2.1. Respiratory Sounds Detection
Lung auscultation provides valuable information regarding the respiratory function of the patient, and it is important to analyze respiratory sounds using an algorithm to give support to medical doctors. There are a few methods in the literature to deal with this challenge. Typically, wheezing is found in asthma and chronic obstructive lung diseases. Wheezes can be so loud you can hear it just by standing next to the patient. Crackles, on the other hand, are only heard using a stethoscope, and they are a sign of too much fluid in the lung. Crackles and wheezes are indications of the pathology.
Islam et al. [
6] detected asthma pathology by basing their research on the fact that asthma detection from lung sound signals rely on the presence of wheeze. They collected lung sounds from 60 subjects in which the 50% had asthma and using a data acquisition system from four different positions on the back of the chest. For the classification step, ANN (Artificial Neural Networks) and SVM (Support Vector Machine) were used with the best results (93.3%) obtained in the SVM scenario.
Other studies based on the detection of wheezes and crackles [
7] used different configurations of a neural network, obtaining results of up to 93% for detecting crackles and 91.7% for wheezes. The same goal is pursued in Reference [
8], but the dataset used in this case consists of seven classes: normal, coarse crackle, fine crackle, monophonic wheeze, polyphonic wheeze, squawk, and stridor. The best results were achieved using a Convolutional Neural Network (CNN). Chen et al. [
