Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Data Preprocessing
2.3. HRV Parameters
- SDNN refers to the standard deviation of IBI. It estimates overall power spectrum of IBI timeseries. The SDNN is defined as the “gold standard” to assess both morbidity and mortality in the population [2].
- rMSSD is the root mean square of the successive IBI differences estimates short-term components of HRV [35].
2.4. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- McCraty, R.; Shaffer, F. Heart rate variability: New perspectives on physiological mechanisms, assessment of self-regulatory capacity, and health risk. Glob. Adv. Health Med. 2015, 4, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, F.; Ginsberg, J. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, A.; Weiser-Bitoun, I.; Billman, G.; Yaniv, Y. Signatures of the autonomic nervous system and the heart’s pacemaker cells in canine electrocardiograms and their applications to humans. Sci. Rep. 2020, 10, 9971. [Google Scholar] [CrossRef] [PubMed]
- Petretta, M.; Bonaduce, B.; Scalfi, L.; De Filippo, E.; Marciano, F.; Migaux, M.; Themistoclakis, S.; Ianniciello, A.; Contaldo, F. Heart rate variability as a measure of autonomic nervous system function in anorexia nervosa. Clin. Cardiol. 1997, 20, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Karemaker, J. An introduction into autonomic nervous function. Physiol. Meas. 2017, 38, 89–118. [Google Scholar] [CrossRef]
- Tsutsui, K.; Monfredi, O.; Sirenko-Tagirova, S.; Maltseva, L.; Bychkov, R.; Kim, M.S.; Ziman, B.D.; Tarasov, K.V.; Tarasova, Y.S.; Wang, M.; et al. A coupled-clock system drives the automaticity of human sinoatrial nodal pacemaker cells. Sci. Signal. 2018, 11, eaap7608. [Google Scholar] [CrossRef]
- Stein, P.; Domitrovich, P.; Hui, N.; Rautaharju, P.; Gottdiener, J. Sometimes higher heart rate variability is not better heart rate variability: Results of graphical and nonlinear analyses. J. Cardiovasc. Electrophysiol. 2005, 16, 954–959. [Google Scholar] [CrossRef]
- Laborde, S.; Mosley, E.; Thayer, J. Heart Rate Variability and Cardiac Vagal Tone in Psychophysiological Research—Recommendations for Experiment Planning, Data Analysis and Data Reporting. Front. Psychol. 2017, 8, 213. [Google Scholar] [CrossRef]
- Camm, A.J.; Malik, M.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, E.L.; Fallen, H.L.; Kennedy, R.E.; Lombardi, F.; et al. Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 1996, 93, 1043–1065. [Google Scholar]
- Kleiger, R.; Miller, J.; Bigger, J.; Moss, A.J. Decreased heart rate variability and its association with increased mortality after acute myocardial infarction. Am. J. Cardiol. 1987, 59, 256–262. [Google Scholar] [CrossRef]
- Karcz, M.; Chojnowska, L.W.Z.R. Prognostic significance of heart rate variability in dilated cardiomyopathy. EP Eur. 2003, 87, 75–81. [Google Scholar] [CrossRef]
- Gao, L.; Chen, Y.; Shi, Y.; Xue, H.; Wang, J. Value of DC and DRs in prediction of cardiovascular events in acute myocardial infarction patients. Zhonghua Yi Xue Za Zhi 2016, 96. [Google Scholar] [CrossRef]
