Comparison of CNN Algorithms on Hyperspectral Image Classification in Agricultural Lands
Abstract
1. Introduction
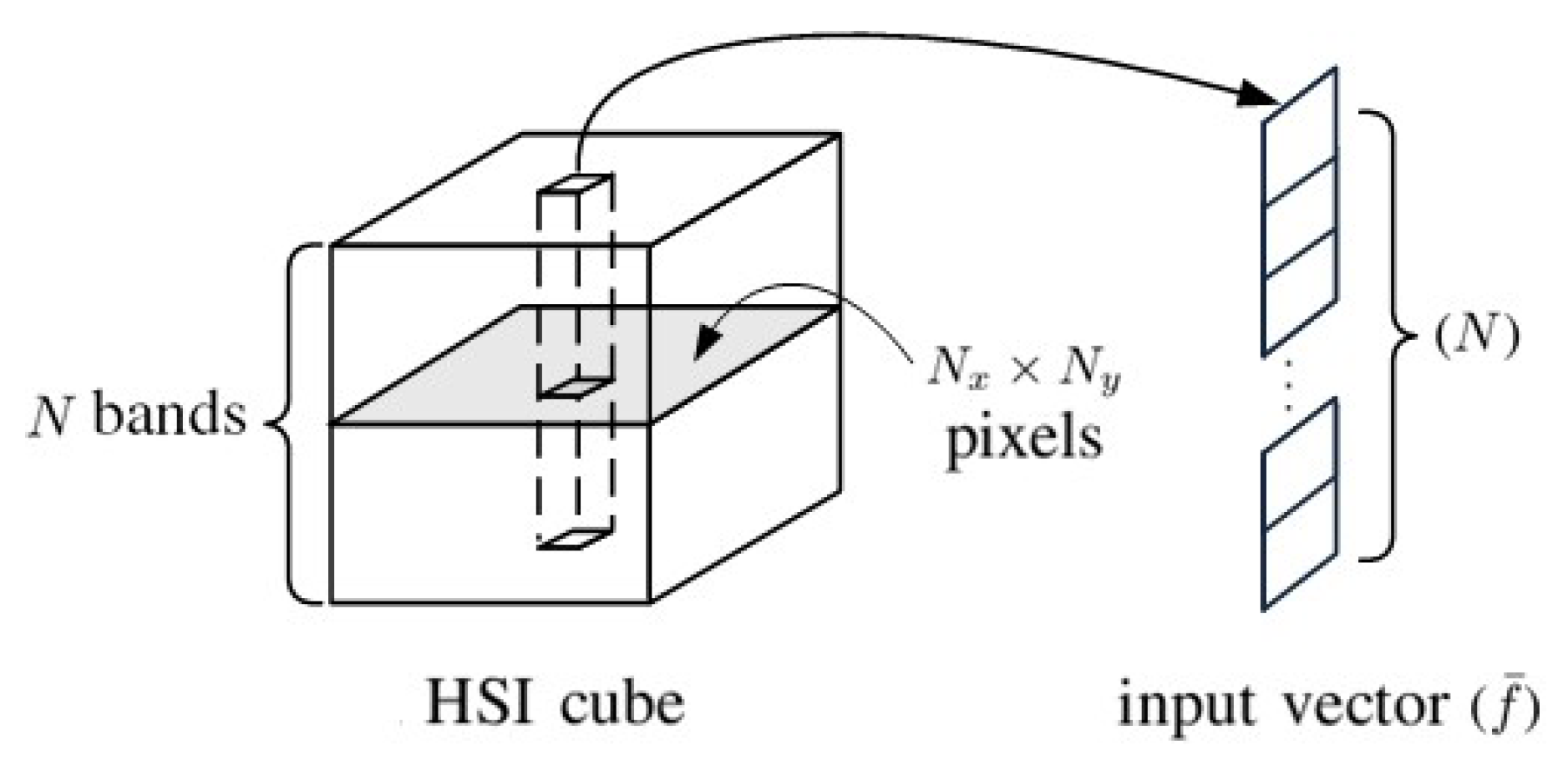
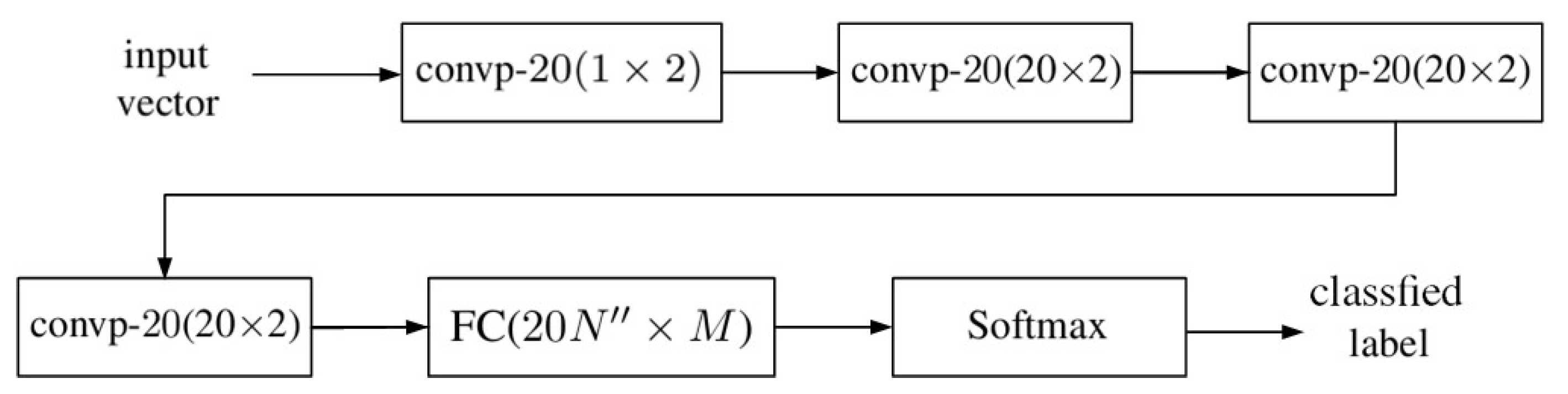
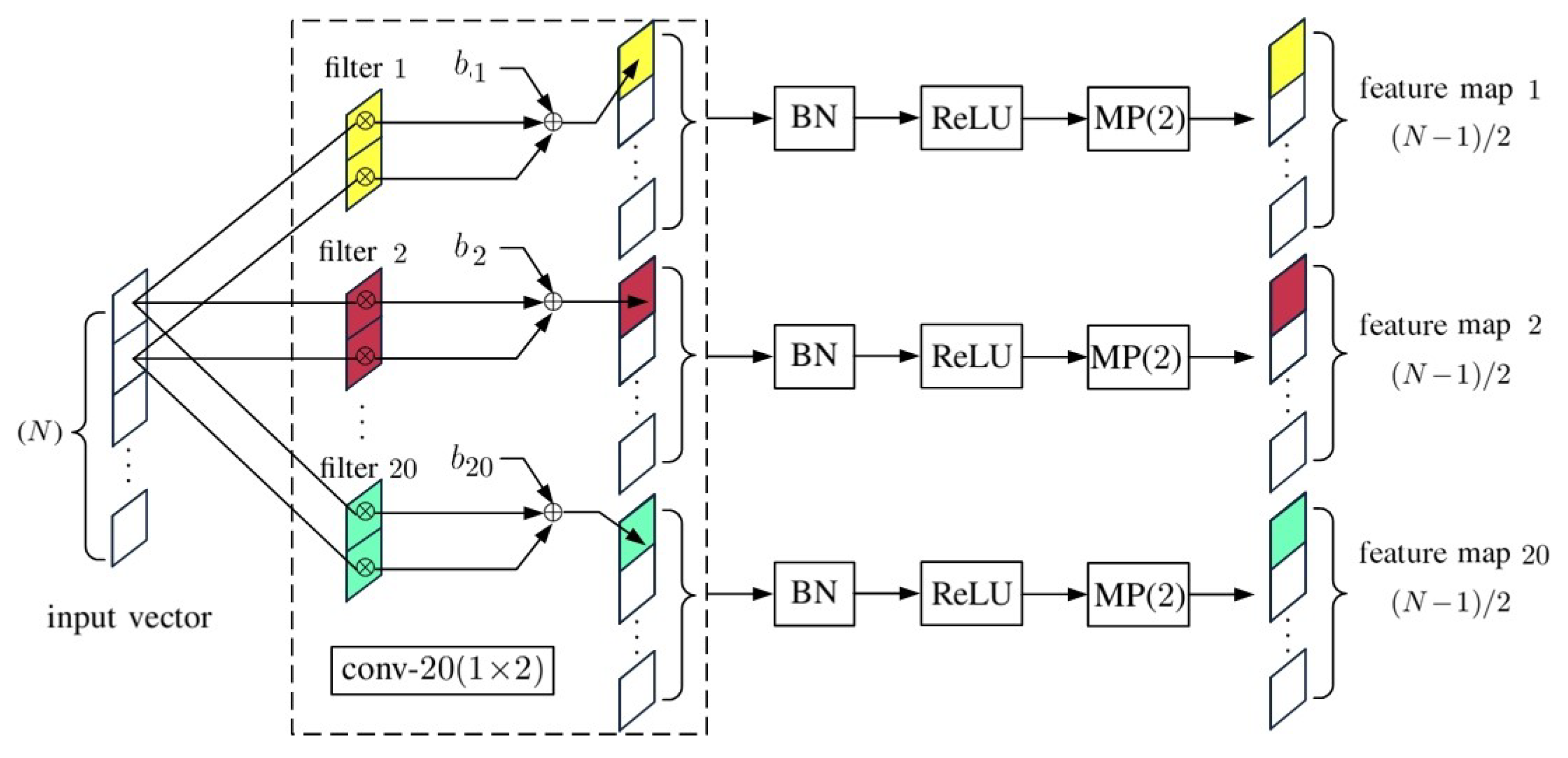
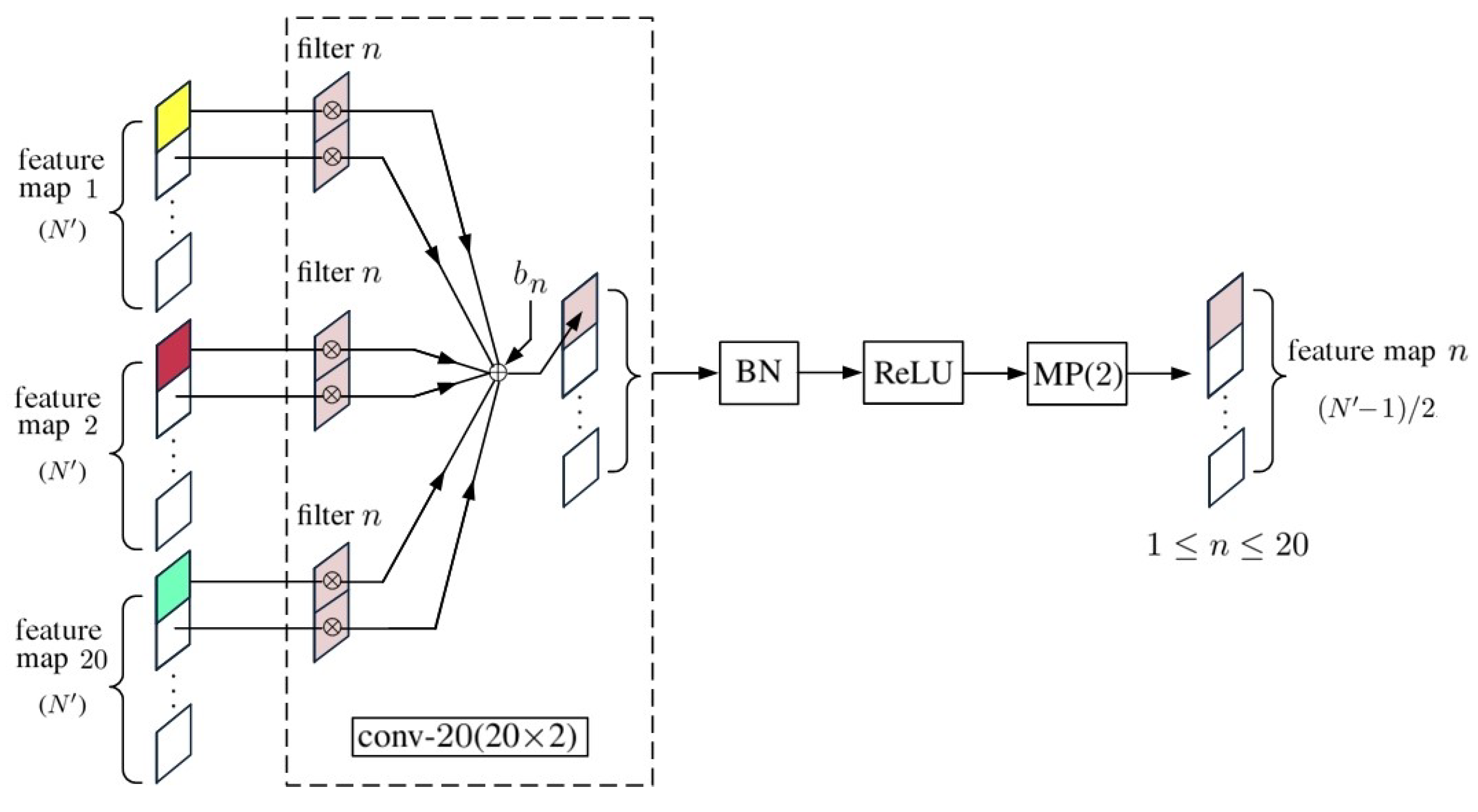
2. 1D-CNN with Pixelwise Spectral Data
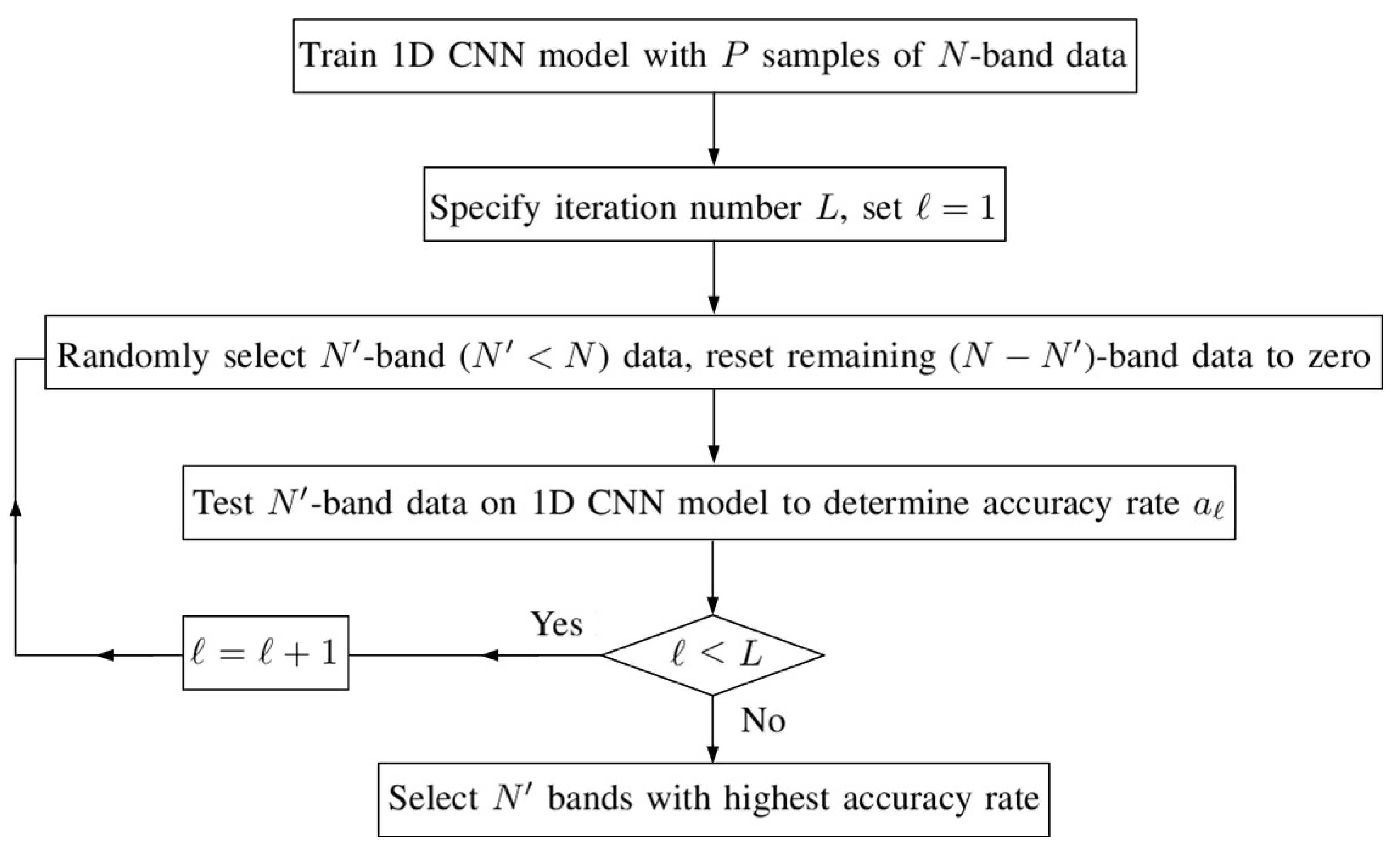
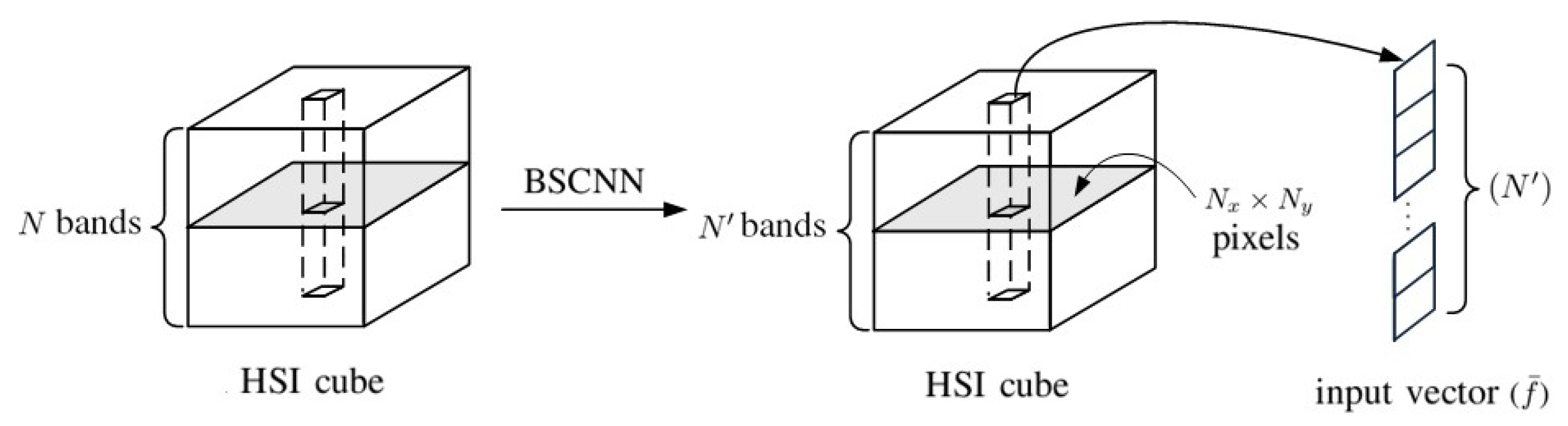
Band Selection Approach
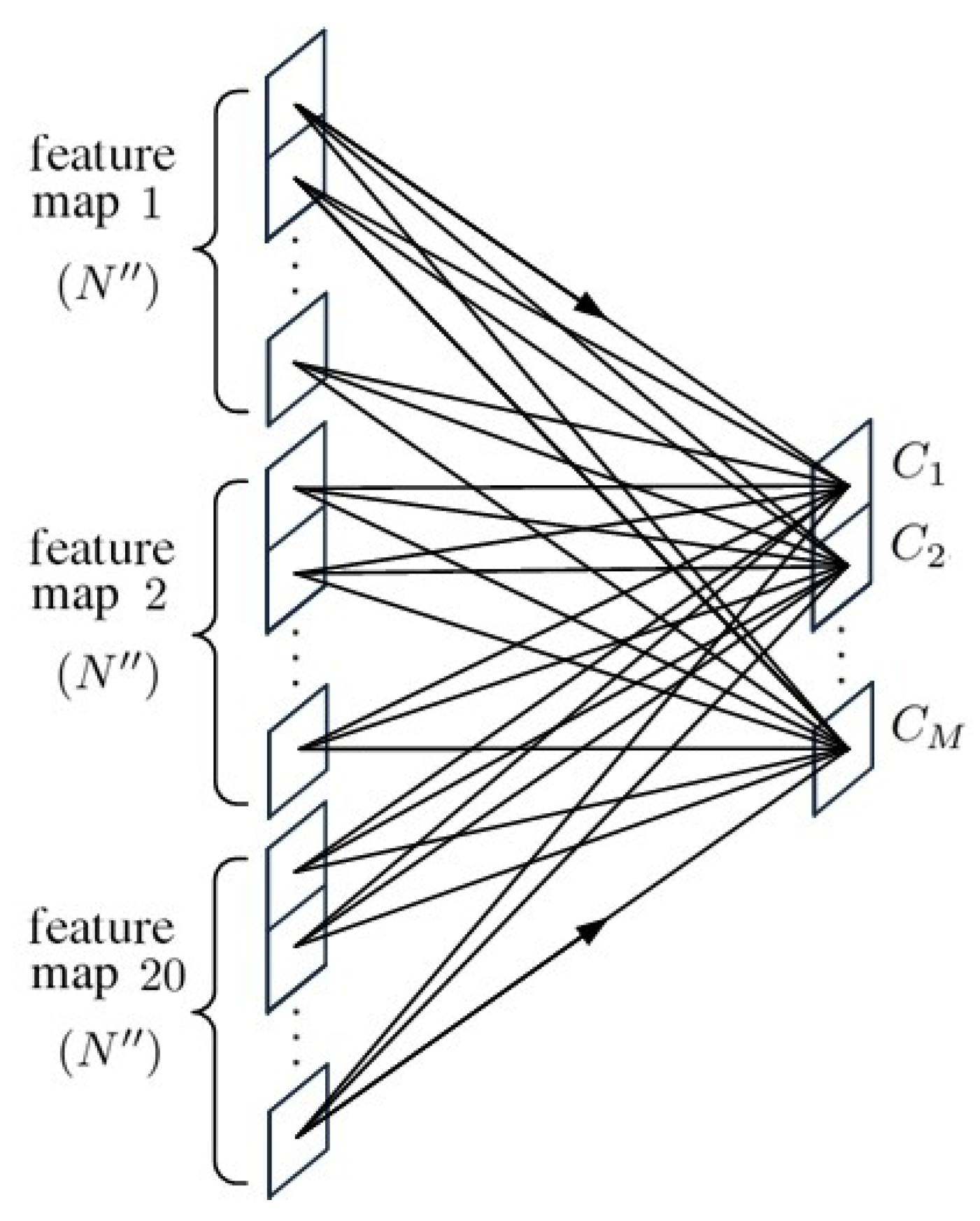
3. 1D-CNN with Spectral-Spatial Data
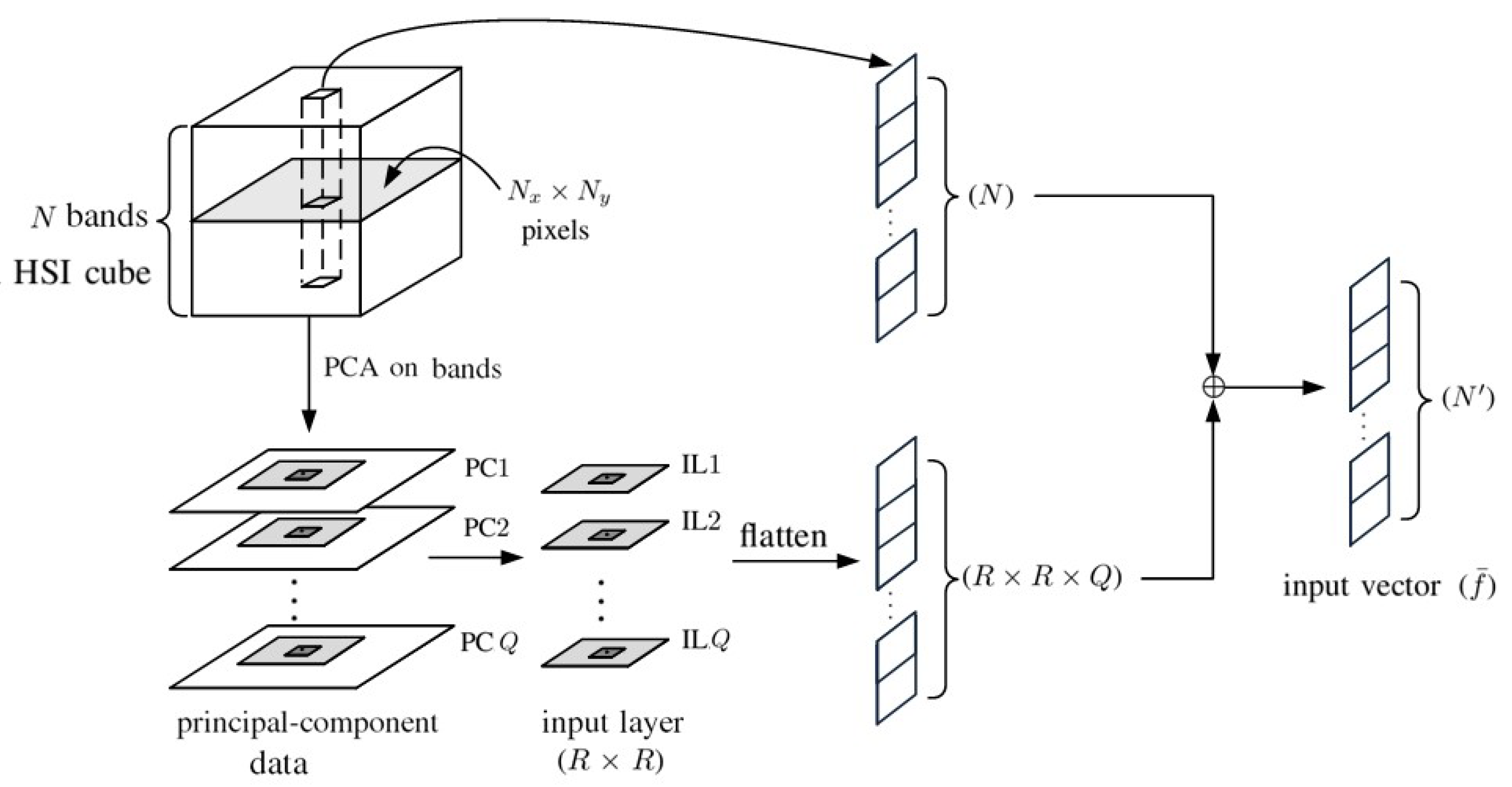
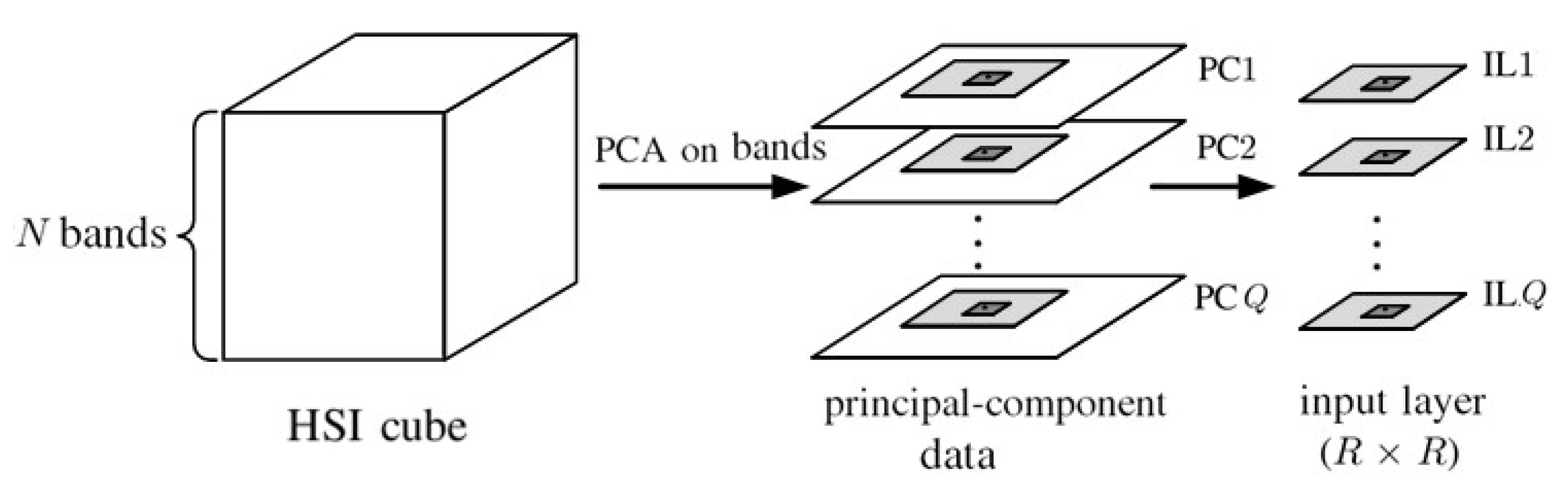
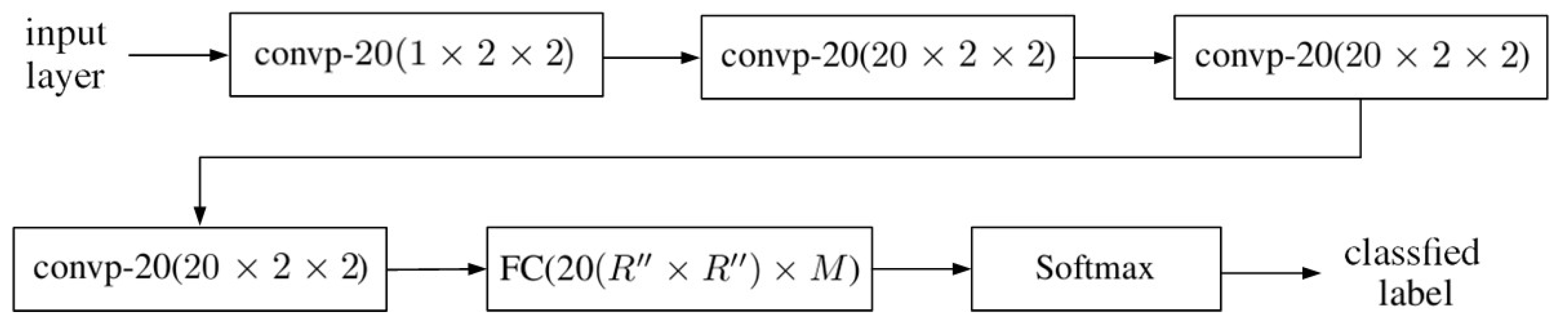
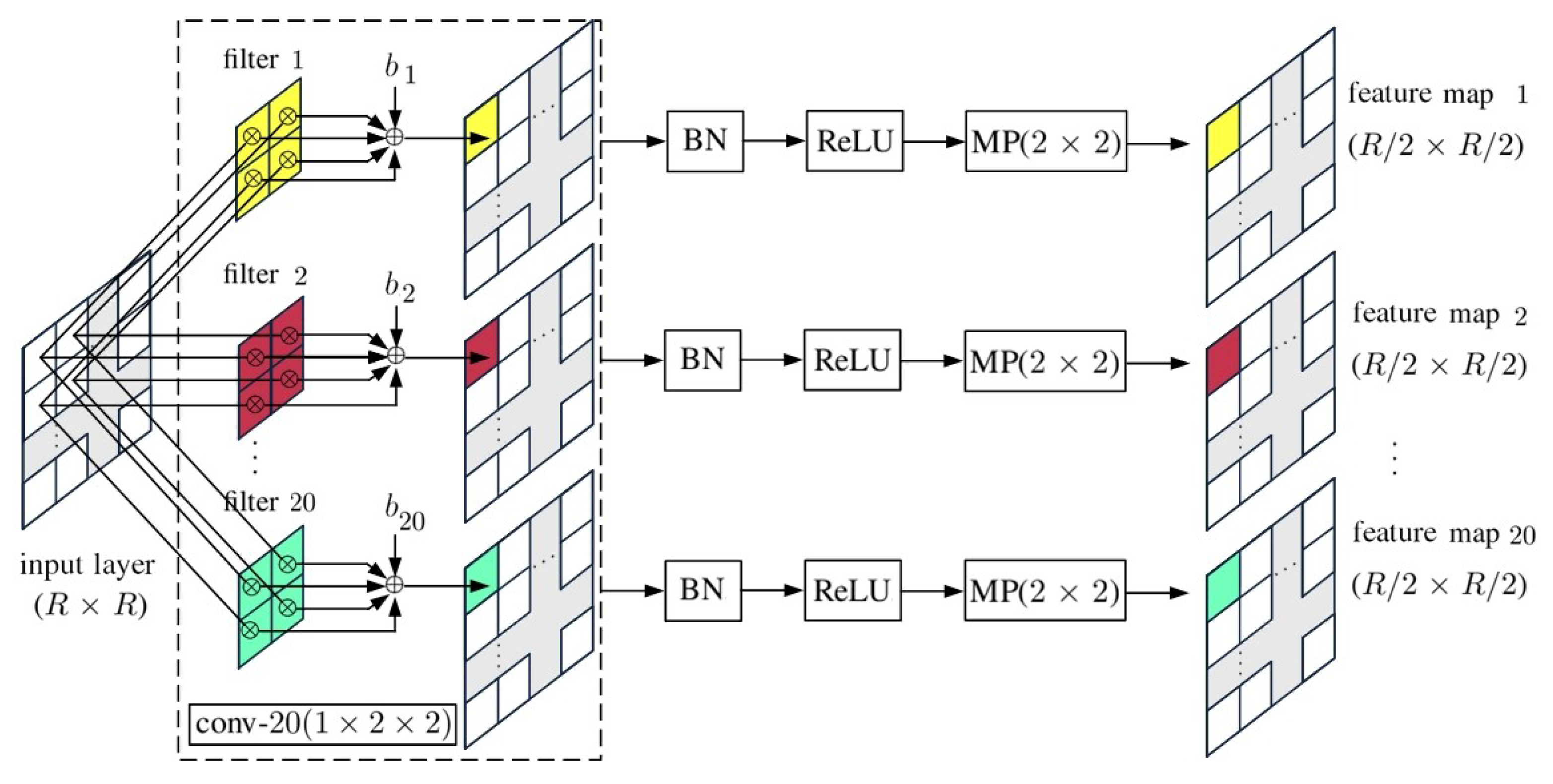
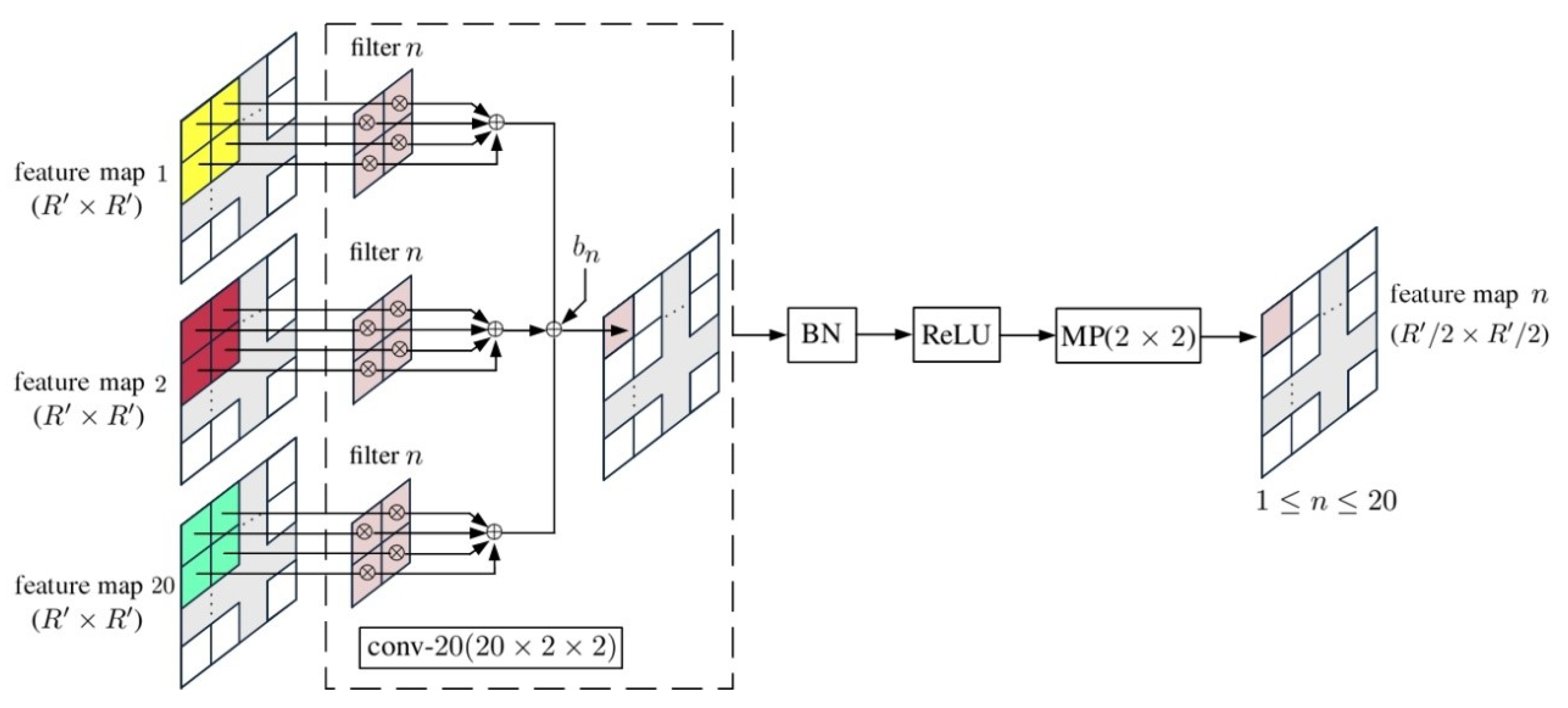
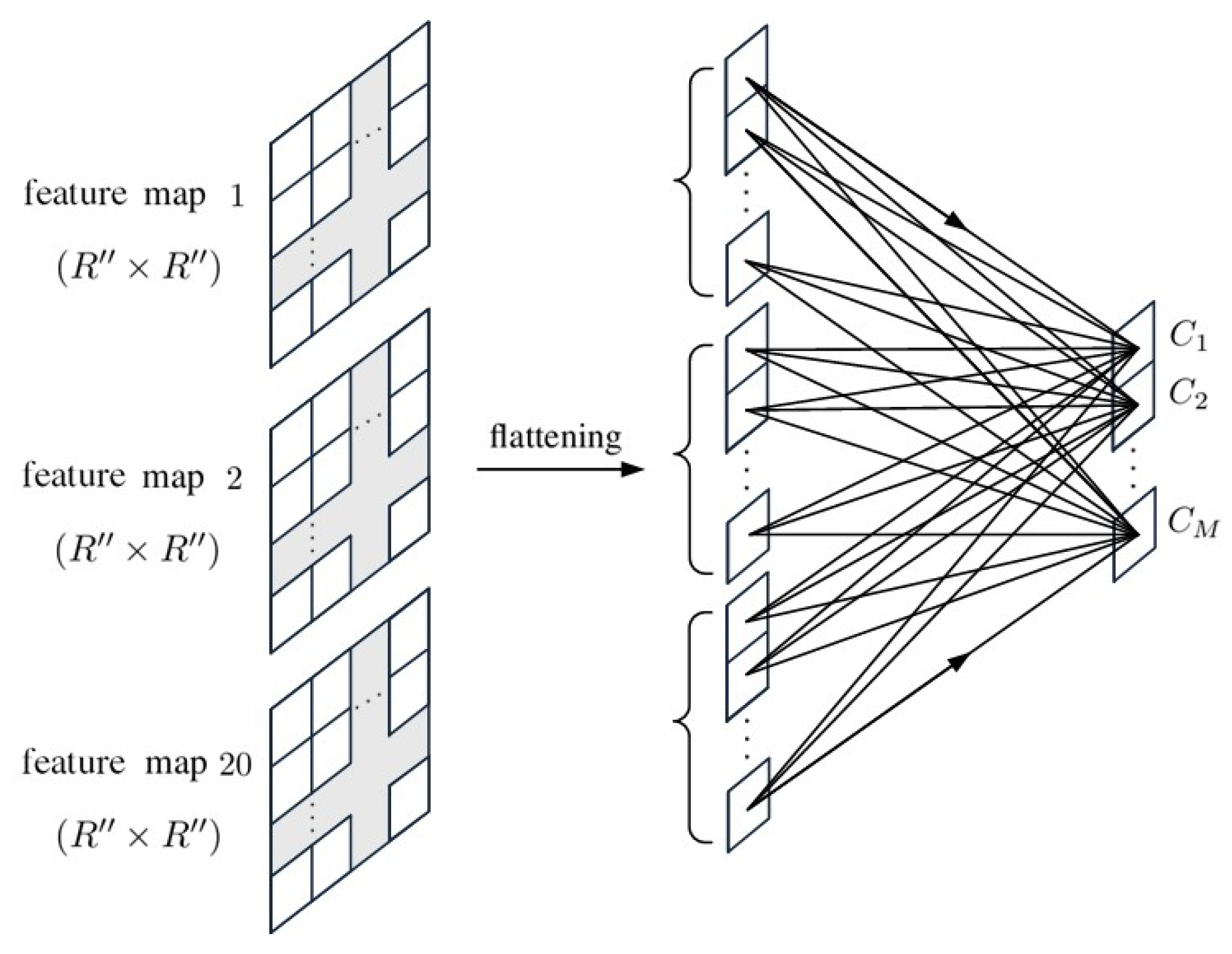
4. 2D-CNN with Principal Components
5. Simulations and Discussions
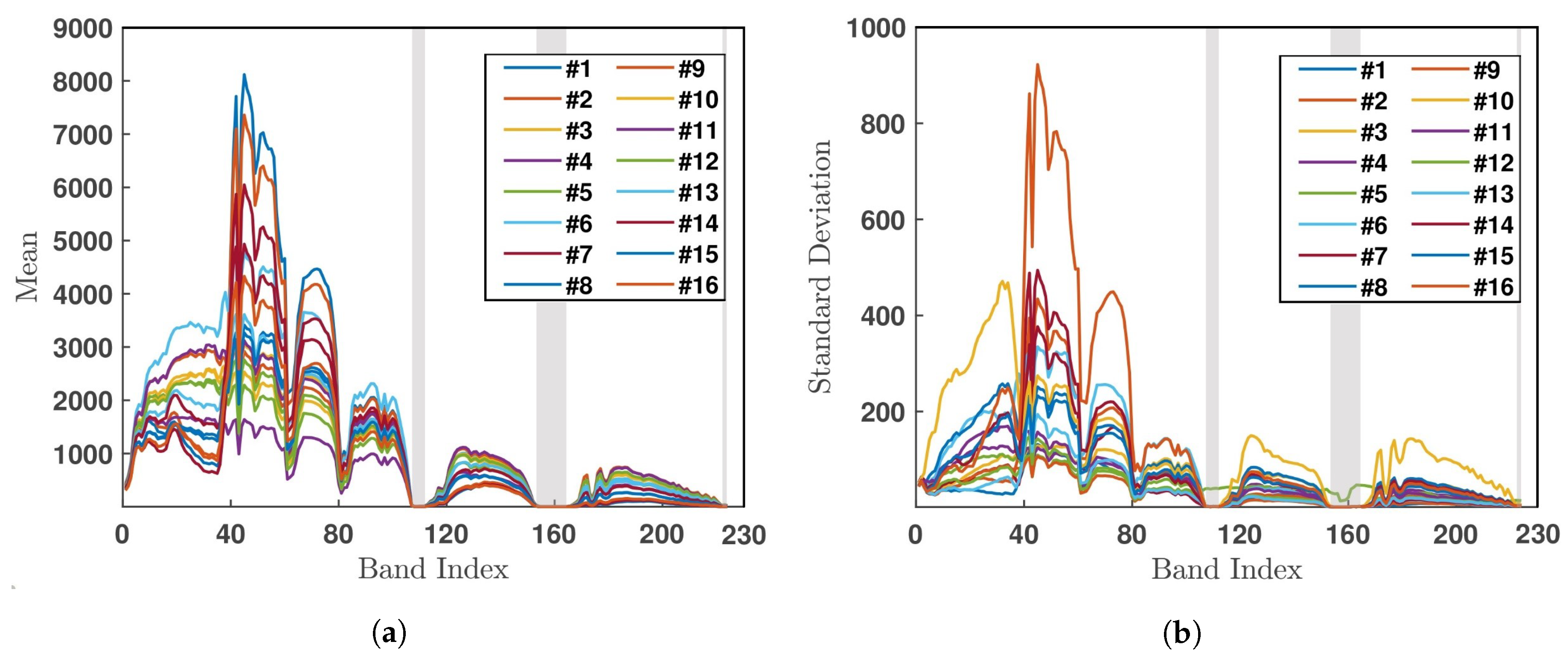
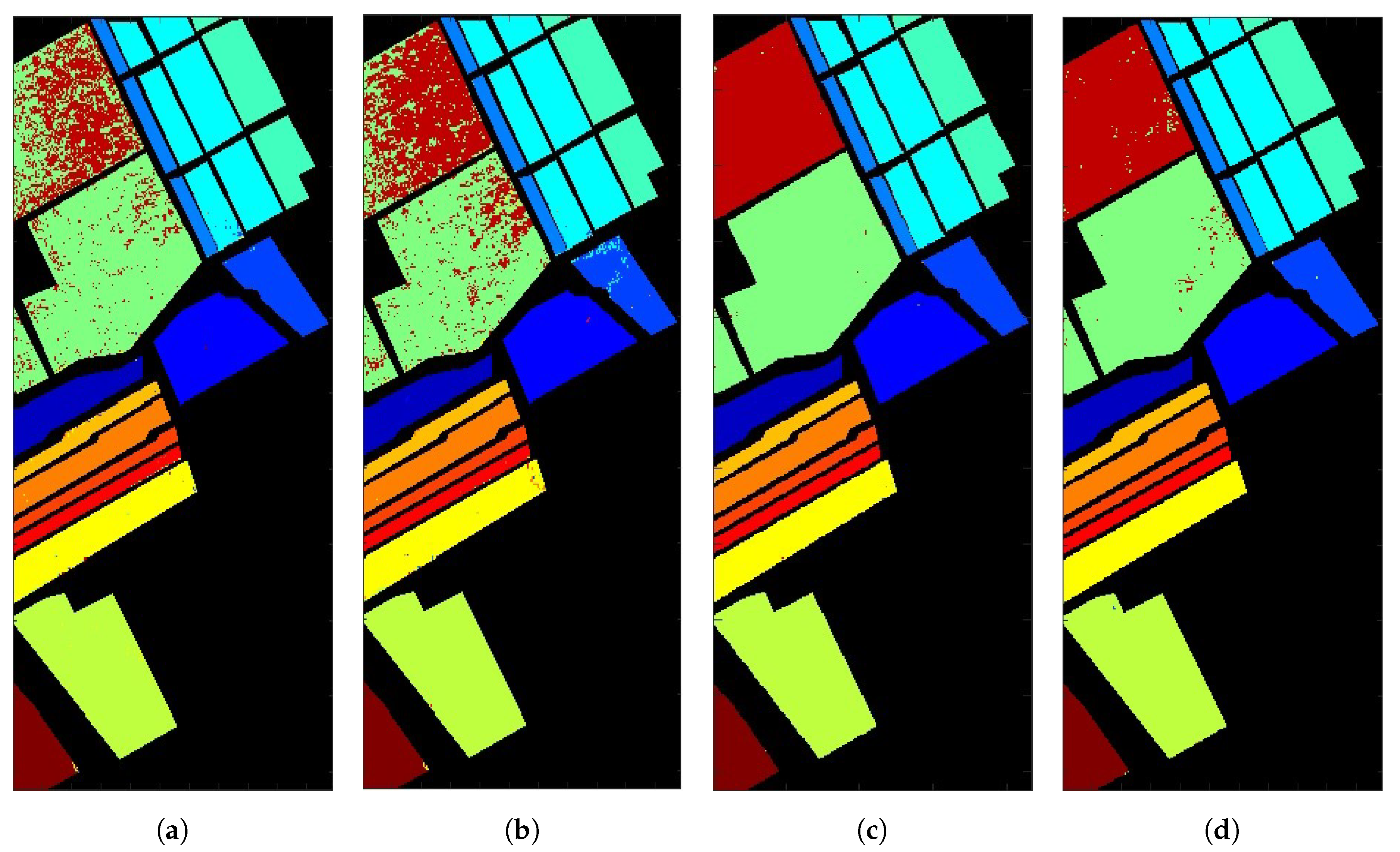
5.1. Salinas Valley HSI
5.2. Indian Pines HSI
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Ham, J.; Chen, Y.; Crawford, M.M.; Ghosh, J. Investigation of the random forest framework for classification of hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 492–501. [Google Scholar] [CrossRef]
- Ma, L.; Crawford, M.M.; Tian, J. Local manifold learning-based k-nearest-neighbor for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4099–4109. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Spectral-spatial hyperspectral image segmentation using subspace multinomial logistic regression and Markov random fields. IEEE Trans. Geosci. Remote Sens. 2012, 50, 809–823. