Development of a Low-Cost Narrow Band Multispectral Imaging System Coupled with Chemometric Analysis for Rapid Detection of Rice False Smut in Rice Seed
Abstract
1. Introduction
2. Materials and Methods
2.1. Rice Seeds and Pathogen Inoculation
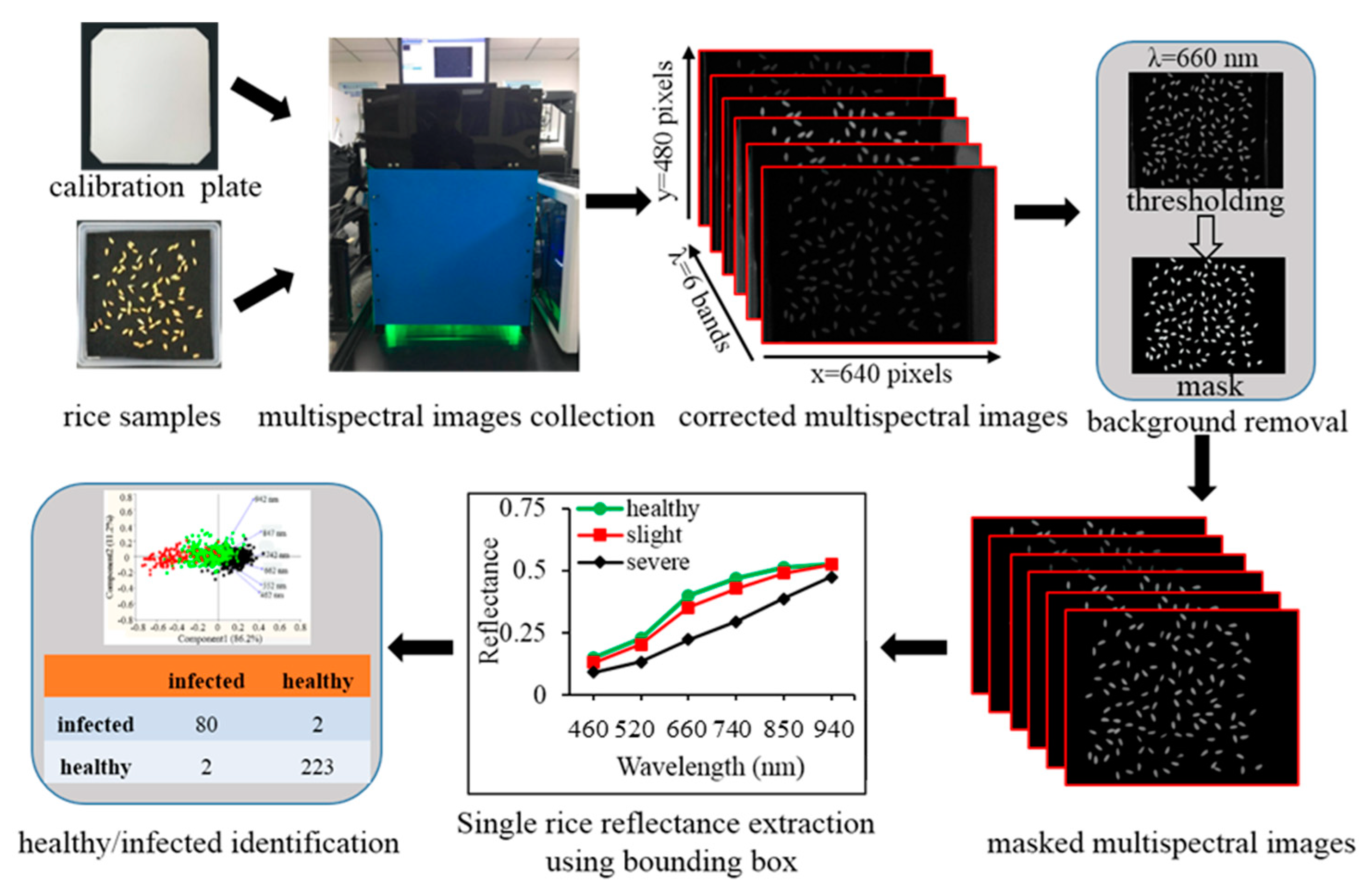
2.2. Multispectral Imaging System Design, Images Acquisition, and Processing
2.3. Principal Component Analysis (PCA)
2.4. Development and Validation of Classification Models for Rice False Smut (RFS) Detection
3. Results and Discussion
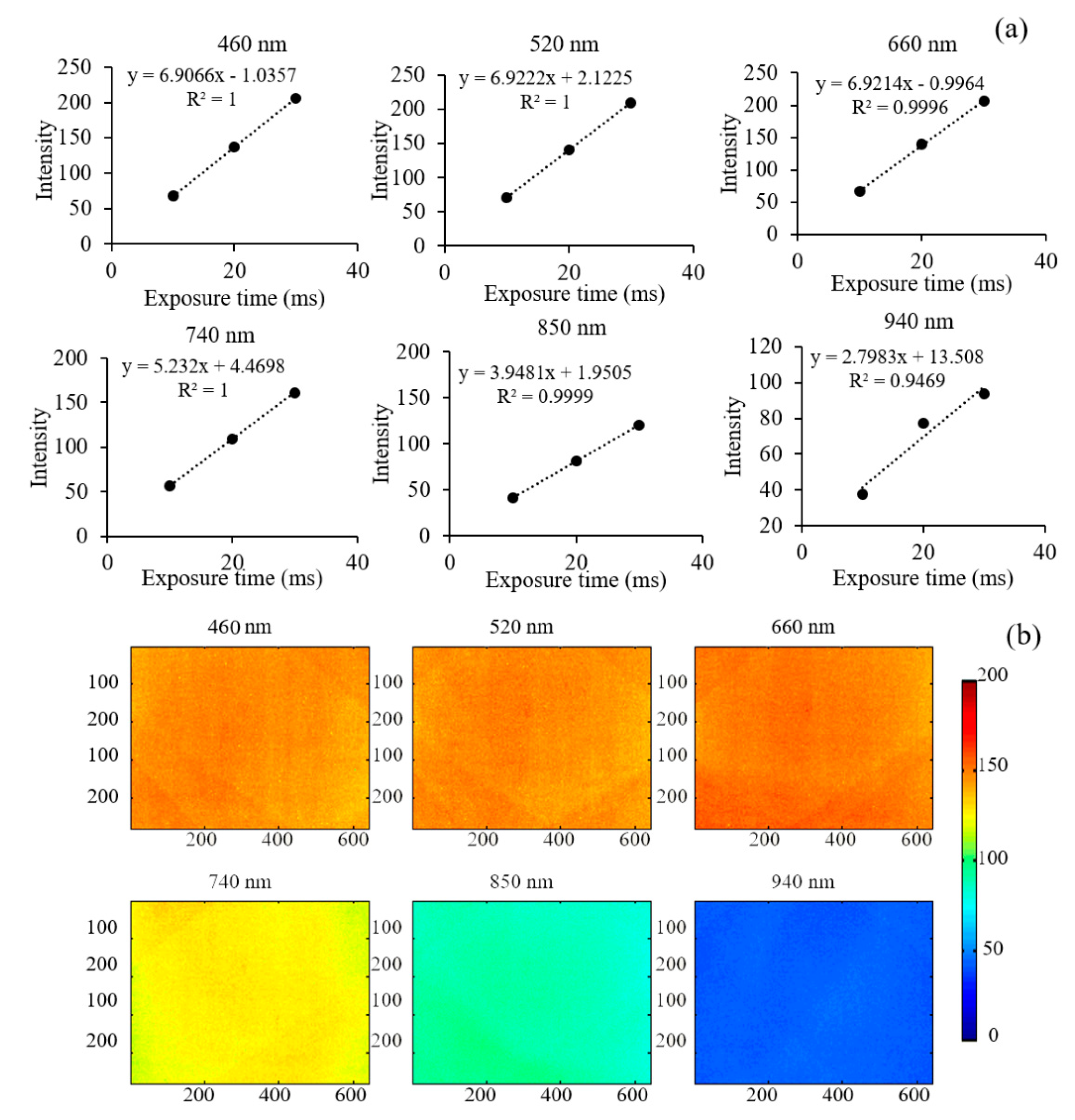
3.1. System Performance Assessment
3.2. Reflectance Spectra of Healthy and Rice False Smut (RFS) Infected Rice Seeds of Zheliang and Xiushui
3.3. Spectral Features for Rice False Smut (RFS) Detection in Rice Seeds
3.4. Validation of the Classification Model Using Model Transfer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tanaka, E.; Ashizawa, T.; Sonoda, R.; Tanaka, C. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 2008, 106, 491–501. [Google Scholar]
- Tang, Y.X.; Jin, J.; Hu, D.W.; Yong, M.L.; Xu, Y.; He, L.P. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) rice spikelets. Plant Pathol. 2013, 62, 1–8. [Google Scholar] [CrossRef]
- Guo, X.; Li, Y.; Fan, J.; Li, L.; Huang, F.; Wan, W. Progress in the study of false smut disease in rice. J. Agric. Sci. Technol. 2012, 2, 1211–1217. [Google Scholar]
- Fu, R.; Ding, L.; Zhu, J.; Li, P.; Zheng, A.P. Morphological structure of propagules and electrophoretic karyotype analysis of false smut Villosiclava virens in rice. J. Microbiol. 2012, 50, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.I.; Izumiyama, N.; Ohtsubo, K.I.; Koiso, Y.; Iwasaki, S.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. “Lupinosis”-like lesions in mice caused by ustiloxin, produced by Ustilaginoieda virens: A morphological study. Nat. Toxins. 1994, 2, 22–28. [Google Scholar] [CrossRef]
