, a movement matrix M is constructed first. The specific formula is shown in (9).
Image normalization: To prevent the information on the low-value area from being concealed by the information on the high-value area, the image is normalized.
The pseudocode for the image enhancement process of this article is shown in Algorithm 1.
| Algorithm 1 Image Enhancement Processing |
| Input: . |
| Output: |
| 1 is the original image matrix, is the enhanced image matrix. |
| 2 FOR p = 1: s //s is the number of image samples |
| 3 Implement random cropping based on grayscale matching for : |
| 4 (i) Perform random cropping, according to Formulas (4) and (5): |
| 5 . |
| 6 (ii) Determine whether the following conditions are met: |
| 7 |
| 8 Randomly shift the randomly cropped image tensor according to Equation (6): |
| 9 |
| 10 Randomly rotate according to Formula (7): |
| 11 |
| 12 Randomly scale according to Formula (8): |
| 13 |
| 14 Randomly adjust the brightness of according to Equation (9): |
| 15 |
| 16 Normalize the enhanced, small-sized image according to Equation (10): |
| 17 |
| 18 END |
| 19 Return |
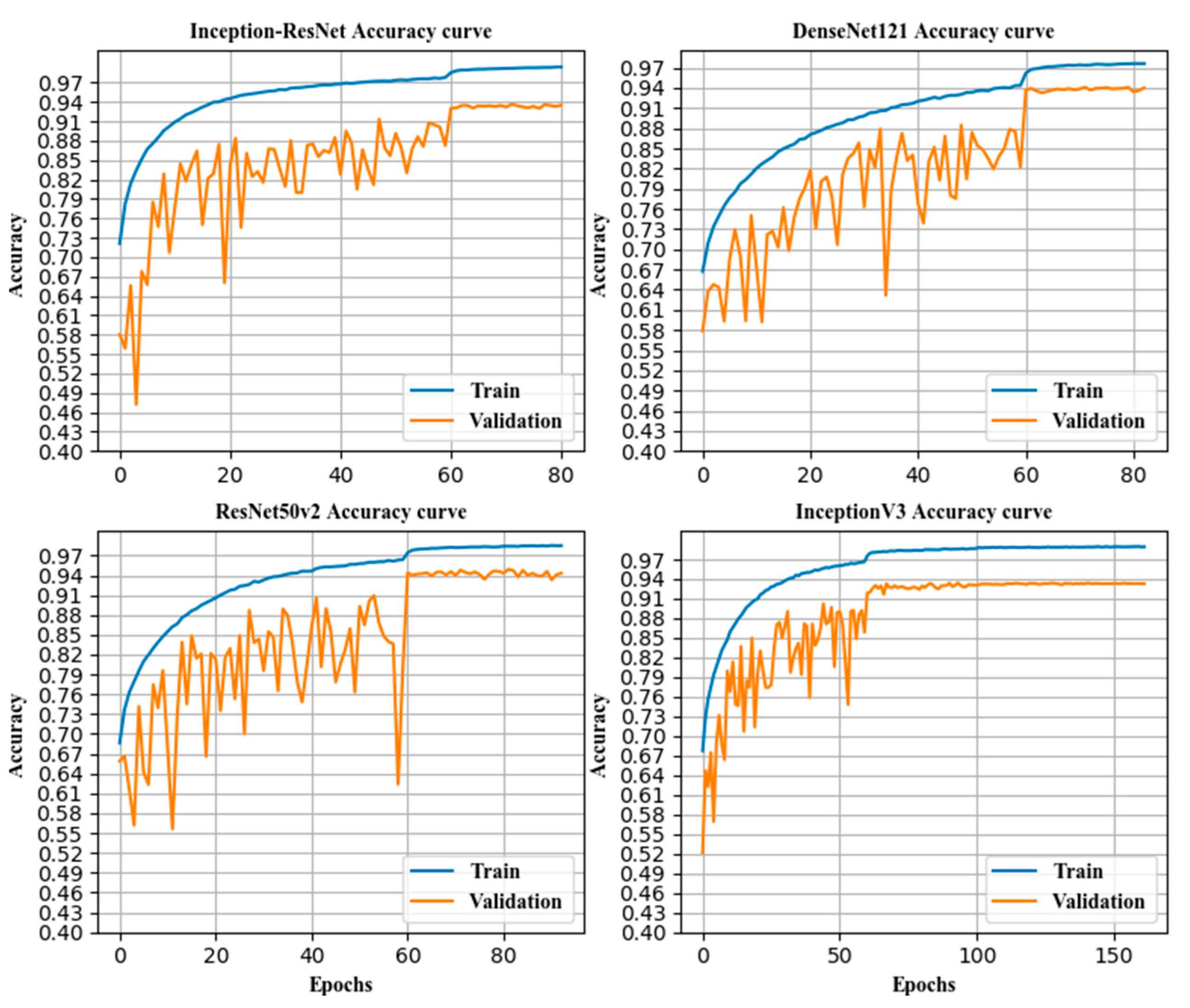
2.3. Fine-Tuned Transfer Model
In this paper, the Calling the Applications module of the deep learning framework Keras, and the DenseNet121, ResNet50 v2, Inception v3, and Inception-ResNet models were pre-trained on ImageNet [
35] data and used for transfer learning. Because the images in ImageNet had a large gap between images of cervical pathology, and the image features recognized by the top convolutional layer were more abstract and specific, this paper used only the weight of the 1st convolutional layer of the pre-trained model.