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Jia, X. Spectral-spatial classification of hyperspectral data based on deep belief network. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Mughees, A.; Tao, L. Efficient deep auto-encoder learning for the classification of hyperspectral images. In Proceedings of the International Conference on Virtual Reality and Visualization, Hangzhou, China, 24–26 September 2016. [Google Scholar]
- Farooq, A.; Hu, J.; Jia, X. Weed classification in hyperspectral remote sensing images via deep convolutional neural network. In Proceedings of the IEEE International Symposium Geoscience and Remote Sensing (IGARSS), Valencia, Spain, 22–27 July 2018. [Google Scholar]
- Chova, L.G.; Tuia, D.; Moser, G.; Valls, G.C. Multimodal classification of remote sensing images: A review and future directions. Proc. IEEE 2015, 103, 1560–1584. [Google Scholar] [CrossRef]
- Martinez-Uso, A.; Pla, F.; Sotoca, J.M.; Garcia-Sevilla, P. Clustering-based hyperspectral band selection using information measures. IEEE Trans. Geosci. Remote Sens. 2007, 45, 4158–4171. [Google Scholar] [CrossRef]
- Chang, C.-I.; Wang, S. Constrained band selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1575–1585. [Google Scholar] [CrossRef]
- Rodarmel, C.; Shan, J. Principal component analysis for hyperspectral image classification. Surv. Land Inf. Sci. 2002, 62, 115. [Google Scholar]
