Rapid Yeast Cell Viability Analysis by Using a Portable Microscope Based on the Fiber Optic Array and Simple Image Processing
Abstract
1. Introduction
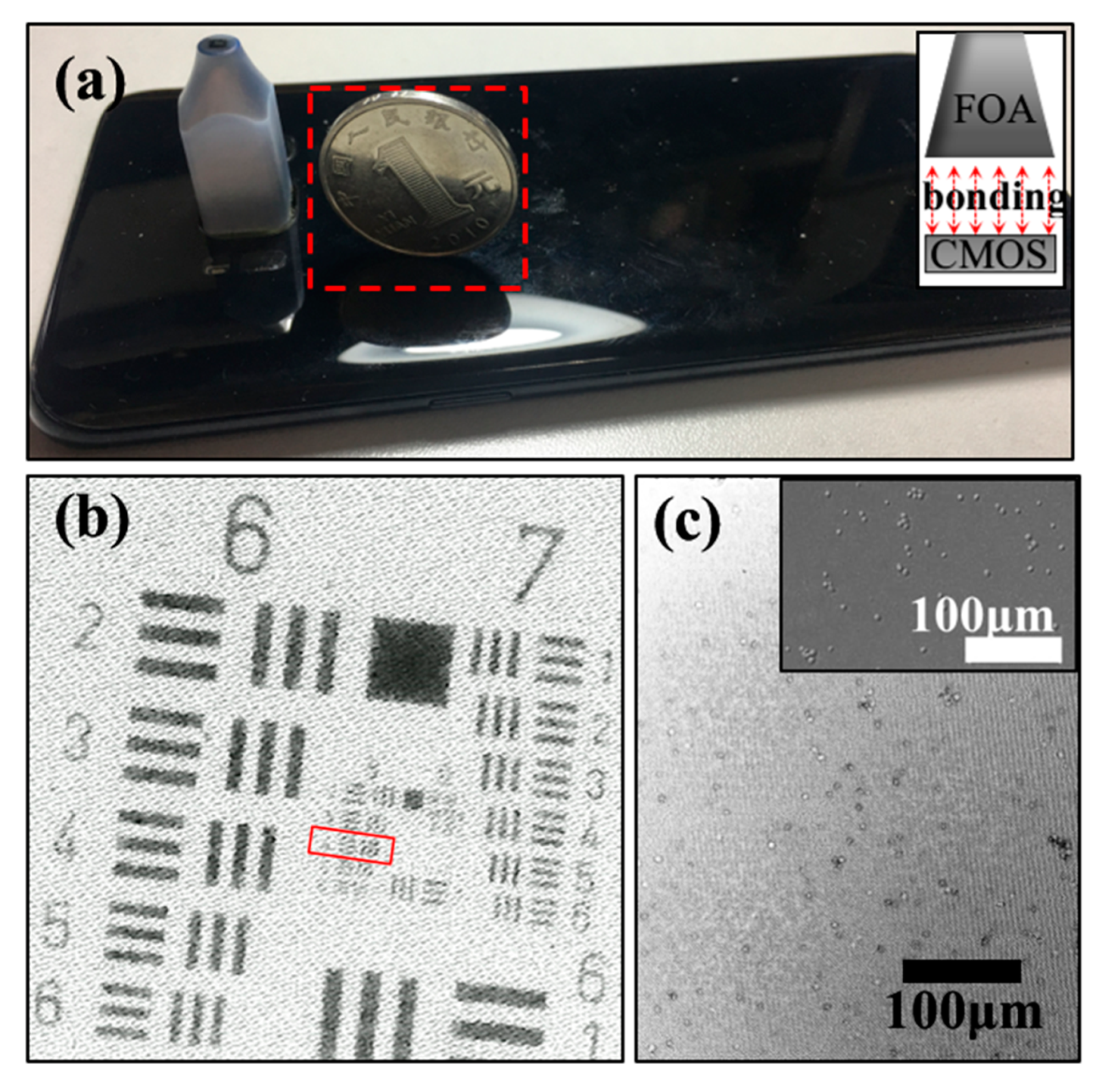
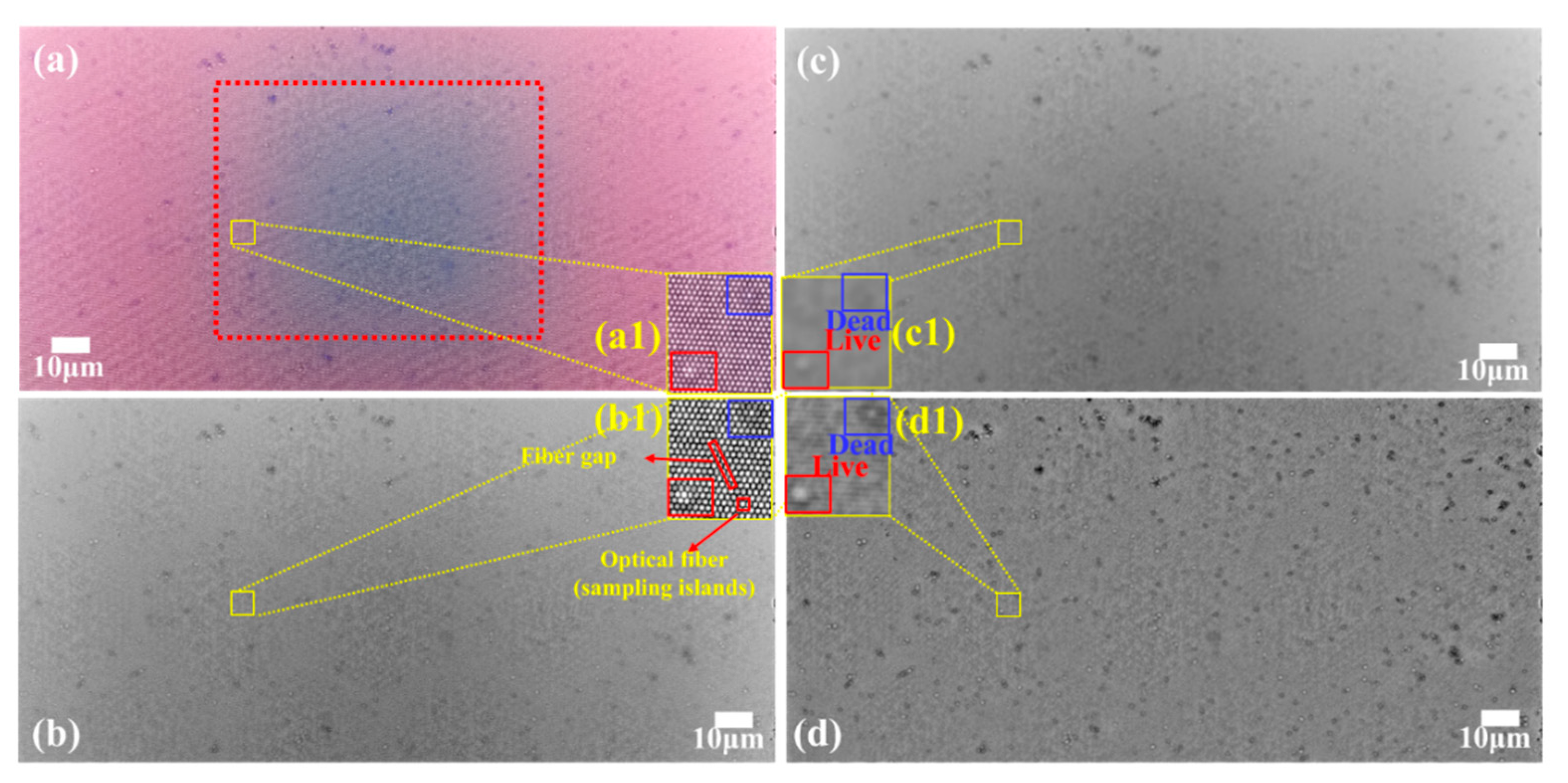
2. Yeast Activity and Counting Detection by the FOA Microscope
2.1. Sample Preparation
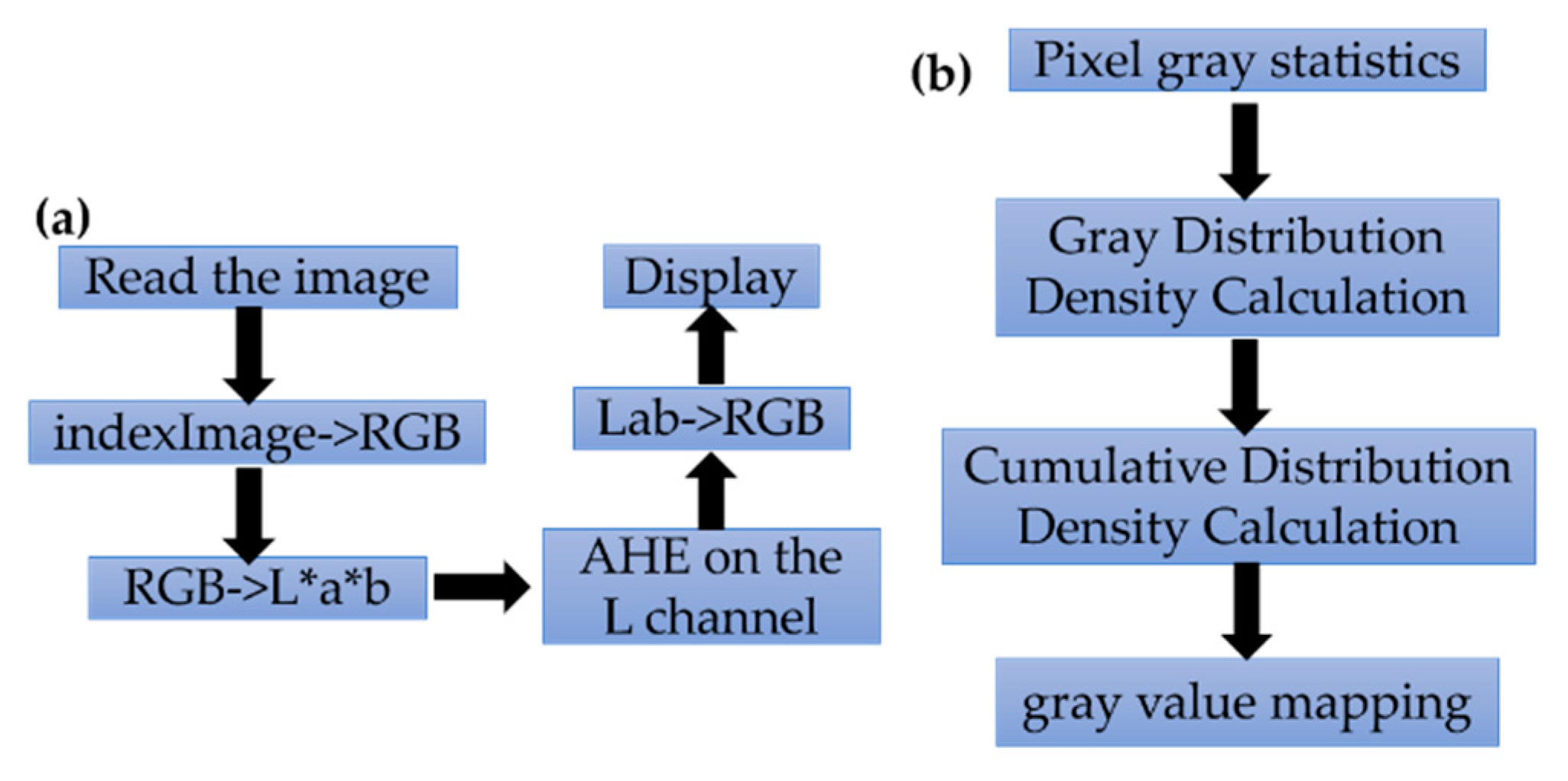
2.2. AHE Processing
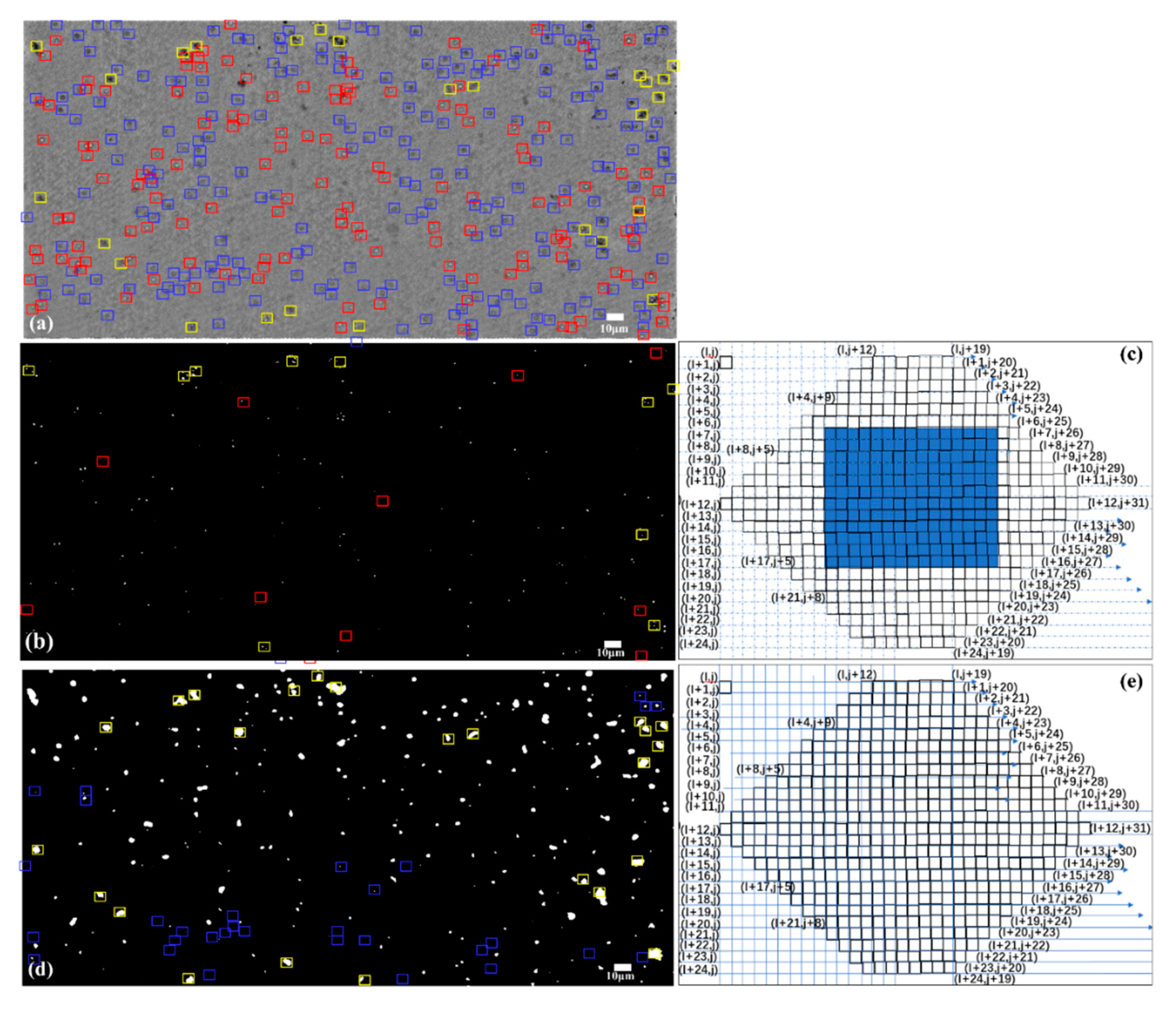
2.3. Scan Window for Cell Detection
3. The Factors That Affect the FOA Microscope’s Performance
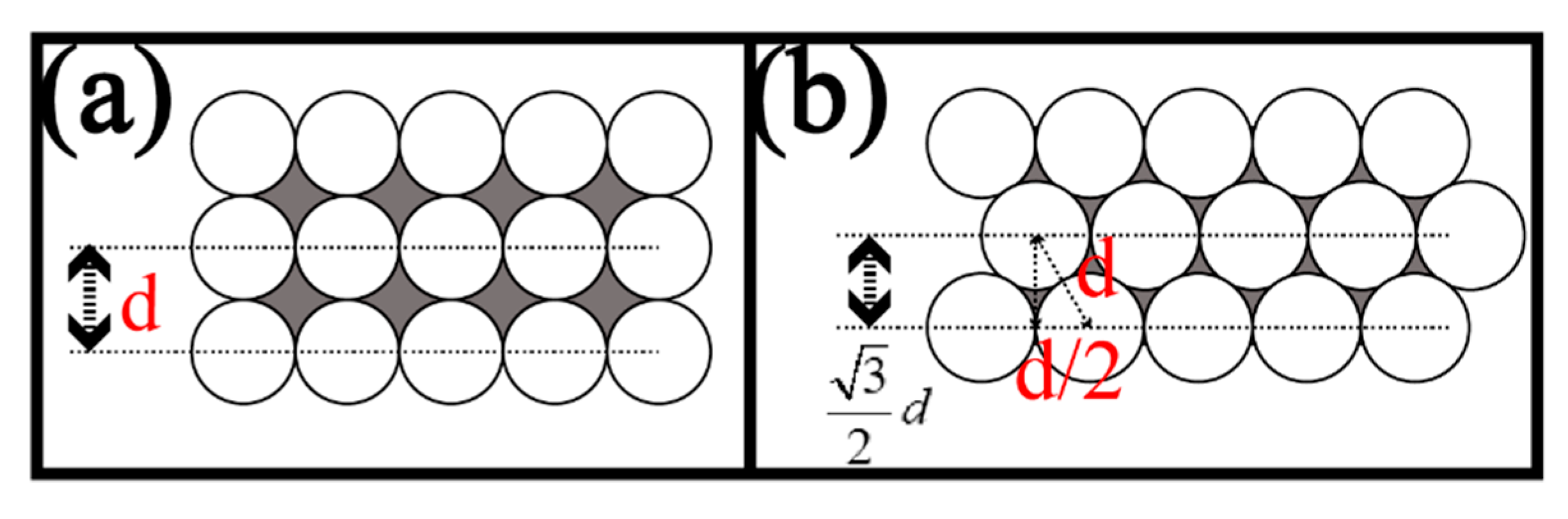
3.1. The Design Parameters of the FOA
3.2. Tapered Fiber Made of Weakly Conducting Fibers Helps to Reduce the Distortion
3.3. Effects of Crosstalk between Fibers, and Core and Cladding Thicknesses on Microscope Resolution
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pérez De León, A.A.; Teel, P.D.; Li, A.; Ponnusamy, L.; Roe, R.M. Advancing integrated tick management to mitigate burden of tick-borne diseases. Outlooks Pest Manag. 2014, 25, 382–389. [Google Scholar] [CrossRef]