- Yong, M.; Deng, Q.; Fan, L.; Miao, J.; Lai, C.; Chen, H.; Yang, X.; Wang, S.; Chen, F.; Jin, L.; et al. The role of Ustilaginoidea virens sclerotia in increasing incidence of rice false smut disease in the subtropical zone in China. European. J. Plant Pathol. 2017, 150, 669–677. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Fan, R.H.; Liu, B.; Li, S.H.; Zheng, D.W.; Zhang, J.Z.; Hu, D.W. Preliminary analysis of rDNA-IGS of Ustilaginoidea virens isolates from different geographical regions in China. Acta Phytopathol. Sin. 2010, 40, 214–216. [Google Scholar]
- Jia, S.; An, D.; Liu, Z.; Gu, J.; Li, S.; Zhang, X.; Zhu, D.; Guo, T.; Yan, Y. Variety identification method of coated maize seeds based on near-infrared spectroscopy and chemometrics. J. Cereal Sci. 2015, 63, 21–26. [Google Scholar] [CrossRef]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using NIR hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef]
- Shrestha, S.; Knapič, M.; Žibrat, U.; Deleuran, L.C.; Gislum, R. Single seed near-infrared hyperspectral imaging in determining tomato (Solanum lycopersicum L.) seed quality in association with multivariate data analysis. Sens. Actuators B Chem. 2016, 237, 1027–1034. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Wu, C. Non-destructive discrimination of paddy seeds of different storage age based on Vis/NIR spectroscopy. J. Stored Prod. Res. 2008, 44, 264–268. [Google Scholar] [CrossRef]
- Wakholi, C.; Kandpal, L.M.; Lee, H.; Bae, H.; Park, E.; Kim, M.S.; Mo, C.; Lee, W.H.; Cho, B.K. Rapid assessment of corn seed viability using short wave infrared line-scan hyperspectral imaging and chemometrics. Sens. Actuators B Chem. 2018, 255, 498–507. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Lohumi, S.; Kim, M.S.; Kang, J.S.; Cho, B.K. Near-infrared hyperspectral imaging system coupled with multivariate methods to predict viability and vigor in muskmelon seeds. Sens. Actuators B Chem. 2016, 229, 534–544. [Google Scholar] [CrossRef]