- Joy, A.A.; Hasan, M.A.M.; Hossain, M.A. A comparison of supervised and unsupervised dimension reduction methods for hyperspectral image classification. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7–9 February 2019. [Google Scholar]
- Liao, D.; Qian, Y.; Tang, Y.Y. Constrained manifold learning for hyperspectral imagery visualization. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2018, 11, 1213–1226. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, Y.; Zhang, L.; Xu, Y. Spatial group sparsity regularized nonnegative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6287–6304. [Google Scholar] [CrossRef]
- Feng, J.; Chen, J.; Liu, L.; Cao, X.; Zhang, X.; Jiao, L.; Yu, T. CNN-based multilayer spatial-spectral feature fusion and sample augmentation with local and nonlocal constraints for hyperspectral image classification. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2019, 12, 1299–1313. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, X.; Cheng, Y. Spectral-spatial feature extraction for HSI classification based on supervised hypergraph and sample expanded CNN. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2018, 11, 4128–4140. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhu, G.; Wang, Q. Hyperspectral band selection by multitask sparsity pursuit. IEEE Trans. Geosci. Remote Sens. 2015, 53, 631–644. [Google Scholar] [CrossRef]
- Zhu, G.; Huang, Y.; Lei, J.; Bi, Z.; Xu, F. Unsupervised hyperspectral band selection by dominant set extraction. IEEE Trans. Geosci. Remote Sens. 2016, 54, 227–239. [Google Scholar] [CrossRef]