9] proposed ResNet with an OST-based (Optimized S-Transform based) feature map to classify wheeze, crackle, and normal sounds. In detail, three RGB -maps (Red-Green-Blue) of the rescaled feature map is fed into ResNet due to the balance between the depth and performance. The input feature map is passed through three steps of the ResNet structure and finally, the output corresponds to the class (wheeze, crackle, or normal). The results are compared with ResNet-STFT (Short Term Fourier Transform) and ResNet-ST (S-Transform), with the best accuracy achieved using their proposal ResNet-OST.
In Reference [
10], the authors propose a methodology to classify the respiratory sounds into wheezes, crackles, both wheezes and crackles, and normal using the same dataset as that used in our work: ICBHI [
2]. The procedure consists of a noise suppression step using spectral subtraction followed by a feature extraction process. Hidden Markov Models were used in the classification step obtaining 39.56% using the score metric, defined as the average of sensitivity and specificity. These results are not promising but in Reference [
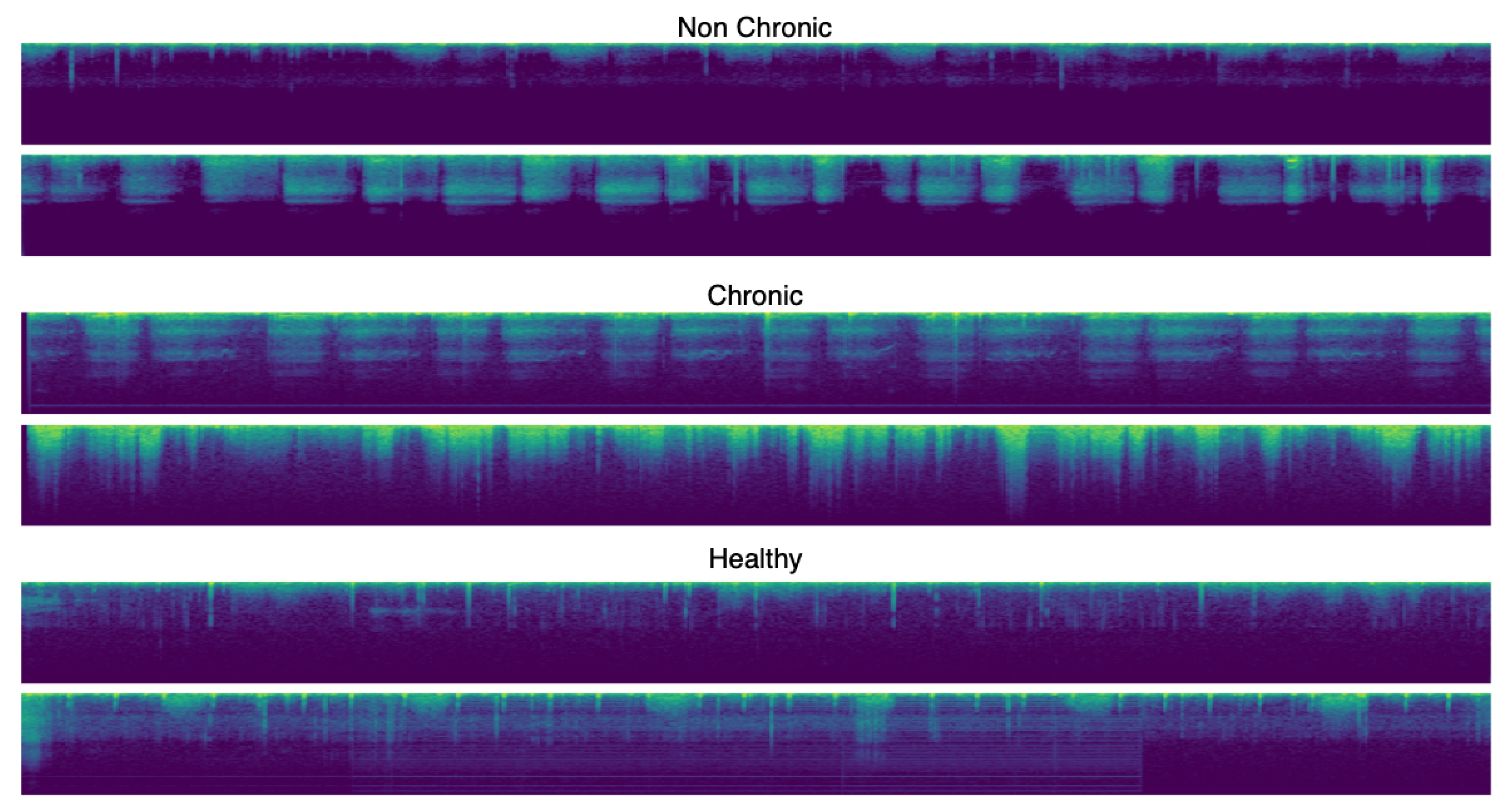
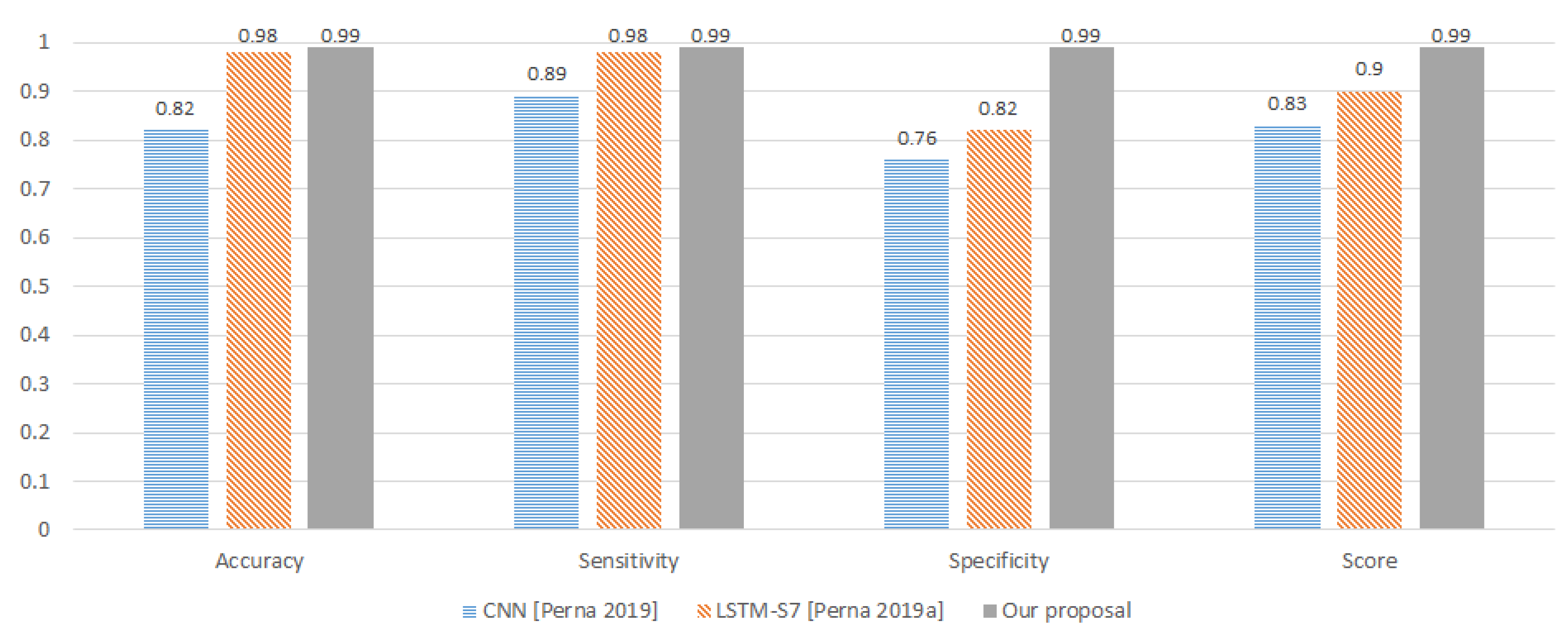
11], Perna et al. proposed a reliable method to classify in healthy, chronic disease, or non-chronic disease based-on wheezes, crackles, or normal sounds using deep learning and, more concretely, recurrent neural networks and again using the ICBHI benchmark [
2].