The last layer of the deep network model pre-trained on ImageNet was excessively specialized, and the last layer (pre-trained models of applications of Keras) was obviously not suitable for transfer learning. Thus, this layer was deleted.
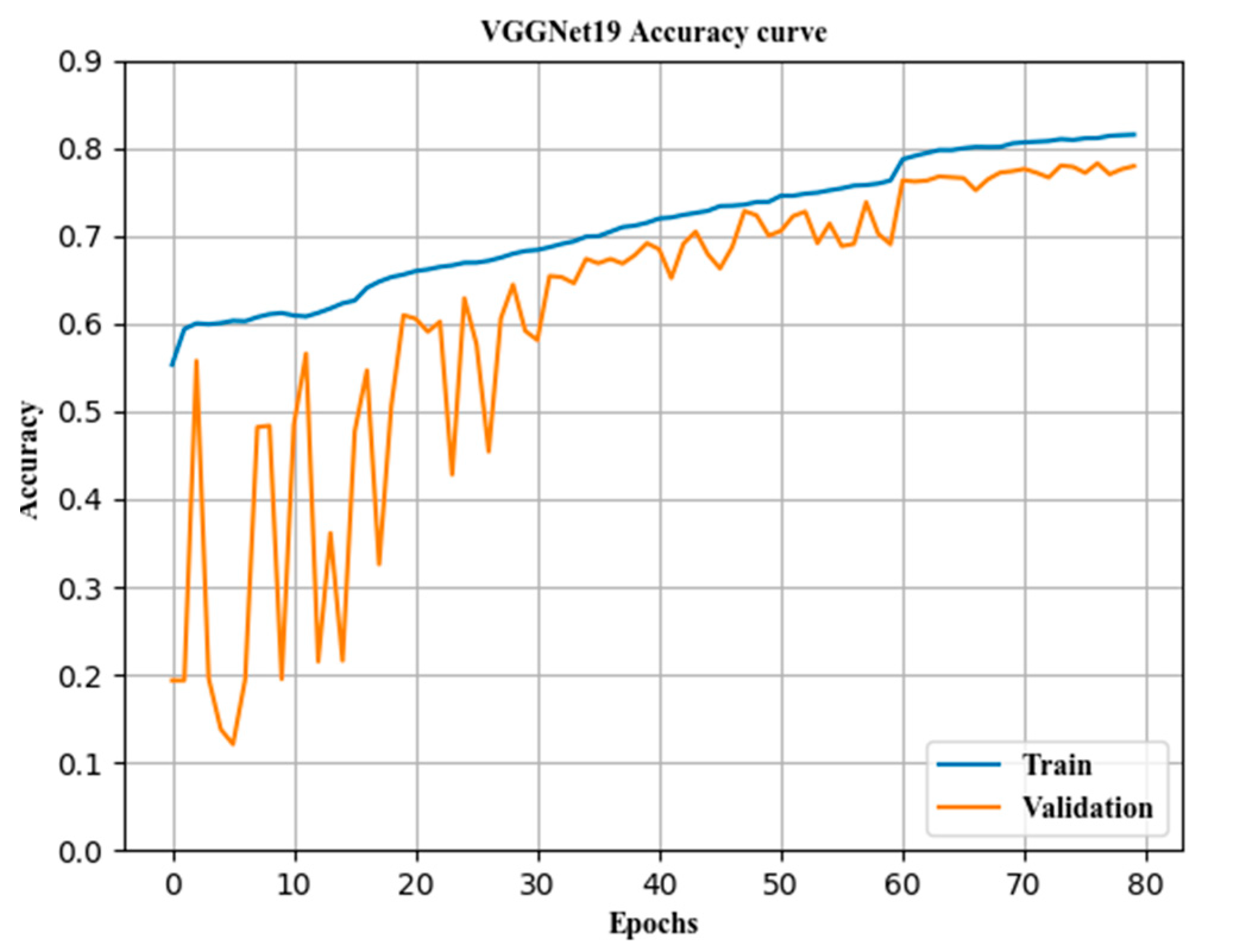
The most important aspects of transfer learning are the setting of the learning rate, the selection of the loss function, the configuration of the optimizer, and the measures for the prevention of overfitting.
The loss function is the objective function in transfer learning, and it is an indicator of the directions of weight changes. The choice of the loss function directly determines the quality of the result of transfer learning. This article used the categorical cross-entropy (CE) [
36] function as the loss function. The basic principle is shown in Equation (11):
where
represents the input sample,
is the expected total number of classifications,
is the
-th true label, and
corresponds to the output value of the model.
The optimizer is also one of the most important parameters in transfer learning. In this paper, stochastic gradient descent (SGD) [
37] was used as the optimization algorithm. It updates only once per epoch without redundancy and is fast. The basic principle is shown in Equation (12).
where η is the learning rate, also known as the step size, which is one of the most important parameters in transfer training. An excessively large learning rate causes the gradient to disappear, so the optimal solution cannot be found or the convergence time is too long. The learning rate was 0.1 for epochs 0–60, 0.01 for epochs 61–120, 0.001 for epochs 121–180 epochs, and 0.0001 for epochs 181 and above in this paper.
represents the sample data onto the i-th epoch.
In transfer learning, overfitting occurs frequently and has a large impact on the training results. The main method to prevent overfitting is data augmentation. In addition, in the top layer designed in this paper, the convolution kernel was regularized, and the dropout layer and batch normalization [
38] layer were added after the full connection in
Figure 5. The relevant parameters of the fully connected layer and the regularization layer are shown in
Table 3.
The regularization and processing algorithm used in this paper was LASSO [
39], and the basic principle implemented in the convolution kernel is shown in Equation (13).
where
represents the predicted label value,
represents the predicted label value,
represents the L1 regularization coefficient, and
represents the L1 regularization processing on the weight.
The forward calculation formulas for the dropout layer [
40] used in the article are shown in Equations (14) and (15):
where
obeys the Bernoulli binomial distribution of probability
and
is the generated 0,1 vector. By setting the activation value of 0, some nodes in layer
of the network stop working, and for the input of layer
, only the nonzero nodes in layer
are considered (in fact, all nodes are considered, but the output is a node of 0 that has no effect on the next layer of the network, and it cannot update its related network weights during backpropagation).