- Patra, S.; Modi, P.; Bruzzone, L. Hyperspectral band selection based on rough set. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5495–5503. [Google Scholar] [CrossRef]
- Bai, X.; Guo, Z.; Wang, Y.; Zhang, Z.; Zhou, J. Semisupervised hyperspectral band selection via spectral-spatial hypergraph model. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2015, 8, 2774–2783. [Google Scholar] [CrossRef]
- Damodaran, B.B.; Courty, N.; Lefevre, S. Sparse Hilbert Schmidt independence criterion and surrogate-kernel-based feature selection for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2385–2398. [Google Scholar] [CrossRef]
- Feng, S.; Itoh, Y.; Parente, M.; Duarte, M.F. Hyperspectral band selection from statistical wavelet models. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2111–2123. [Google Scholar] [CrossRef]
- Chang, C.-I.; Du, Q.; Sun, T.-L.; Althouse, M.L.G. A joint band prioritization and band-decorrelation approach to band selection for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2631–2641. [Google Scholar] [CrossRef]
- Cao, X.; Xiong, T.; Jiao, L. Supervised band selection using local spatial information for hyperspectral image. IEEE Geosci. Remote Sens. Lett. 2016, 13, 329–333. [Google Scholar] [CrossRef]
- Mei, S.; He, M.; Wang, Z.; Feng, D. Spatial purity based endmember extraction for spectral mixture analysis. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3434–3445. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Ayerdi, B.; Graña, M. Hyperspectral image analysis by spectral-spatial processing and anticipative hybrid extreme rotation forest classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 2627–2639. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef]