In Reference [
12], Jacome et al. also proposed a CNN to deal with respiratory sounds for detecting breathing phase with a 97% of success in inspiration detection and a 87% in expiration.
Early models of RNNs suffered from both exploding and vanishing gradient problems. Long Short Term Memory (LSTM) and Gated Recurrent Unit (GRU) were designed to address the gradient problems successfully. The authors exploited the LSTM and GRU advantages and obtained promising results of up to 91% of the ICBHI Score calculated as the average value of sensitivity and specificity [
11].
Deep learning techniques have also been used to detect some kinds of pathologies such as bronchiolitis, URTI, pneumonia, etc., which supposed a more challenging problem than classifying wheezes and crackles. In Reference [
13], the authors try to distinguish between pathological and non-pathological voice over the Saarbrücken Voice Database (SVD) using the MultiFocal toolkit for a discriminative calibration and fusion. The authors carry out a feature extraction step, and these features (Mel-frequency cepstral coefficients, harmonics-to-noise ratio, normalized noise energy and glottal-to-noise excitation ratio) are used to train a generative Gaussian Mixture Models (GMM) model [
14].
2.2. Deep Learning Techniques
In the literature, many deep learning techniques have been used to resolve all kinds of problem. This, gives us an idea of how useful artificial intelligence is.
Today, one of the most used techniques for all kinds of purposes are autoencoders and CNNs. Sugimoto et al. [
15] try to detect myocardial infarction using the ECG (electrocardiogram) information using a CNN. In their experiments, the classification performance was evaluated using 353,640 beats obtained from the ECG data of MI (myocardial infarction) patients and healthy subjects. ECG data was extremely imbalanced, and the minority class, including abnormal ECG data, may not be learned adequately. To solve this problem, the authors proposed to use the convolutional autoencoder in the following way: The CAE model is constructed for each lead and outputs reconstructed input ECG data if normal ECG data is inputted. Otherwise, the waveform is distorted and outputted. After this process, k-Nearest Neighbors (kNN) is used as a classifier.
A CAE (Convolutional autoencoder) is also used in Reference [
16] to restore the corrupted laser stripe images of the depth sensor by denoising the data.
In Reference [
17], Kao et al. propose a method of classifying Lycopersicons based on three levels of maturity (immature, semi-mature, and mature). Their method includes two artificial neural networks, a convolutional autoencoder (CAE), and a backpropagation neural network. With the first one, the ROI in the Lycopersicon is detected (instead of doing it manually). Then, using the extracted features, the neural network employs self-learning mechanisms to determine Lycopersicon maturity obtaining an accuracy rate of 100%.
A variational autoencoder is used in Reference [
18] for video anomaly detection and localization using only normal samples. The method is based on Gaussian Mixture Variational Autoencoder, which can learn the feature representations of the normal samples as a Gaussian Mixture Model trained using deep learning. A Fully Convolutional Network (FCN) is employed for the encoder-decoder structure to preserve relative spatial coordinates between the input image and the output feature map.
In Reference [
19], a non-linear surrogate model based on deep learning is proposed using a variational autoencoder with deep convolutional layers and a deep neural network with batch normalization (VAEDC-DNN) for a real-time analysis of the probability of death in toxic gas scenarios.
Advances in indoor positioning technologies can generate large volumes of spatial trajectory data on the occupants. These data can reveal the distribution of the occupants. In Reference [
20], the authors propose a method of evaluating similarities in occupant trajectory data using a convolutional autoencoder (CAE).
In Reference [
21], deep autoencoders are proposed to produce efficient bimodal features from the audio and visual stream inputs. The authors obtained an average relative reduction of 36.9% for a range of different noisy conditions, and also, a relative reduction of 19.2% for the clean condition in terms of the Phoneme Error Rates (PER) in comparison with the baseline method.
In 2019, variational autoencoders have been widely used to analyze different kind of signals and monitoring them [
22,
23]. In addition, in Zemouri et al. [
24], variational autoencoders have been used for train a model as a 2D visualization tool for partial discharge source classification.