- Scherr, T.F.; Gupta, S.; Wright, D.W.; Haselton, F.R. An embedded barcode for “connected” malaria rapid diagnostic tests. Lab Chip 2017, 17, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Scherr, T.F.; Gupta, S.; Wright, D.W.; Haselton, F.R. Mobile phone imaging and cloud-based analysis for standardized malaria detection and reporting. Sci. Rep. 2016, 6, 28645. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Bhatia, K. Smart phone as an adjunctive imaging tool to visualize scolex in orbital myocysticercosis. Int. Ophthalmol. 2013, 33, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, J.T.; Ouattara, M.; D Ambrosio, M.V.; Fletcher, D.A.; Keiser, J.; Utzinger, J.; N’Goran, E.K.; Andrews, J.R.; Bogoch, I.I. Accuracy of mobile phone and handheld light microscopy for the diagnosis of schistosomiasis and intestinal protozoa infections in Côte d’Ivoire. PLoS Negl. Trop. Dis. 2016, 10, e0004768. [Google Scholar] [CrossRef]
- Ephraim, R.K.; Duah, E.; Cybulski, J.S.; Prakash, M.; D’Ambrosio, M.V.; Fletcher, D.A.; Keiser, J.; Andrews, J.R.; Bogoch, I.I. Diagnosis of Schistosoma haematobium infection with a mobile phone-mounted Foldscope and a reversed-lens CellScope in Ghana. Am. J. Trop. Med. Hyg. 2015, 92, 1253–1256. [Google Scholar] [CrossRef]
- Bogoch, I.I.; Coulibaly, J.T.; Andrews, J.R.; Speich, B.; Keiser, J.; Stothard, J.R.; N’Goran, E.K.; Utzinger, J. Evaluation of portable microscopic devices for the diagnosis of Schistosoma and soil-transmitted helminth infection. Parasitology 2014, 141, 1811–1818. [Google Scholar] [CrossRef]
- Xiaotong, L.; Zhaofeng, C. Geometrical Optics, Aberrations and Optical Design; Zhejiang University Press: Hangzhou, China, 2006. [Google Scholar]
- Lee, S.A.; Yang, C. A smartphone-based chip-scale microscope using ambient illumination. Lab Chip 2014, 14, 3056–3063. [Google Scholar] [CrossRef]
- Navruz, I.; Coskun, A.F.; Wong, J.; Mohammad, S.; Tseng, D.; Nagi, R.; Phillips, S.; Ozcan, A. Smart-phone based computational microscopy using multi-frame contact imaging on a fiber-optic array. Lab Chip 2013, 13, 415–423. [Google Scholar] [CrossRef]