- Zarco-Tejada, P. Chlorophyll fluorescence effects on vegetation apparent reflectance: I. Leaf-level measurements and model simulation. Remote Sens. Environ. 2000, 74, 582–595. [Google Scholar] [CrossRef]
- Penuelas, J.; Filella, I.; Biel, C.; Save, R.; Serrano, L. The reflectance at the 950–970 nm region as an indicator of plant water status. Int. J. Remote Sens. 1993, 14, 1887–1905. [Google Scholar] [CrossRef]
- Galvao, L.S.; Pizarro, M.A.; Epiphanio, J.C.N. Variations in reflectance of tropical soils: Spectral-chemical composition relationships from AVIRIS data. Remote Sens. Environ. 2001, 75, 245–255. [Google Scholar] [CrossRef]
- Summers, D.; Lewis, M.; Ostendorf, B.; Chittleborough, D. Visible near-infrared reflectance spectroscopy as a predictive indicator of soil properties. Ecol. Indicators 2011, 11, 123–131. [Google Scholar] [CrossRef]
- Strachan, I.; Pattey, E.; Boisvert, J.B. Impact of nitrogen and environmental conditions on corn as detected by hyperspectral reflectance. Remote Sens. Environ. 2002, 80, 213–224. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Evangelista, P.; Stohlgren, T.J.; Morisette, J.T.; Kumar, S. Mapping invasive tamarisk (tamarix): A comparison of single-scene and time-series analyses of remotely sensed data. Remote Sens. 2009, 1, 519–533. [Google Scholar] [CrossRef]
- Wang, L. Invasive species spread mapping using multi-resolution remote sensing data. Int. Arch. Photogram. Remote Sens. Spatial Info. Sci. 2008, 37, 135–142. [Google Scholar]