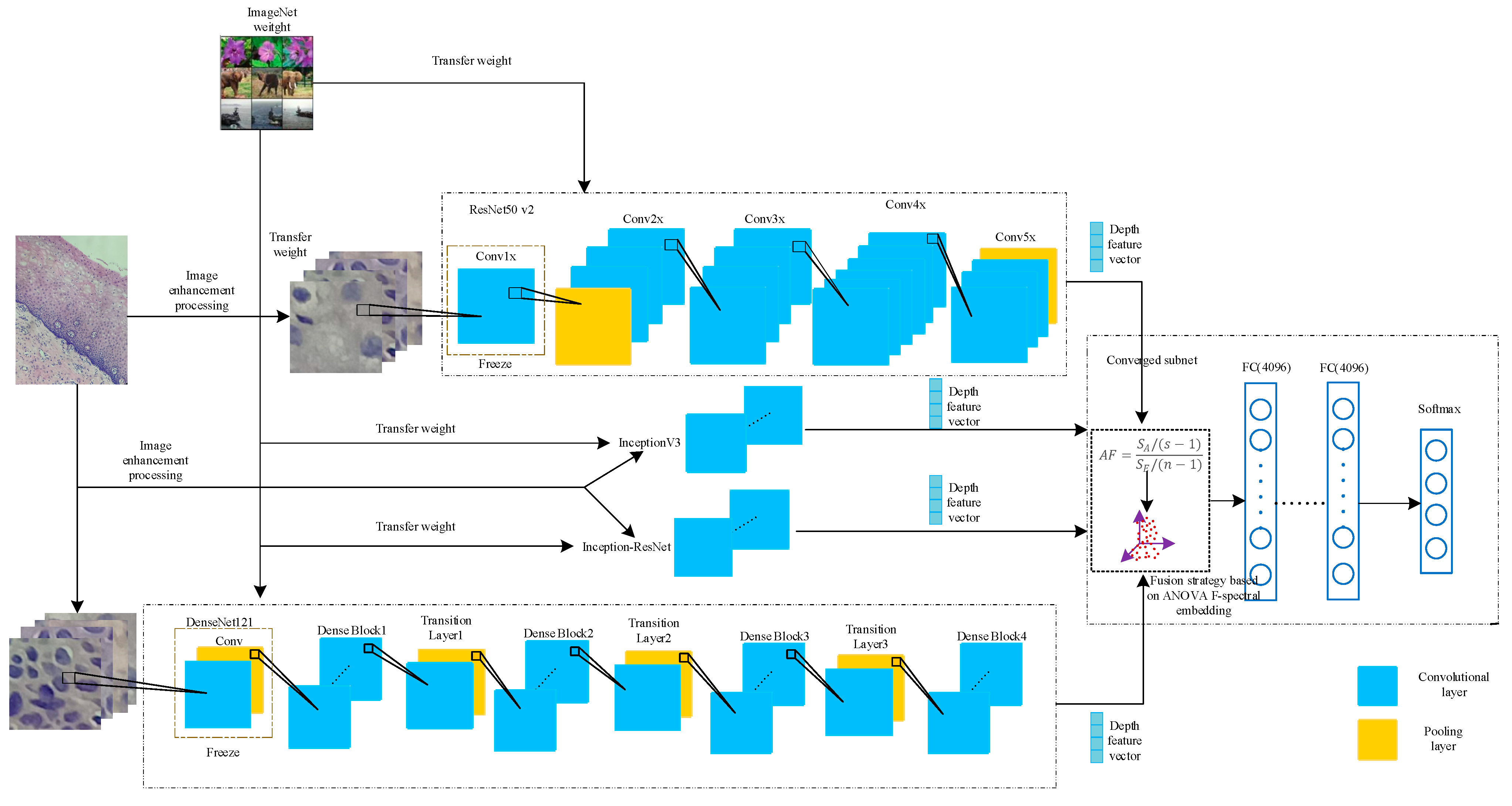
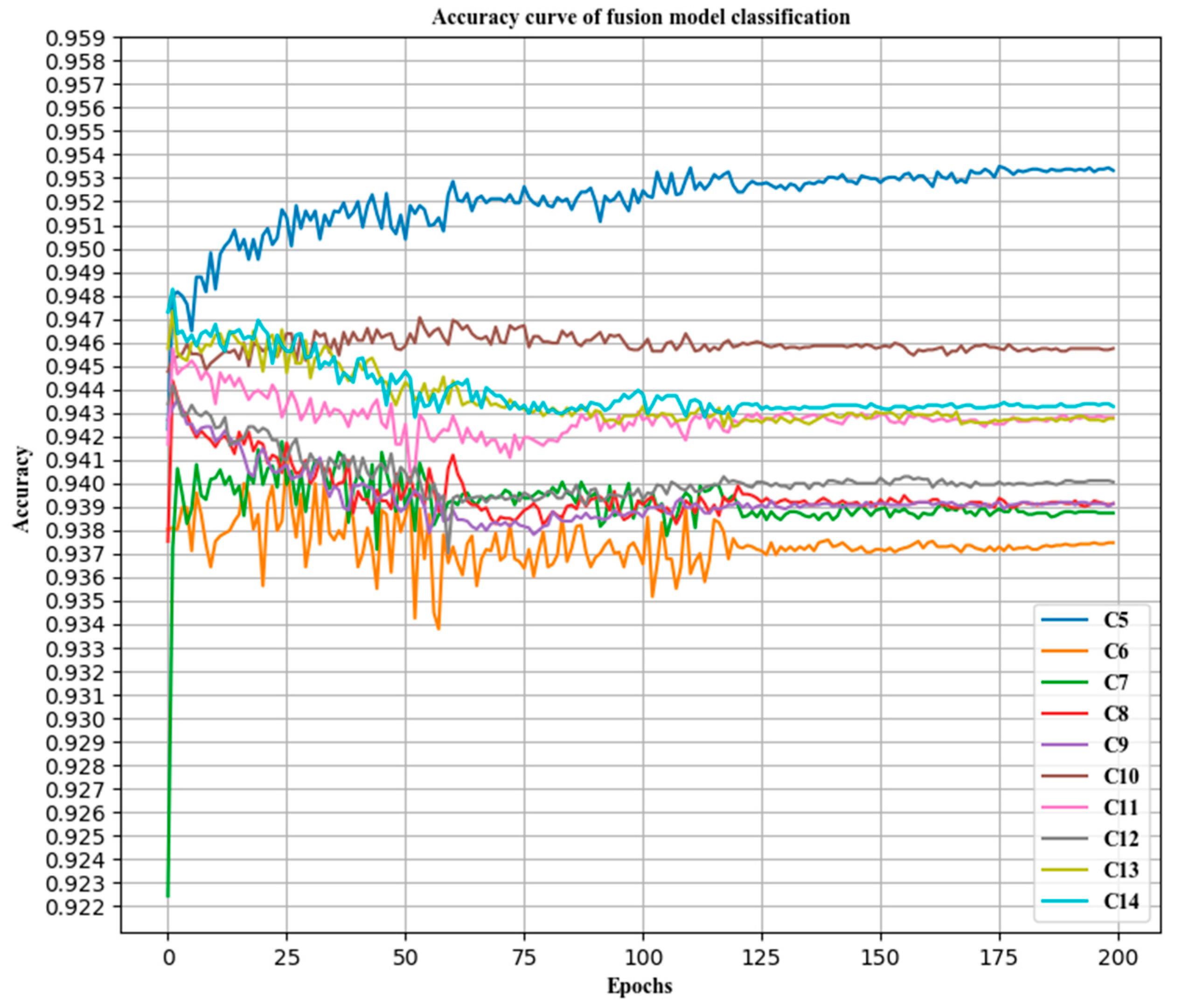
2.4. Deep Convolution Feature Fusion Mechanism Based on Analysis of Variance-F Value-Spectral Embedding (AF-SENet)
In this paper, the DenseNet121, ResNet50 v2, Inception v3, and Inception-ResNet models were pre-trained on the ImageNet dataset and then transferred to the pathological cervical tissue image dataset for further fine-tuning [
41]. The different trained models may contain complementary information. To explore this possible information complementarity, this paper proposed the use of the analysis of variance-F value (ANOVA F)-spectral embedding strategy to analyze the changes in the ANOVA F values for different fusion combinations. Spectral embedding [
42] was then used for fusion mapping. The softmax classifier was used for classification. The fused subnet is own in
Figure 5.
In this section, the deep convolutional network feature tensor extracted from a single model after migration fine-tuning is represented by
, where
represents the number of samples and
is the length of the row in the depth feature matrix (the feature-length of the sample). In this section, ANOVA F was used to evaluate the redundancy and correlation between different combinations of deep convolutional features. Analysis of variance (ANOVA) mainly explores the contributions to features of the between-group variance and within-group variance in datasets. The definition of the variance value is shown in Equations (16) and (17).