- Wittenstein, W.; Fontanella, J.C.; Newbery, A.R.; Baars, J. The Definition of the OTF and the Measurement of Aliasing for Sampled Imaging Systems. Opt. Acta Int. J. Opt. 1982, 29, 41–50. [Google Scholar] [CrossRef]
- Xin, F.X. Optical fiber coupling technique of ICCD. Infrared Laser Eng. 2001, 30, 200–213. [Google Scholar]
- King, N.S.P.; Albright, K.L.; Jaramillo, S.A.; McDonald, T.E.; Yates, G.J.; Turko, B.T. High-frame-rate CCD cameras with fast optical shutters for military and medical imaging applications. In Ultrahigh-and High-Speed Photography, Videography, and Photonics’ 94; SPIE: Bellingham, DC, USA, 1994; pp. 56–60. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Z. X-Ray Image Device Based on Optical Fiber Taper Coupling. Acta Opt. Sin. 2010, 30, 1478–1482. [Google Scholar]
- Wang, W.; Yu, Y.; Huang, H.; Ou, J. Portable microscopy platform for the clinical and environmental monitoring. In Proceedings of the SPIE Smart Structures and Materials + Nondestructive Evaluation and Health Monitoring, Las Vegas, NV, USA, 20–24 March 2016. [Google Scholar] [CrossRef]