- Lee, H.; Kim, M.S.; Lim, H.S.; Park, E.; Lee, W.H.; Cho, B.K. Detection of cucumber green mottle mosaic virus-infected watermelon seeds using a near-infrared (NIR) hyperspectral imaging system: Application to seeds of the “Sambok Honey” cultivar. Biosyst. Eng. 2016, 148, 138–147. [Google Scholar] [CrossRef]
- Senthilkumar, T.; Jayas, D.S.; White, N.D.G.; Fields, P.G.; Gräfenhan, T. Detection of ochratoxin A contamination in stored wheat using near-infrared hyperspectral imaging. Infrared Phys. Technol. 2017, 81, 228–235. [Google Scholar] [CrossRef]
- Wu, N.; Jiang, H.B.; Bao, Y.D.; Zhang, C.; Zhang, J.Z.; Song, W.J.; Zhao, Y.Y.; Mia, C.X.; He, Y.; Liu, F. Practicability investigation of using near-infrared hyperspectral imaging to detect rice kernels infected with rice false smut in different conditions. Sens. Actuators B Chem. 2020, 308, 127696. [Google Scholar] [CrossRef]
- Wang, S.; Li, M.; Dong, H.; Liu, X.Z.; Bai, Y.J.; Yang, H. Sporulation, Inoculation Methods and Pathogenicity of Ustilaginoidea albicans, the Cause of White Rice False Smut in China. J. Phytopathol. 2008, 156, 755–757. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, K.; Fang, A.; Han, Y.; Yang, J.; Xue, M.; Bao, J.; Hu, D.; Zhou, B.; Sun, X.; et al. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 2014, 5, 3849. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Naomi Altman, N. Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef]
- Liu, F.; He, Y. Application of successive projections algorithm for variable selection to determine organic acids of plum vinegar. Food Chem. 2009, 115, 1430–1436. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Chu, B.Q.; Fan, Y.Y.; Tao, X.Y.; Yin, W.X.; He, Y. Hyperspectral Imaging for Predicting the Internal Quality of Kiwifruits Based on Variable Selection Algorithms and Chemometric Models. Sci. Rep. 2017, 7, 7845. [Google Scholar] [CrossRef] [PubMed]