According to Formulas (16) and (17), the test statistic
can be constructed as follows:
where
represents the sum of variance values between different samples in the depth feature sample matrix and
represents the sum of variance values between different features in the depth feature sample matrix.
This paper proposed the ANOVA F-spectral embedding algorithm to select sample features and reduce the dimension to reduce the time complexity of training the subnets under the premise of ensuring high classification accuracy. First, a selection was performed by using the test statistic
. The
-value of each feature of the sample image feature matrix
was calculated. Second, the
-values of the sample features were summed to obtain the total value defined as
. The average
-value (
) was constructed to measure the importance of each feature of the entire feature set;
is shown in Equation (19).
According to the size of , the features were sorted in descending order, and the of the first sample features was calculated. If > 99.9%, the feature selection process was stopped, and the subsequent features were eliminated.
Based on the above selection of features, there were many redundant features. The effect of traditional feature selection methods of objective functions (labels) is not ideal, and linear methods, such as Principal Components Analysis (PCA) and Linear Discriminant Analysis (LDA), can be easily used to perform feature space transformations. The loss of the nonlinear relationships to samples is meant to avoid these problems. The ANOVA F-spectral embedding algorithm is shown in Algorithm 2.
| Algorithm 2 ANOVA F-spectral Embedding |
| Input: . |
| Output: |
| 1 is the sample image feature matrix, is the selected and transformed sample image feature matrix. |
| 2 FOR i = 1: n //n is the dimension of a feature in the feature matrix of the image sample. |
| 3 Calculate the -value of each feature according to Formula (18). |
| 4 |
| 5 // is the sum of the f-values of the sample features, is the f-value of the i-th feature. |
| 6 END |