- Breslauer, D.N.; Maamari, R.N.; Switz, N.A.; Lam, W.A.; Fletcher, D.A. Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS ONE 2009, 4, e6320. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, H.; Cong, F.; Yu, Y.; Cui, A. Fiber optic array as an alternative to the optical lens in microscopy: A proof-of-concept study. Measurement 2019, 148, 106959. [Google Scholar] [CrossRef]
- Lodolo, E.J.; Kock, J.L.; Axcell, B.C.; Brooks, M. The yeast Saccharomyces cerevisiae–the main character in beer brewing. Fems Yeast Res. 2008, 8, 1018–1036. [Google Scholar] [CrossRef]
- Nielsen, J. Yeast cell factories on the horizon. Science 2015, 349, 1050–1051. [Google Scholar] [CrossRef]
- Caspeta, L.; Buijs, N.A.; Nielsen, J. The role of biofuels in the future energy supply. Energ Environ. Sci. 2013, 6, 1077–1082. [Google Scholar] [CrossRef]
- Amorim, H.V.; Lopes, M.L.; de Castro Oliveira, J.V.; Buckeridge, M.S.; Goldman, G.H. Scientific challenges of bioethanol production in Brazil. Appl. Microbiol. Biot. 2011, 91, 1267. [Google Scholar] [CrossRef]