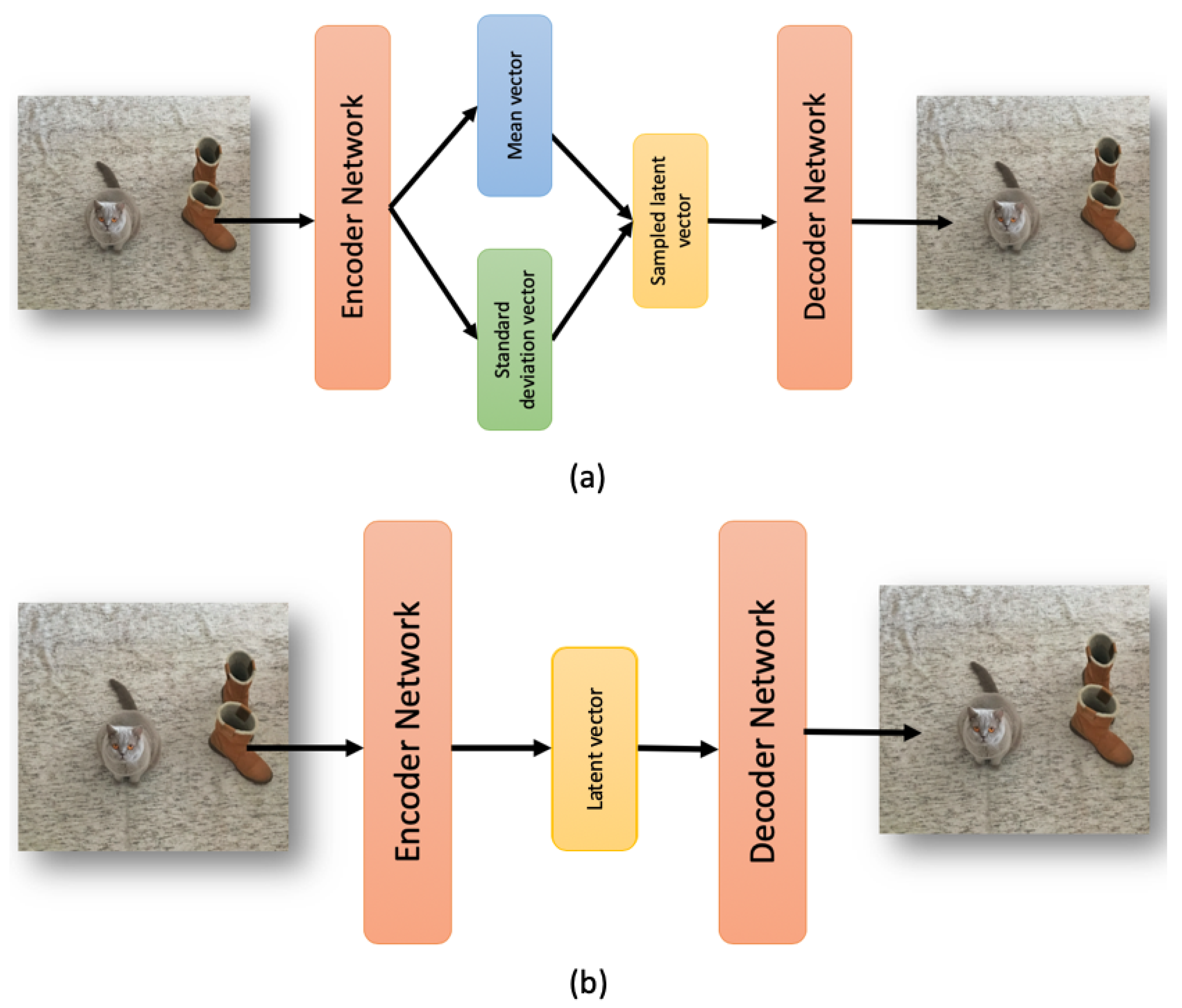
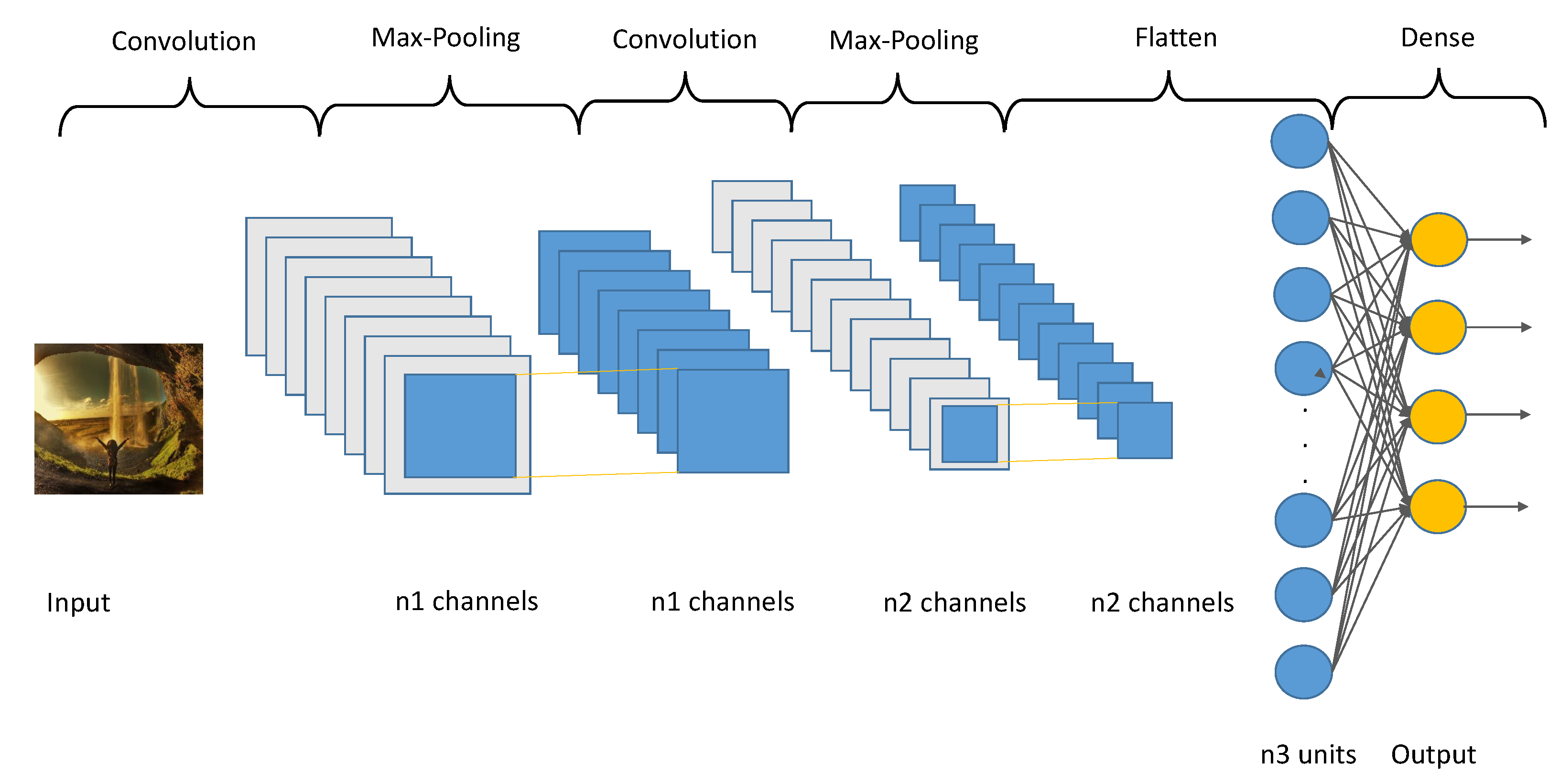
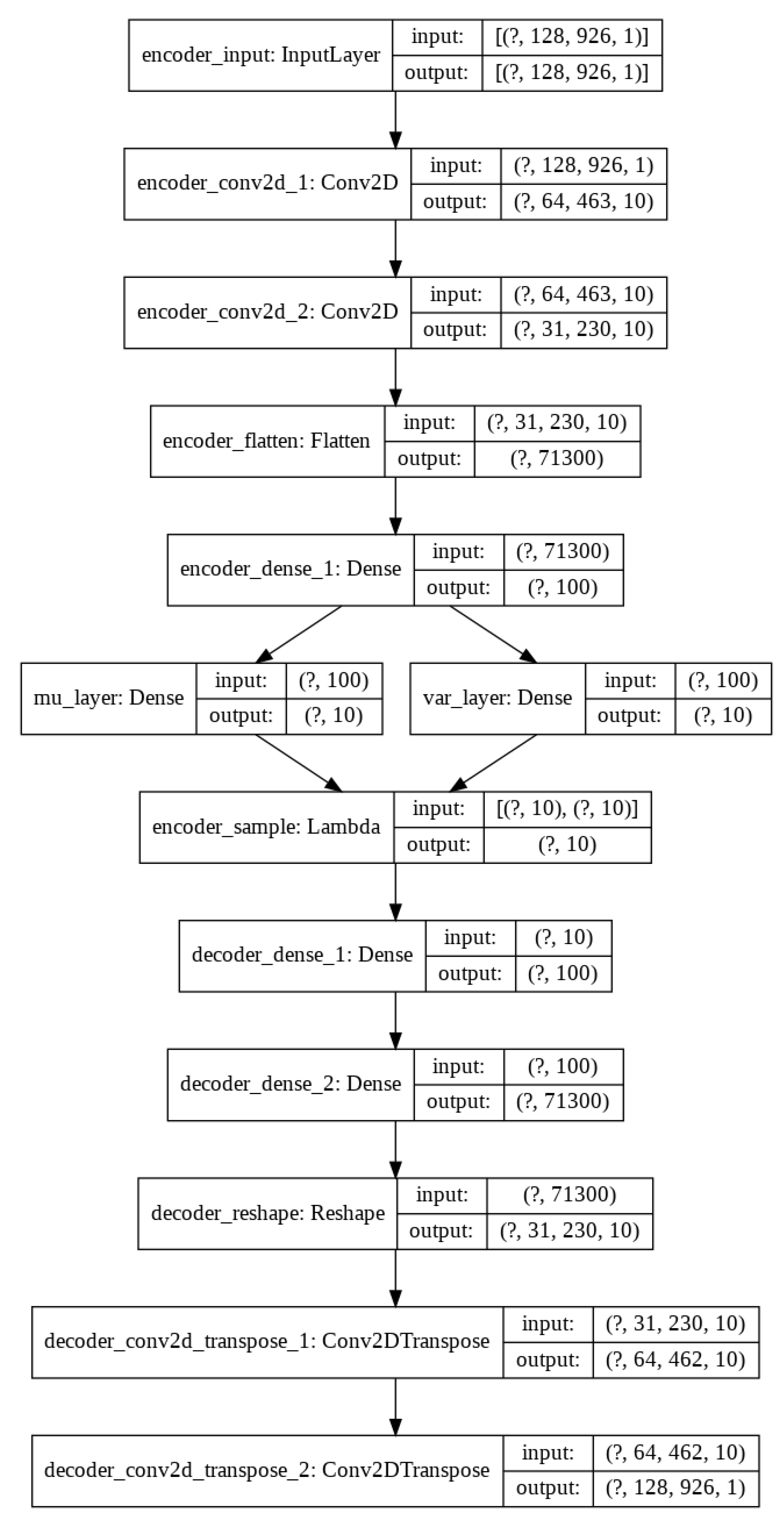
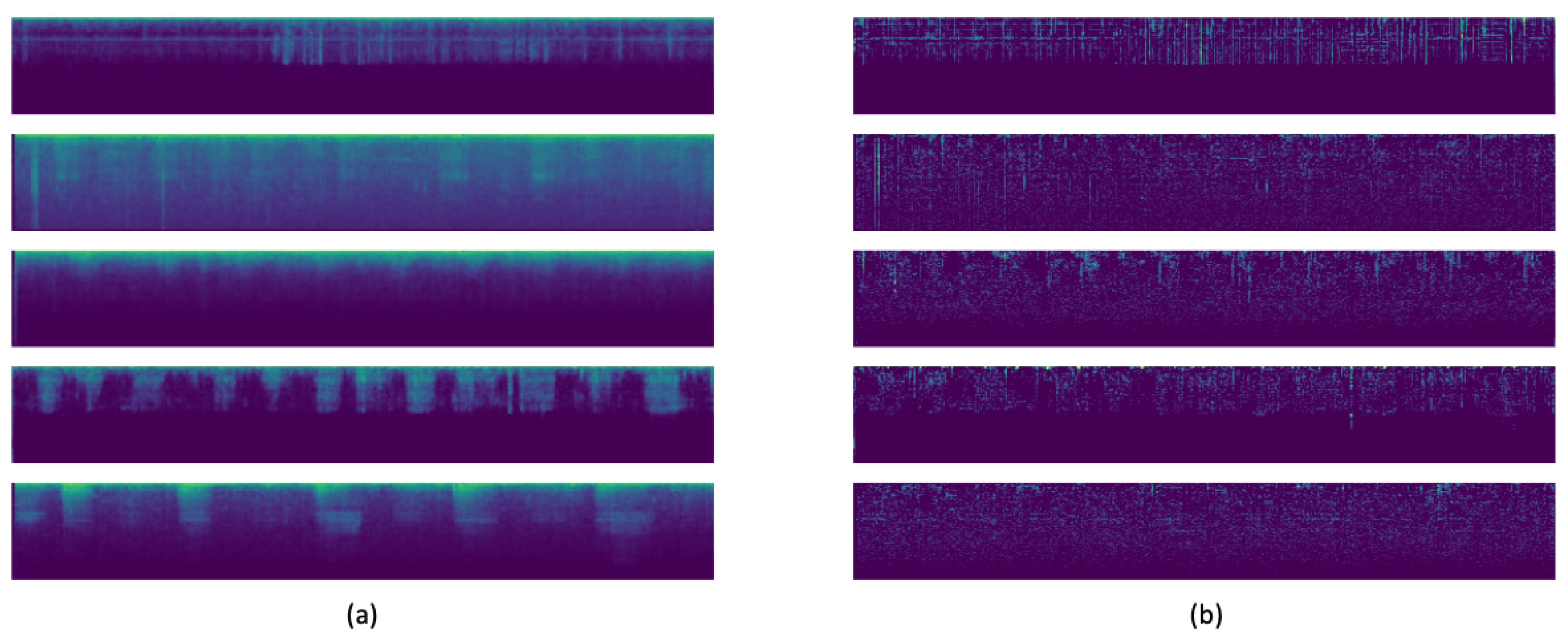
So, today autoencoders and CNNs are widely used in the literature to solve all kinds of problems. We take advantage of both methods and proposed the use of a variational convolutional autoencoder to balance the data, as well as a CNN to carry out the classification step.