- Shaffer, F.; McCraty, R.; Zerr, C. A healthy heart is not a metronome: An integrative review of the heart’s anatomy and heart rate variability. Front. Psychol. 2014, 5, 1040. [Google Scholar] [CrossRef] [PubMed]
- Zygmunt, A.; Stanczyk, J. Methods of evaluation of autonomic nervous system function. Arch. Med. Sci. 2010, 6, 11–18. [Google Scholar] [CrossRef]
- Plews, D.; Laursen, P.; Kilding, A.; Buchheit, M. Heart rate variability and training intensity distribution in elite rowers. Int. J. Sport. Physiol. Perform. 2014, 9, 1026–1032. [Google Scholar] [CrossRef]
- Chen, J.; Yeh, D.; Lee, J.; Chen, C.; Huang, C.; Lee, S.D.; Chen, C.C.; Kuo, T.B.J.; Kao, C.L.; Kuo, C.H. Parasympathetic nervous activity mirrors recovery status in weightlifting performance after training. J. Strength Cond. Res. 2011, 25, 1546–1552. [Google Scholar] [CrossRef]
- Edmonds, R.; Sinclair, W.; Leicht, A. Effect of a training week on heart rate variability in elite youth rugby league players. Int. J. Sport. Med. 2013, 34, 1087–1092. [Google Scholar] [CrossRef]
- Jennings, J.; Sheu, L.; Kuan, D.; Manuck, S.; Gianaros, P. Resting state connectivity of the medial prefrontal cortex covaries with individual differences in high-frequency heart rate variability. Psychophysiology 2016, 53, 444–454. [Google Scholar] [CrossRef]
- Kemp, A.; Quintana, D.; Gray, M.; Felmingham, K.; Brown, K.; Gatt, J. Impact of depression and antidepressant treatment on heart rate variability: A review and meta-analysis. Biol. Psychiatry 2010, 67, 1067–1074. [Google Scholar] [CrossRef]
- Agelink, M.; Majewski, T.; Wurthmann, C.; Postert, T.; Linka, T.; Rotterdam, S. Autonomic neurocardiac function in patients with major depression and effects of antidepressive treatment with nefazodone. J. Affect. Disord. 2001, 62, 187–198. [Google Scholar] [CrossRef]
- Haghi, M.; Thurow, K.; Stoll, R. Wearable Devices in Medical Internet of Things: Scientific Research and Commercially Available Devices. Healthc. Inf. Res 2017, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Hörnsten, R.; Rydberg, A.; Wiklund, U. Automatic filtering of outliers in RR intervals before analysis of heart rate variability in Holter recordings: A comparison with carefully edited data. Biomed. Eng. 2012, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Morelli, D.; Rossi, A.; Cairo, M.; Clifton, D. Analysis of the Impact of Interpolation Methods of Missing RR-Intervals Caused by Motion Artifacts on HRV Features Estimations. Sensors 2019, 19, 3163. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lim, Y.; Kim, J.; Park, K. Effect of missing RR-interval data on heart rate variability analysis in the time domain. Physiol. Meas. 2007, 28, 1485–1494. [Google Scholar] [CrossRef]
- Munoz, M.; Van Roon, A.; Riese, H.; Thio, C.; Oostenbroek, E.; Westrik, I.; De Geus, E.J.C.; Gansevoort, R.; Lefrandt, J.; Nolte, I.; et al. Validity of (Ultra-)Short Recordings for Heart Rate Variability Measurements. PLoS ONE 2015, 10, e0138921. [Google Scholar] [CrossRef]
- Beak, H.; Cho, C.; Cho, J.; Woo, J. Reliability of Ultra-Short-Term Analysis as a Surrogate of Standard 5-Min Analysis of Heart Rate Variability. Telemed. e-Health 2015, 21, 404–414. [Google Scholar] [CrossRef]