- White, C.; Zainasheff, J. Yeast: The Practical Guide to Beer Fermentation; Brewers Publications: Boulder, CO, USA, 2010. [Google Scholar]
- Feizi, A.; Zhang, Y.; Greenbaum, A.; Guziak, A.; Luong, M.; Chan, R.Y.L.; Berg, B.; Ozkan, H.; Luo, W.; Wu, M.; et al. Rapid, portable and cost-effective yeast cell viability and concentration analysis using lensfree on-chip microscopy and machine learning. Lab Chip 2016, 16, 4350–4358. [Google Scholar] [CrossRef]
- Linder, E.; Grote, A.; Varjo, S.; Linder, N.; Lebbad, M.; Lundin, M.; Diwan, V.; Hannuksela, J.; Lundin, J. On-chip imaging of Schistosoma haematobium eggs in urine for diagnosis by computer vision. PLoS Negl. Trop. Dis. 2013, 7. [Google Scholar] [CrossRef]
- Kim, T.K.; Paik, J.K.; Kang, B.S. Contrast enhancement system using spatially adaptive histogram equalization with temporal filtering. IEEE Trans. Consum. Electron. 1998, 44, 82–87. [Google Scholar] [CrossRef]
- Deseng, L. Fiber Optics; Science Press: Beijing, China, 1987. [Google Scholar]