- Suykens, J.A.K.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Wu, W.; Mallet, Y.; Walczak, B.; Penninckx, W.; Massart, D.L.; Heuerding, S. Comparison of regularized discriminant analysis linear discriminant analysis and quadratic discriminant analysis applied to NIR data. Anal. Chim. Acta 1996, 329, 257–265. [Google Scholar] [CrossRef]
- Ravikanth, L.; Chelladurai, V.; Jayas, D.S.; White, N.D.G. Detection of broken kernels content in bulk wheat samples using near-infrared hyperspectral imaging. Agric. Res. 2016, 5, 285–292. [Google Scholar] [CrossRef]
- Lu, H.; Yu, X.; Zhou, L.; He, Y. Selection of spectral resolution and scanning speed for detecting green jujubes chilling injury based on hyperspectral reflectance imaging. Appl. Sci. 2018, 8, 523. [Google Scholar] [CrossRef]
- Chelladurai, V.; Karuppiah, K.; Jayas, D.S.; Fields, P.G.; White, N.D.G. Detection of Callosobruchus maculatus (F.) infestation in soybean using soft X-ray and NIR hyperspectral imaging techniques. J. Stored Prod. Res. 2014, 57, 43–48. [Google Scholar] [CrossRef]
- Bo, C.J.; Lu, H.C.; Wang, D. Spectral-spatial K-Nearest Neighbor approach for hyperspectral image classification. Multimed. Tools Appl. 2018, 77, 10419–10436. [Google Scholar] [CrossRef]
- Macho, S.; Rius, A.; Callao, M.P.; Larrechi, M.S. Monitoring ethylene content in heterophasic copolymers by near-infrared spectroscopy standardisation of the calibration model. Anal. Chim. Acta 2001, 455, 213–220. [Google Scholar] [CrossRef]
- Feudale, R.N.; Woody, N.A.; Tan, H.W.; Myles, A.J.; Brown, S.D.; Ferré. Transfer of multivariate calibration models: A review. Chemom. Intell. Lab. Syst. 2002, 64, 181–192. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]



| Wavelengths (nm) | Zheliang | Xiushui | ||
|---|---|---|---|---|
| Component 1 | Component 2 | Component 1 | Component 2 | |
| 460 nm | 0.400 | −0.456 | 0.397 | −0.473 |
| 520 nm | 0.419 | −0.353 | 0.419 | −0.357 |
| 660 nm | 0.431 | −0.157 | 0.427 | −0.134 |
| 740 nm | 0.433 | 0.028 | 0.431 | 0.043 |