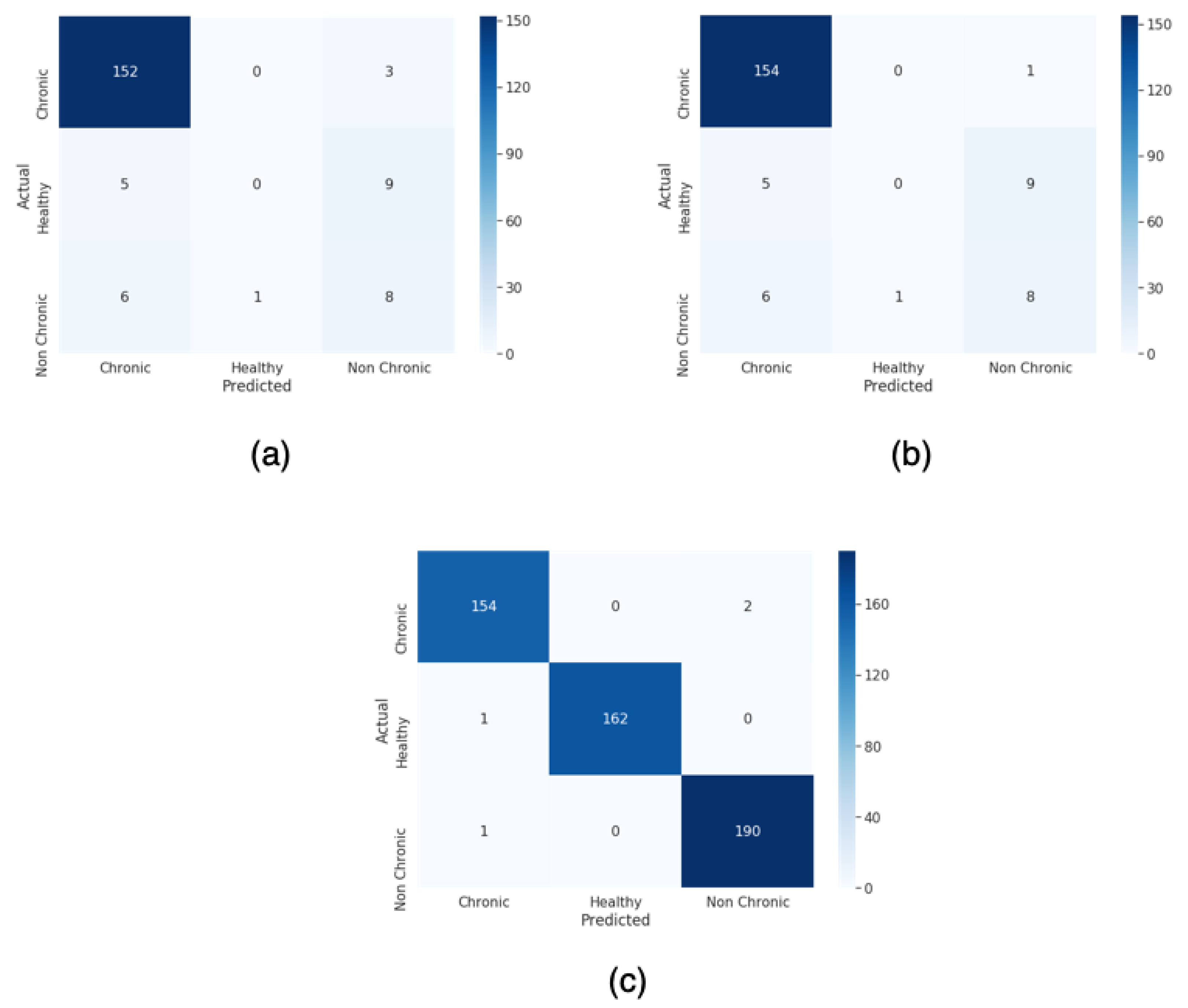
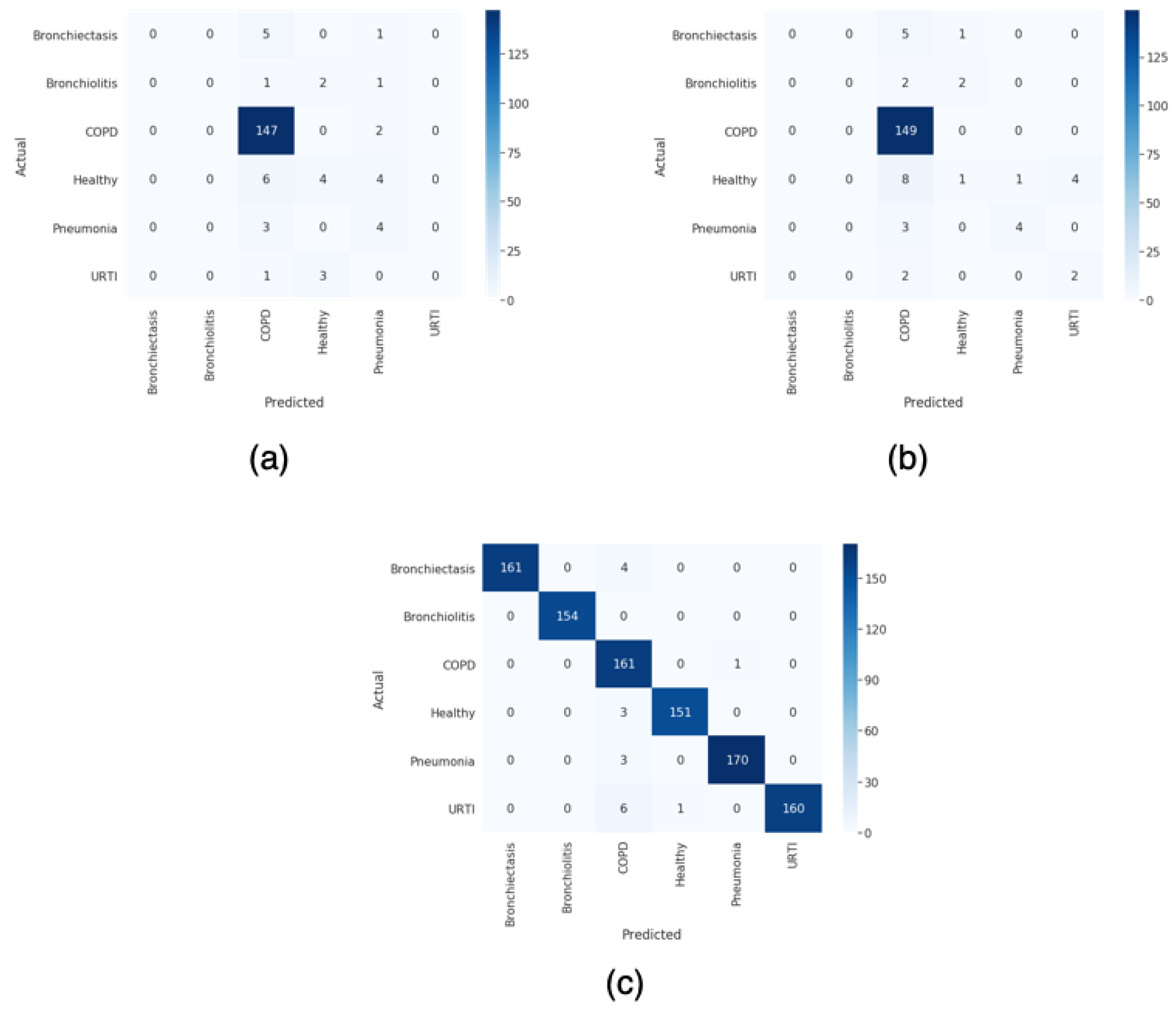
For this paper, we proposed a technique to classify healthy, chronic disease, and non-chronic disease and six different pathology classes: Chronic obstructive pulmonary disease (COPD), upper respiratory tract infection (URTI), bronchiectasis, pneumonia, bronchiolitis, and healthy. Our procedure outperforms the state-of-the-art proposals.
The rest of the paper is organized as follows: In
Section 3, we describe the methodology, including data preprocessing, data normalization, and data augmentation, using our Variational Convolutional autoencoder and finally data classification using a CNN. Experiments and results are detailed in
Section 4, taking into account two types of classification, and finally, we conclude in
Section 5.