- Li, L.; Huang, Y. Basis of Fibre Optical Communication; National Defense Industry Press: Beijing, China, 1991. [Google Scholar]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Liu, H.; Yu, Y.; Cong, F.; Yu, J. Rapid Yeast Cell Viability Analysis by Using a Portable Microscope Based on the Fiber Optic Array and Simple Image Processing. Sensors 2020, 20, 2092. https://doi.org/10.3390/s20072092
Wang W, Liu H, Yu Y, Cong F, Yu J. Rapid Yeast Cell Viability Analysis by Using a Portable Microscope Based on the Fiber Optic Array and Simple Image Processing. Sensors. 2020; 20(7):2092. https://doi.org/10.3390/s20072092
Chicago/Turabian StyleWang, Weiming, Hang Liu, Yan Yu, Fengyu Cong, and Jun Yu. 2020. "Rapid Yeast Cell Viability Analysis by Using a Portable Microscope Based on the Fiber Optic Array and Simple Image Processing" Sensors 20, no. 7: 2092. https://doi.org/10.3390/s20072092
APA StyleWang, W., Liu, H., Yu, Y., Cong, F., & Yu, J. (2020). Rapid Yeast Cell Viability Analysis by Using a Portable Microscope Based on the Fiber Optic Array and Simple Image Processing. Sensors, 20(7), 2092. https://doi.org/10.3390/s20072092