- Lawrence, R.; Labus, M. Early detection of Douglas-Fir beetle infestation with subcanopy resolution hyperspectral imagery. Western J. Appl. For. 2003, 18, 202–206. [Google Scholar] [CrossRef]
- Sampson, P.H.; Zarco-Tejada, P.J.; Mohammed, G.H.; Miller, J.R.; Noland, T.L. Hyperspectral remote sensing of forest condition: Estimating chlorophyll content in tolerant hardwoods. For. Sci. 2003, 49, 381–391. [Google Scholar]
- Carter, G.A.; Miller, R.L. Early detection of plant stress by digital imaging within narrow stress-sensitive wavebands. Remote Sens. Environ. 1994, 50, 295–302. [Google Scholar] [CrossRef]
- Smith, K.L.; Steven, M.D.; Colls, J.J. Use of hyperspectral derivative ratios in the red-edge region to identify plant stress responses to gas leaks. Remote Sens. Environ. 2004, 92, 207–217. [Google Scholar] [CrossRef]
- Bellante, G.J.; Powell, S.L.; Lawrence, R.L.; Repasky, K.S.; Dougher, T.A. Aerial detection of a simulated CO2 leak from a geologic sequestration site using hyperspectral imagery. Int. J. Greenhouse Gas Control 2013, 13, 124–137. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Phil. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Hyperspectral Remote Sensing Dataset. Available online: http://www.ehu.eus/ccwintco/index.php/Hyperspectral_Remote_Sensing_Scenes (accessed on 15 August 2019).
- Airborne Visible/Infrared Imaging Spectrometer. Available online: https://aviris.jpl.nasa.gov/ (accessed on 15 August 2019).