| 7 Calculate the value of each feature according to Formula (18), sort in descending order |
| 8 FOR i = 1: n |
| 9 IF (sum + = ) < 99.9% |
| 10 = () |
| 11 END |
| 12 END |
| 13 Transform into a graph representation using the affinity (adjacency) matrix representation. |
| 14 Construct an unnormalized Laplacian graph as and a normalized graph as . |
| 15 Perform eigenvalue decomposition on the Laplacian graph after performing the above treatment on . |
| 16 Return |
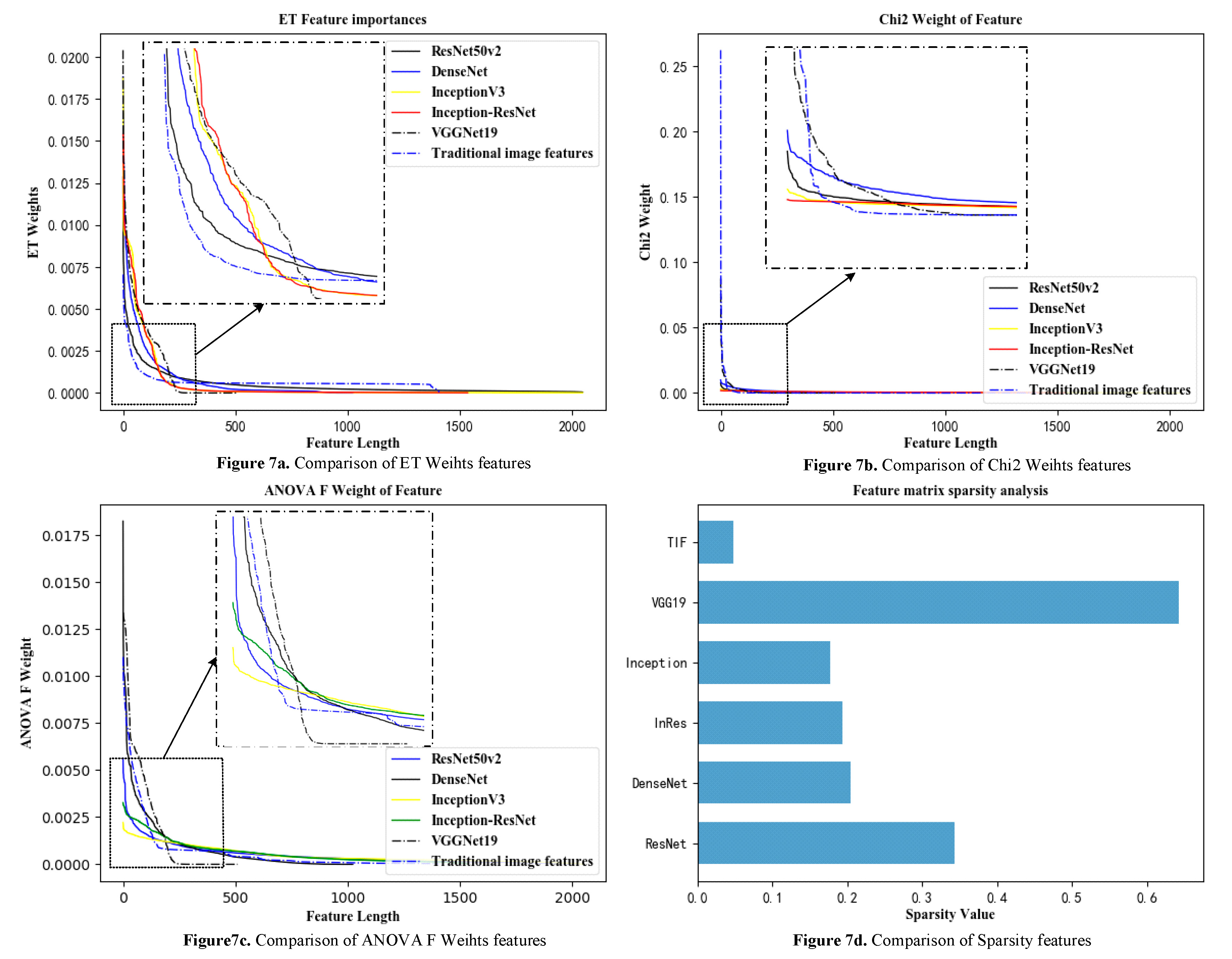
2.5. Feature Analysis
For the problem of feature classification, there are a variety of indicators to evaluate the pros and cons of features, including the correlation between features and categories, the redundancy of the features themselves, and the sparsity of features in the feature matrix. In this paper, to explore the advantages and disadvantages of deep convolutional network features and traditional image features of cervical cancer as well as the ability to represent image samples, the chi-square test (Chi2) is shown in Equation (20), and the ANOVA F(AF) test is shown in Equations (16)–(18). These tests explore the redundancy and correlation of features, using the average tree attributes of extremely randomized trees (ETs) [
43] to measure the importance of each feature.
where
is the actual value, and
is the predicted value.
Based on the introduction to
Section 2.1,
Section 2.2,
Section 2.3,
Section 2.4 and
Section 2.5, this article drew the overall implementation in
Figure 6.Block diagram of the AF-SENet algorithm, as shown in
Figure 6. The complete process includes using the DenseNet121, ResNet50 v2, Inception v3, and Inception-ResNet models pre-trained on ImageNet data and freezing the lowest layers. The pre-training model was used to extract the deep convolution features, and the ANOVA F-spectral embedding algorithm was used for dimension reduction. Serial fusion was performed, and the training subnet was input. The training subnet had two fully connected layers (the number of neurons is 4096) and an output layer, which contained a four-class softmax classifier.