- Castaldo, R.; Montesinos, P.; Melillo, C.; Pecchia, L. Ultra-short term HRV features as surrogates of short term HRV: A case study on mental stress detection in real life. BMC Med. Inform. Decis. Mak. 2019, 19, 12. [Google Scholar] [CrossRef]
- Pecchia, L.; Castaldo, R.; Montesinos, L.; Melillo, P. Are ultra-short heart rate variability features good surrogates of short-term ones? State-of-the-art review and recommendations. Healthc. Technol. Lett. 2018, 5, 94–100. [Google Scholar] [CrossRef]
- Rossi, A.; Da Pozzo, E.; Menicagli, D.; Tremolanti, C.; Priami, C.; Sirbu, A.; Clifton, D.; Martini, C.; Morelli, D. A Public Dataset of 24-h Multi-Levels Psycho-Physiological Responses in Young Healthy Adults. Data 2020, 5, 91. [Google Scholar] [CrossRef]
- Rossi, A.; Da Pozzo, E.; Menicagli, D.; Tremolanti, C.; Priami, C.; Sirbu, A.; Clifton, D.; Martini, C.; Morelli, D. Multilevel Monitoring of Activity and Sleep in Healthy People (version 1.0.0). PhysioNet 2020. [Google Scholar] [CrossRef]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.; Mark, R.; Stanley, H. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Plews, D.; Scott, B.; Altini, M.; Wood, M.; Kilding, A.; Laursen, P. Comparison of Heart-Rate-Variability Recording With Smartphone Photoplethysmography, Polar H7 Chest Strap, and Electrocardiography. Int. J. Sport. Physiol. Perform. 2017, 12, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Hideki, I. Essentials of Error-Control Coding Techniques; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Task Force of the European Society of Cardiology the North American Society of Pacing Electrophysiology. Heart Rate Variability: Standards of Measurment, Physiological Interpretation, and Clinical Use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef]
- Liao, D.; Cai, J.; Rosamond, W.; Barnes, R.; Hutchinson, R.; Whitsel, E. Cardiac autonomic function and incident coronary heart disease: A population-based case-cohort study. The ARIC study. Am. J. Epidemiol. 1997, 145, 696–706. [Google Scholar] [CrossRef]
- Lomb, N. Least-squares frequency analysis of unequally spaced data. Astrophys. Space Sci. 1976, 39, 447–462. [Google Scholar] [CrossRef]
- Scargle, J.D. Studies in Astronomical Time Series Analysis. V. Bayesian Blocks, a New Method to Analyze Structure in Photon Counting Data. Am. Astron. Soc. 1998, 504, 405. [Google Scholar] [CrossRef]
- Kleiger, R.; Stein, P.; Bigger, J. Heart rate variability: Measurement and clinical utility. Ann. Noninvasive Electrocardiol. 2005, 10, 88–101. [Google Scholar] [CrossRef]
- Baek, H.; Lee, H.; Kim, J.; Choi, J.; Kim, K.; Park, K. Non intrusive biological signal monitoring in a car to evaluate a driver’s stress and health state. Telemed. J. e-Health 2009, 15, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Gouin, J.; Wenzel, K.; Deschenes, S.; Dang-Vu, T. Heart rate variability predicts sleep efficiency. Sleep Med. 2013, 14, e142. [Google Scholar] [CrossRef]
- Patel, M.; Lal, S.; Kavanagh, D.; Rossiter, P. Applying neural network analysis on heart rate variability data to assess deriver fatigue. Expert Syst. Appl. 2011, 38, 7235–7242. [Google Scholar] [CrossRef]