- Zhu, Z.; Woodcock, C.E.; Rogan, J.; Kellndorfer, J. Assessment of spectral, polarimetric, temporal, and spatial dimensions for urban and peri-urban land cover classification using Landsat and SAR data. Remote Sens. Environ. 2012, 117, 72–82. [Google Scholar] [CrossRef]
- Baumgardner, M.F.; Biehl, L.L.; Landgrebe, D.A. 220 Band AVIRIS Hyperspectral Image Data Set: June 12, 1992 Indian Pine Test Site 3; Purdue University Research Repository: West Lafayette, ID, USA, 2015. [Google Scholar]
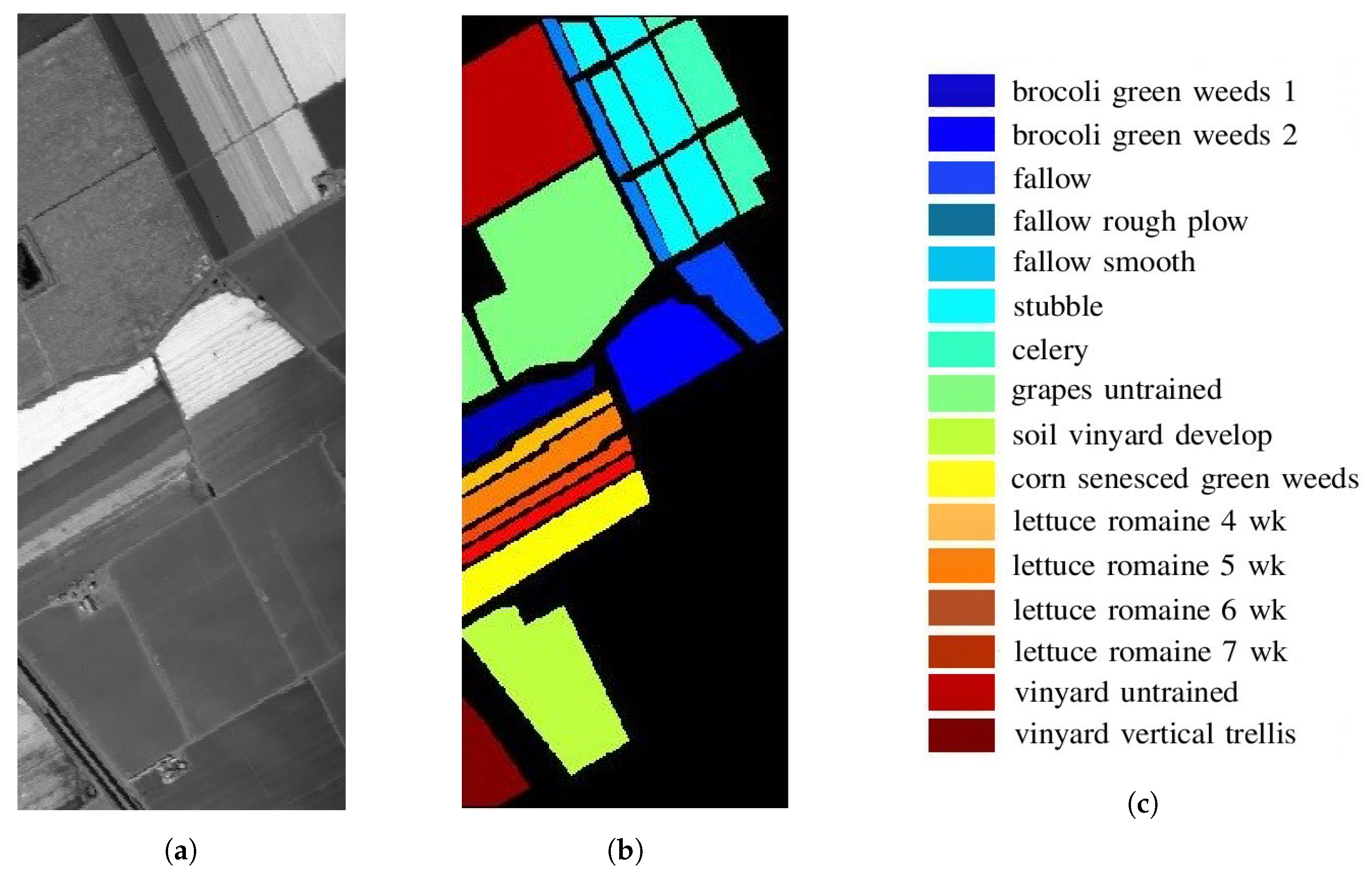

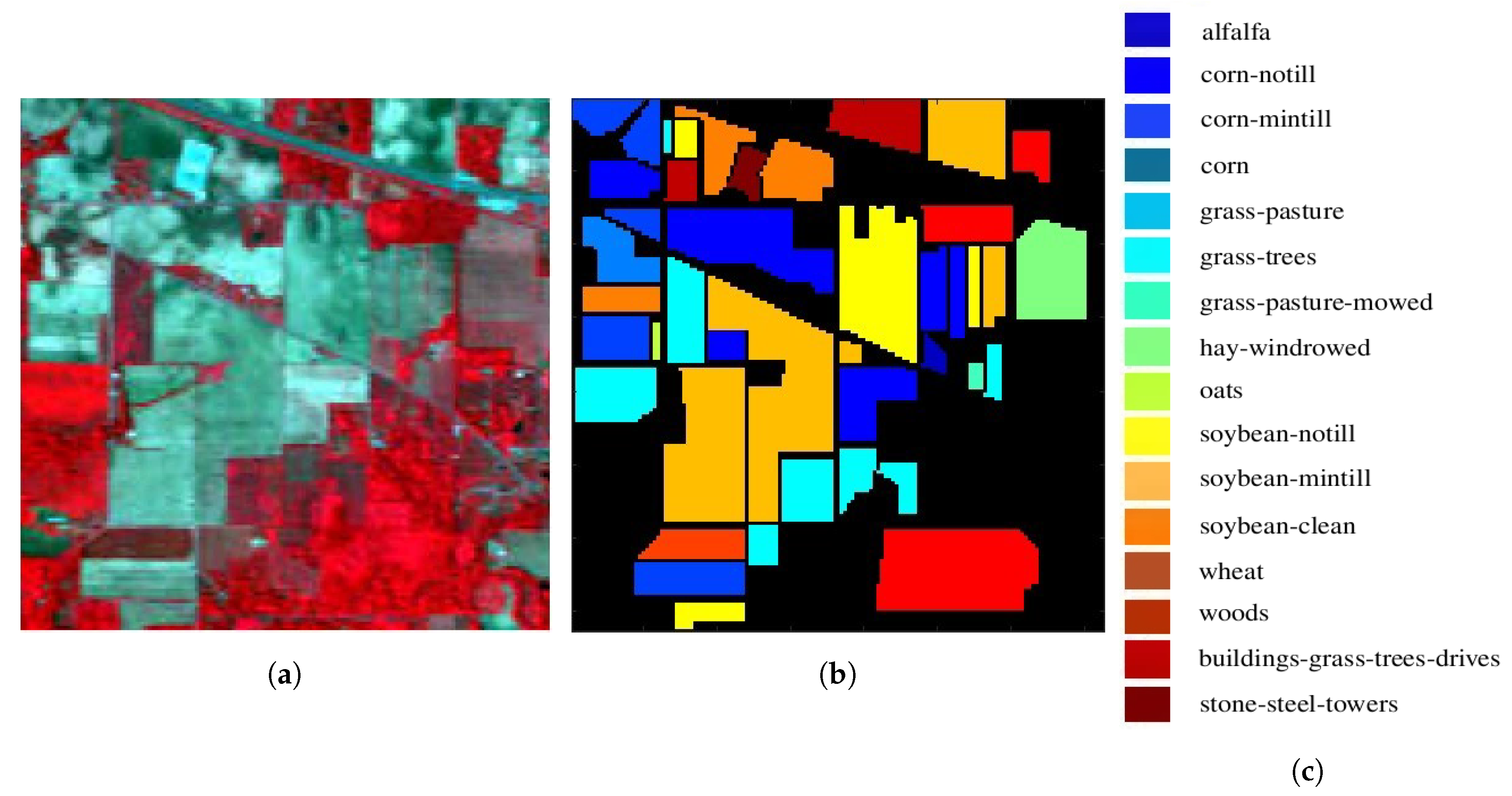
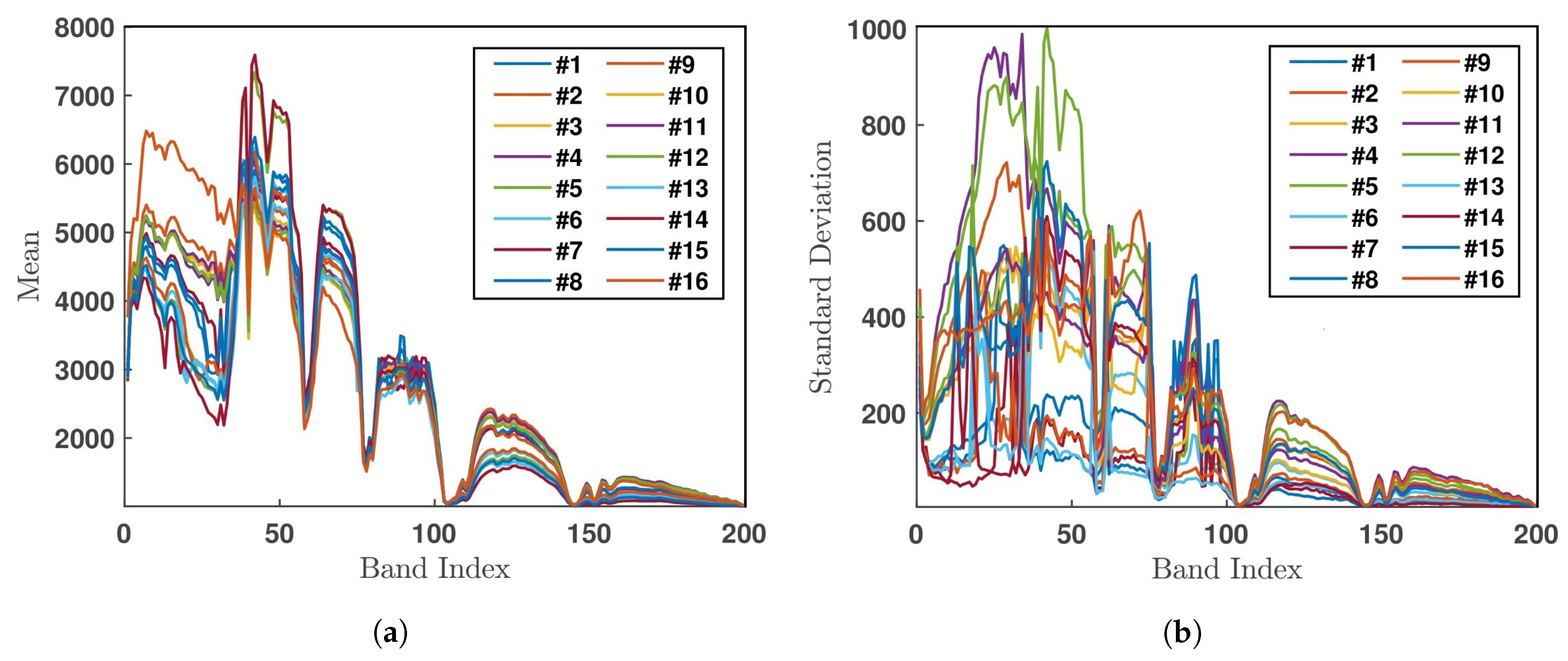

| # | Class | Sample Number |
|---|---|---|
| 1 | broccoli green weeds 1 | 2009 |
| 2 | broccoli green weeds 2 | 3726 |
| 3 | fallow | 1976 |
| 4 | fallow rough plow | 1394 |
| 5 | fallow smooth | 2678 |
| 6 | stubble | 3959 |
| 7 | celery | 3579 |
| 8 | grapes untrained | 11,271 |
| 9 | soil vineyard develop | 6203 |
| 10 | corn senesced green weeds | 3278 |
| 11 | lettuce romaine 4 weeks | 1068 |
| 12 | lettuce romaine 5 weeks | 1927 |
| 13 | lettuce romaine 6 weeks | 916 |
| 14 | lettuce romaine 7 weeks | 1070 |
| 15 | vineyard untrained | 7268 |
| 16 | vineyard vertical trellis | 1807 |
| #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | #13 | #14 | #15 | #16 | OA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | 99.8 | 99.8 | 99.8 | 99.9 | 98.1 | 100 | 99.8 | 91.9 | 99.6 | 98.2 | 92 | 99.8 | 100 | 97.5 | 54.8 | 99.7 | 91.8 |
| (b) | 99.8 | 99.8 | 95.6 | 99.4 | 98.3 | 99.9 | 99.8 | 86.1 | 100 | 97.5 | 99.3 | 100 | 99.1 | 99.1 | 74.8 | 99.3 | 93.2 |
| (c) | 99.8 | 99.9 | 100 | 99.9 | 99.9 | 100 | 99.9 | 99.8 | 99.9 | 99.7 | 100 | 99.9 | 99.8 | 100 | 99.7 | 99.9 | 99.8 |
| (d) | 99.7 | 100 | 99.8 | 100 | 99.6 | 100 | 100 | 97.6 | 99.7 | 99.9 | 100 | 100 | 100 | 100 | 97.1 | 99.8 | 99 |
| # | Class | Sample Number |
|---|---|---|
| 1 | alfalfa | 46 |
| 2 | corn-notill | 1428 |
| 3 | corn-mintill | 830 |
| 4 | corn | 237 |
| 5 | grass-pasture | 483 |
| 6 | grass-trees | 730 |
| 7 | grass-pasture-mowed | 28 |
| 8 | hay-windrowed | 478 |
| 9 | oats | 20 |
| 10 | soybean-notill | 972 |
| 11 | soybean-mintill | 2455 |