2.6. Evaluation Criteria
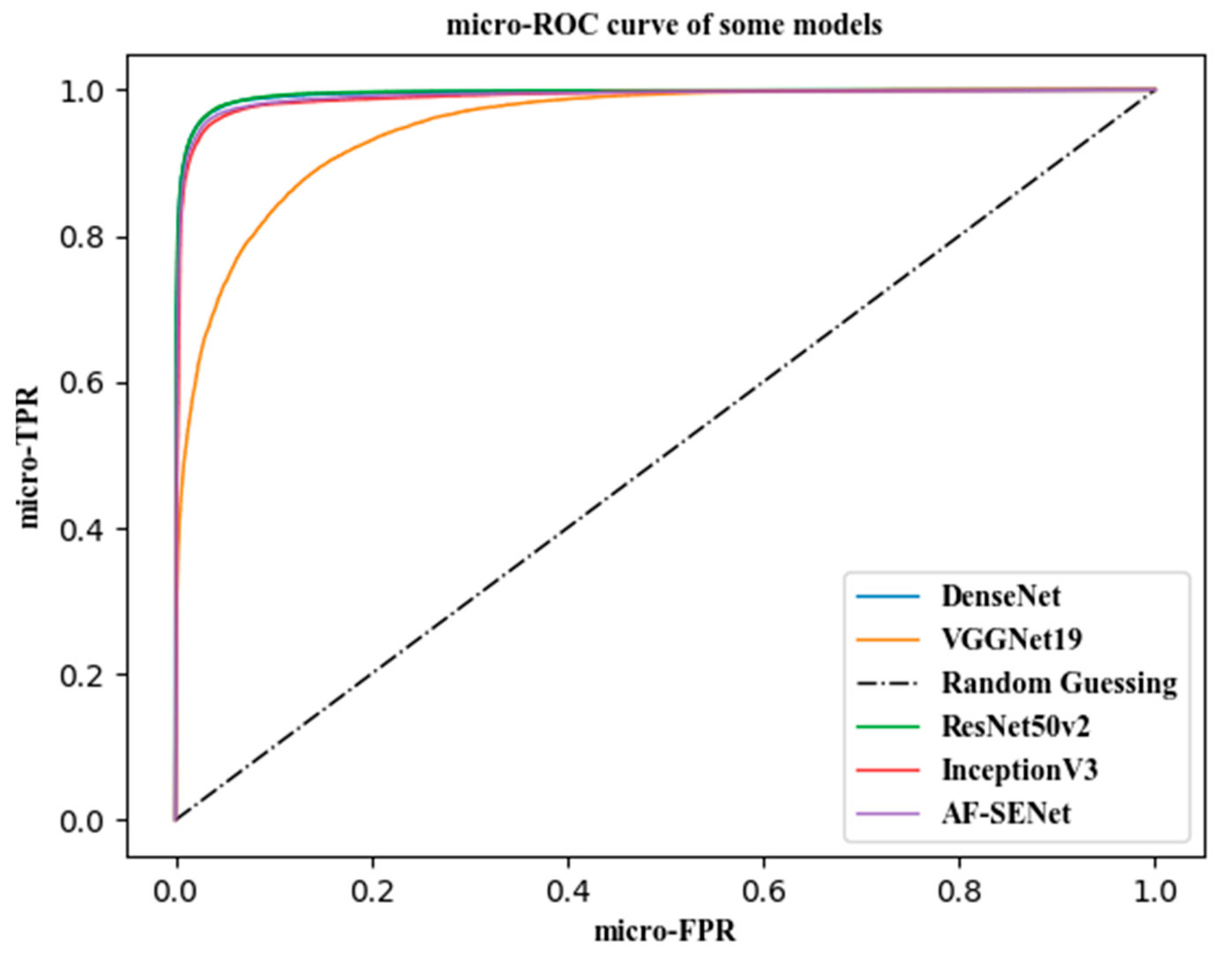
In this article, the Receiver Operating Characteristic (ROC) curve was used to evaluate the generalization ability of the model. The ROC curve is one of the most commonly used indicators in the evaluation of artificial intelligence models. The true-positive rate (TPR) was calculated, as expressed in Formula (22), and the false-positive rate (FPR) was calculated, as expressed in Formula (23) each time, with the TPR as the vertical axis of the ROC curve and the FPR as the horizontal axis of the ROC curve.
where TP, FN, FP, and TN are the true-positive examples, false-negative examples, false-positive examples, and true-negative examples, respectively, in the confusion matrix of the classification results.
Suppose that the sample size of the data to be analyzed by ROC is m, and the number of classifications of the sample is n, so a label matrix L of [m, n] can be obtained. The value is 0 or 1. Correspondingly, if we predicted the probability that each sample would fall into each category of the outcome of the statistical model, we could also obtain a probability matrix p of [m, n], and the value of the matrix p was 0–1.
Micro method: Expand the matrices L and p by rows, and form two columns of length mxn after transposing. In this way, the multicategory outcome can be converted into a two-category situation, followed by the classic two-category outcome. ROC analysis is sufficient.