- Kaikkonen, P.; Hynynen, E.; Mann, T.; Rusko, H.; Nummela, A. Heart rate variability is related to training load variables in interval running exercises. Eur. J. Appl. Physiol. 2012, 112, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.; Pu, Y. Heart rate variability, sleep and sleep disorders. Sleep Med. Rev. 2012, 16, 47–66. [Google Scholar] [CrossRef] [PubMed]
- Thayer, J.; Ahs, F.; Fredrikson, M.; Sollers, J.; Wager, T. A meta-analysis of heartrate variability and neuromonitoring studies: Implications for heart rate variability as a marker of stress and health. Neurosci. Biobehav. Rev. 2012, 36, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.; Bigger, J.; Camm, A.; Kleiger, R.; Malliani, A.; Moss, A.; Schwartz, P. Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Circulation 1996, 93, 927–934. [Google Scholar] [CrossRef]
| Time Window | Missing Values | SDNN | r | RMSE | Bias | Systematic Error | ES |
|---|---|---|---|---|---|---|---|
| 5 min | — | 72.70 ± 30.23 | — | — | — | — | — |
| 4 min | 0% | 69.32 ± 35.83 | 0.79 | 23.17 | −6.92 ± 22.11 | 0.08 | 0.20 * |
| 5% | 69.15 ± 36.30 | 0.79 | 23.20 | −6.94 ± 22.17 | 0.08 | 0.20 * | |
| 10% | 69.29 ± 36.42 | 0.79 | 23.25 | −7.02 ± 22.31 | 0.08 | 0.20 * | |
| 15% | 69.18 ± 36.36 | 0.78 | 23.31 | −7.08 ± 22.45 | 0.08 | 0.21 * | |
| 30% | 69.02 ± 36.16 | 0.78 | 23.87 | −7.20 ± 22.75 | 0.08 | 0.21 * | |
| 50% | 68.62 ± 36.26 | 0.77 | 24.71 | −7.48 ± 23.55 | 0.09 | 0.21 * | |
| 70% | 67.64 ± 36.66 | 0.74 | 26.02 | −8.27 ± 24.67 | 0.10 | 0.23 * | |
| 3 min | 0% | 65.96 ± 31.99 | 0.76 | 22.88 | −7.64 ± 21.57 | 0.04 | 0.26 * |
| 5% | 65.92 ± 31.97 | 0.76 | 22.95 | −7.66 ± 21.59 | 0.04 | 0.26 * | |
| 10% | 65.9 ± 31.79 | 0.76 | 22.98 | −7.69 ± 21.61 | 0.04 | 0.26 * | |
| 15% | 65.79 ± 31.95 | 0.76 | 23.06 | −7.73 ± 21.65 | 0.04 | 0.26 * | |
| 30% | 65.72 ± 32.18 | 0.76 | 23.19 | −7.84 ± 21.82 | 0.05 | 0.26 * | |
| 50% | 65.31 ± 32.33 | 0.75 | 23.71 | −8.19 ± 22.25 | 0.05 | 0.27 * | |
| 70% | 64.54 ± 32.98 | 0.73 | 24.65 | −8.76 ± 23.04 | 0.08 | 0.29 * | |
| 2 min | 0% | 63.57 ± 31.39 | 0.71 | 25.15 | −9.51 ± 23.28 | 0.03 | 0.34 * |
| 5% | 63.52 ± 31.42 | 0.71 | 25.19 | −9.56 ± 23.51 | 0.03 | 0.34 * | |
| 10% | 63.5 ± 31.37 | 0.71 | 25.25 | −9.59 ± 23.57 | 0.03 | 0.35 * | |
| 15% | 63.4 ± 31.44 | 0.71 | 25.31 | −9.60 ± 23.55 | 0.04 | 0.35 * | |
| 30% | 63.22 ± 31.56 | 0.70 | 25.66 | −9.76 ± 23.73 | 0.04 | 0.35 * | |
| 50% | 62.82 ± 31.85 | 0.70 | 25.98 | −10.60 ± 23.95 | 0.05 | 0.37 * | |
| 70% | 61.94 ± 32.35 | 0.69 | 26.84 | −10.70 ± 24.62 | 0.07 | 0.38 * | |
| 1 min | 0% | 58.95 ± 30.35 | 0.63 | 29.03 | −13.50 ± 25.70 | 0.01 | 0.45 * |
| 5% | 58.9 ± 31.09 | 0.63 | 29.16 | −13.59 ± 25.78 | 0.01 | 0.45 * | |
| 10% | 58.83 ± 30.93 | 0.63 | 29.21 | −13.64 ± 25.81 | 0.01 | 0.45 * | |
| 15% | 58.69 ± 31.15 | 0.63 | 29.28 | −13.70 ± 25.84 | 0.01 | 0.45 * | |
| 30% | 58.54 ± 30.53 | 0.63 | 29.46 | −13.84 ± 26.00 | 0.02 | 0.45 * | |
| 50% | 58.04 ± 30.78 | 0.62 | 29.98 | −14.18 ± 26.42 | 0.02 | 0.47 * | |
| 70% | 56.61 ± 31.31 | 0.60 | 30.98 | −15.28 ± 26.94 | 0.04 | 0.52 | |
| 30 s | 0% | 51.41 ± 28.62 | 0.53 | 34.18 | −19.97 ± 27.74 | −0.01 | 0.67 |
| 5% | 51.36 ± 28.7 | 0.53 | 34.21 | −20.01 ± 27.88 | −0.01 | 0.67 | |
| 10% | 51.14 ± 28.54 | 0.53 | 34.36 | −20.06 ± 27.91 | −0.01 | 0.69 | |