| 12 | soybean-clean | 593 |
| 13 | wheat | 205 |
| 14 | woods | 1265 |
| 15 | buildings-grass-trees-drives | 386 |
| 16 | stone-steel-towers | 93 |
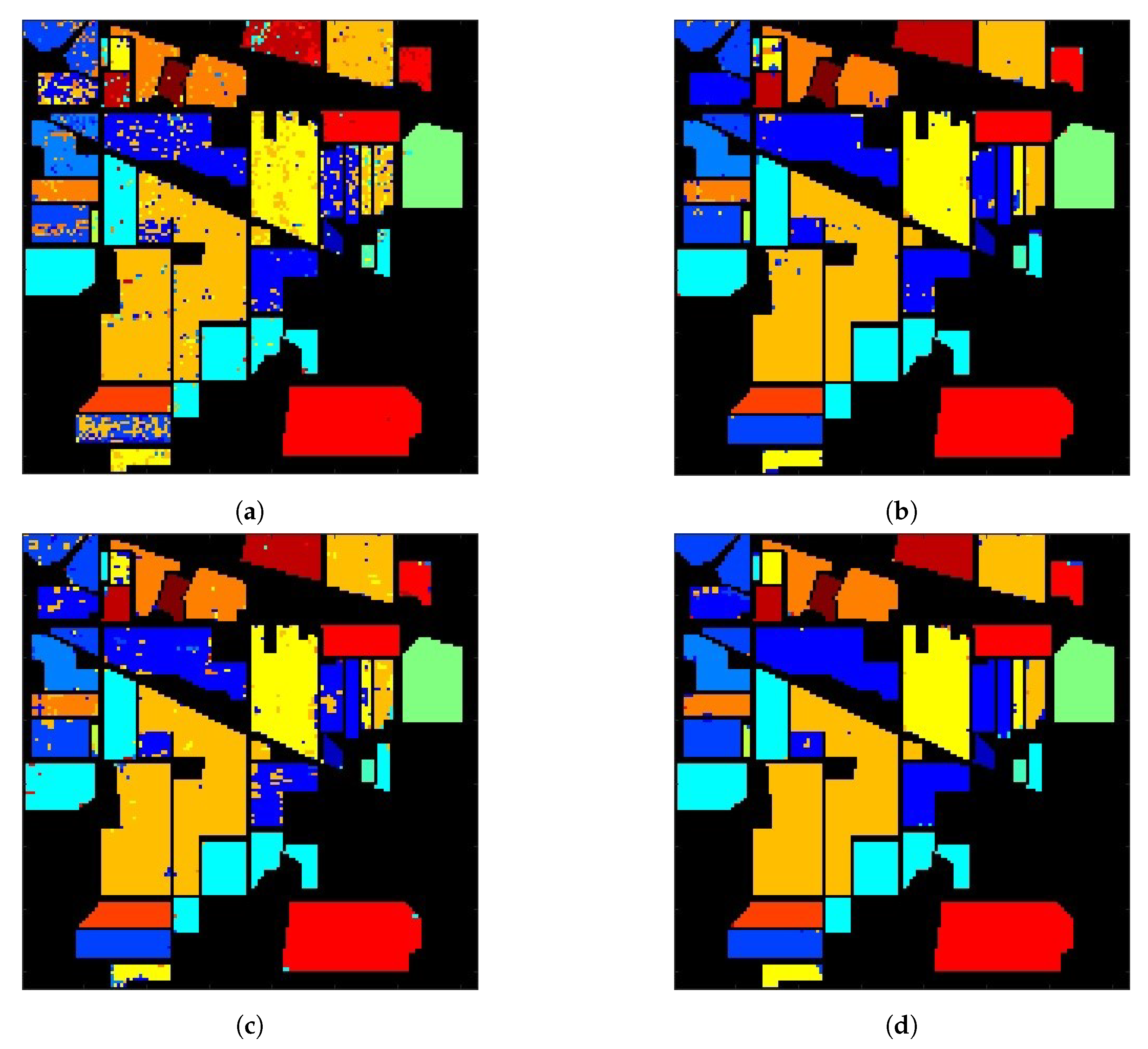
| #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | #13 | #14 | #15 | #16 | OA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | 83.3 | 76.1 | 55.2 | 82 | 94.4 | 96.5 | 57.1 | 98.7 | 77.8 | 73.3 | 87.2 | 84.9 | 99 | 96.6 | 62.2 | 91.7 | 83.4 |
| (b) | 95.2 | 92.8 | 94 | 97.5 | 89.1 | 99.2 | 94.7 | 99.6 | 71.4 | 89.7 | 97.5 | 88.4 | 100 | 98.9 | 99.5 | 100 | 95.4 |
| (c) | 90.5 | 82.2 | 87.4 | 92 | 90.7 | 96.5 | 100 | 100 | 58.3 | 82.1 | 95.1 | 88 | 100 | 97.7 | 96.9 | 97.7 | 91.5 |
| (d) | 100 | 94.9 | 98.6 | 97.5 | 99.2 | 99.7 | 100 | 100 | 90.9 | 96.7 | 98.8 | 97.2 | 99 | 99.2 | 99.5 | 98.2 | 98.1 |
| HSI on Salinas Valley | CPU Time | HSI on Indian Pines | CPU Time |
|---|---|---|---|
| 1D-CNN with 204 bands | 1 h 43 min | 1D-CNN with 200 bands | 18 min |
| BSCNN with 70 selected bands | 1 h 35 min | 1D-CNN with augmented input vectors of 641 bands | 26 min |
| 1D-CNN with augmented input vectors of 645 bands | 6 h 58 min | 2D-CNN with input layer composed of one principal component from each pixel | 17 min |
| 2D-CNN with input layer composed of one principal component from each pixel | 1 h 32 min | 1D-CNN with augmented input vectors of 1964 bands | 1 h 27 min |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, T.-H.; Kiang, J.-F. Comparison of CNN Algorithms on Hyperspectral Image Classification in Agricultural Lands. Sensors 2020, 20, 1734. https://doi.org/10.3390/s20061734
Hsieh T-H, Kiang J-F. Comparison of CNN Algorithms on Hyperspectral Image Classification in Agricultural Lands. Sensors. 2020; 20(6):1734. https://doi.org/10.3390/s20061734
Chicago/Turabian StyleHsieh, Tien-Heng, and Jean-Fu Kiang. 2020. "Comparison of CNN Algorithms on Hyperspectral Image Classification in Agricultural Lands" Sensors 20, no. 6: 1734. https://doi.org/10.3390/s20061734
APA StyleHsieh, T.-H., & Kiang, J.-F. (2020). Comparison of CNN Algorithms on Hyperspectral Image Classification in Agricultural Lands. Sensors, 20(6), 1734. https://doi.org/10.3390/s20061734