| 850 nm | 0.417 | 0.322 | 0.421 | 0.275 |
| 940 nm | 0.343 | 0.734 | 0.349 | 0.744 |
| Cultivars | Zheliang | Xiushui | |||||
|---|---|---|---|---|---|---|---|
| Models | Predicted Class | Actual Class | |||||
| Infected | Healthy | Accuracy (%) | Infected | Healthy | Accuracy (%) | ||
| LDA | infected | 70 | 12 | 85.4 | 150 | 28 | 84.3 |
| healthy | 4 | 221 | 98.2 | 8 | 150 | 94.9 | |
| Overall accuracy (%) | 94.8 | 89.3 | |||||
| KNN | infected | 80 | 2 | 97.6 | 170 | 8 | 95.5 |
| healthy | 14 | 211 | 93.8 | 36 | 122 | 77.2 | |
| Overall accuracies (%) | 94.8 | 86.9 | |||||
| LS-SVM * | infected | 80 | 2 | 97.6 | 166 | 12 | 93.3 |
| healthy | 2 | 223 | 99.1 | 17 | 141 | 89.2 | |
| Overall accuracy (%) | 98.7 | 91.4 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weng, H.; Tian, Y.; Wu, N.; Li, X.; Yang, B.; Huang, Y.; Ye, D.; Wu, R. Development of a Low-Cost Narrow Band Multispectral Imaging System Coupled with Chemometric Analysis for Rapid Detection of Rice False Smut in Rice Seed. Sensors 2020, 20, 1209. https://doi.org/10.3390/s20041209
Weng H, Tian Y, Wu N, Li X, Yang B, Huang Y, Ye D, Wu R. Development of a Low-Cost Narrow Band Multispectral Imaging System Coupled with Chemometric Analysis for Rapid Detection of Rice False Smut in Rice Seed. Sensors. 2020; 20(4):1209. https://doi.org/10.3390/s20041209
Chicago/Turabian StyleWeng, Haiyong, Ya Tian, Na Wu, Xiaoling Li, Biyun Yang, Yiping Huang, Dapeng Ye, and Renye Wu. 2020. "Development of a Low-Cost Narrow Band Multispectral Imaging System Coupled with Chemometric Analysis for Rapid Detection of Rice False Smut in Rice Seed" Sensors 20, no. 4: 1209. https://doi.org/10.3390/s20041209
APA StyleWeng, H., Tian, Y., Wu, N., Li, X., Yang, B., Huang, Y., Ye, D., & Wu, R. (2020). Development of a Low-Cost Narrow Band Multispectral Imaging System Coupled with Chemometric Analysis for Rapid Detection of Rice False Smut in Rice Seed. Sensors, 20(4), 1209. https://doi.org/10.3390/s20041209