| 15% | 51.23 ± 28.86 | 0.52 | 34.48 | −20.08 ± 27.93 | −0.01 | 0.69 | |
| 30% | 50.77 ± 28.80 | 0.52 | 34.73 | −20.49 ± 28.04 | −0.01 | 0.70 | |
| 50% | 49.95 ± 29.05 | 0.51 | 35.42 | −21.16 ± 28.41 | 0.00 | 0.71 | |
| 70% | 48.11 ± 29.63 | 0.48 | 36.85 | −22.63 ± 29.08 | 0.02 | 0.77 |
| Time Window | Missing Values | rMSSD | r | RMSE | Bias | Systematic Error | ES |
|---|---|---|---|---|---|---|---|
| 5 min | — | 42.49 ± 20.75 | — | — | — | — | — |
| 4 min | 0% | 42.34 ± 22.12 | 0.93 | 7.87 | −0.56 ± 7.85 | 0.16 | 0.03 ** |
| 5% | 42.18 ± 22.08 | 0.92 | 8.12 | −0.58 ± 7.91 | 0.16 | 0.03 ** | |
| 10% | 42.11 ± 22.06 | 0.92 | 8.48 | −0.62 ± 8.18 | 0.18 | 0.03 ** | |
| 15% | 42.07 ± 22.46 | 0.91 | 8.55 | −0.67 ± 8.54 | 0.19 | 0.04 ** | |
| 30% | 42.02 ± 22.96 | 0.90 | 9.97 | −0.87 ± 9.93 | 0.21 | 0.05 ** | |
| 50% | 41.35 ± 24.06 | 0.85 | 12.78 | −1.50 ± 12.69 | 0.26 | 0.07 ** | |
| 70% | 39.04 ± 26.09 | 0.72 | 18.34 | −3.69 ± 17.96 | 0.32 | 0.16 ** | |
| 3 min | 0% | 42.15 ± 22.36 | 0.92 | 8.82 | −0.57 ± 8.81 | 0.19 | 0.04 ** |
| 5% | 42.05 ± 22.54 | 0.90 | 9.01 | −0.66 ± 8.98 | 0.22 | 0.04 ** | |
| 10% | 41.94 ± 23.01 | 0.89 | 9.56 | −0.71 ± 9.06 | 0.23 | 0.04 ** | |
| 15% | 41.78 ± 23.07 | 0.89 | 9.88 | −0.85 ± 9.81 | 0.25 | 0.04 ** | |
| 30% | 41.73 ± 23.21 | 0.88 | 10.97 | −0.96 ± 10.93 | 0.24 | 0.05 ** | |
| 50% | 41.08 ± 24.45 | 0.83 | 13.86 | −1.56 ± 13.76 | 0.29 | 0.08 ** | |
| 70% | 39.00 ± 26.89 | 0.71 | 19.21 | −3.48 ± 18.89 | 0.36 | 0.16 ** | |
| 2 min | 0% | 41.83 ± 22.70 | 0.90 | 9.85 | −0.82 ± 9.82 | 0.19 | 0.04 ** |
| 5% | 41.79 ± 22.78 | 0.88 | 10.51 | −1.16 ± 10.05 | 0.25 | 0.05 ** | |
| 10% | 41.81 ± 22.18 | 0.88 | 10.36 | −1.12 ± 9.98 | 0.25 | 0.05 ** | |
| 15% | 41.79 ± 22.81 | 0.86 | 11.76 | −1.16 ± 10.80 | 0.27 | 0.06 ** | |
| 30% | 41.29 ± 23.60 | 0.86 | 12.05 | −1.31 ± 11.98 | 0.25 | 0.07 ** | |
| 50% | 40.40 ± 24.75 | 0.80 | 14.80 | −2.17 ± 14.64 | 0.28 | 0.11 ** | |
| 70% | 38.38 ± 27.60 | 0.68 | 20.63 | −4.00 ± 20.24 | 0.38 | 0.17 ** | |
| 1 min | 0% | 41.26 ± 23.47 | 0.86 | 12.02 | −1.22 ± 11.96 | 0.24 | 0.06 ** |
| 5% | 40.97 ± 23.55 | 0.85 | 12.48 | −1.50 ± 12.87 | 0.24 | 0.07 ** | |
| 10% | 40.90 ± 23.61 | 0.84 | 12.69 | −1.53 ± 13.13 | 0.26 | 0.07 ** | |
| 15% | 40.69 ± 23.77 | 0.84 | 13.17 | −1.71 ± 13.24 | 0.26 | 0.09 ** | |
| 30% | 40.54 ± 24.57 | 0.81 | 14.42 | −1.90 ± 14.29 | 0.29 | 0.09 ** | |
| 50% | 39.52 ± 26.23 | 0.75 | 17.72 | −2.84 ± 17.50 | 0.35 | 0.13 ** | |
| 70% | 36.73 ± 29.70 | 0.62 | 24.01 | −5.37 ± 23.40 | 0.44 | 0.22 * | |
| 30 s | 0% | 38.73 ± 24.13 | 0.79 | 15.13 | −2.34 ± 14.95 | 0.30 | 0.11 ** |
| 5% | 38.71 ± 24.18 | 0.79 | 15.38 | −2.55 ± 15.86 | 0.31 | 0.13 ** | |
| 10% | 38.52 ± 24.67 | 0.78 | 15.99 | −2.57 ± 15.97 | 0.31 | 0.13 ** | |
| 15% | 37.74 ± 24.98 | 0.76 | 17.05 | −3.13 ± 17.11 | 0.33 | 0.13 ** | |
| 30% | 37.66 ± 25.34 | 0.73 | 17.83 | −3.37 ± 17.51 | 0.33 | 0.15 ** | |
| 50% | 36.07 ± 27.20 | 0.65 | 21.41 | −4.83 ± 20.86 | 0.39 | 0.19 ** | |
| 70% | 32.63 ± 30.47 | 0.53 | 27.14 | −7.80 ± 26.00 | 0.47 | 0.31 * |
| SDNN | rMSSD | |
|---|---|---|
| VLF | 0.62 | 0.19 |
| LF | 0.51 | 0.43 |
| HF | 0.45 | 0.80 |
| LF/HF | −0.04 | −0.47 |
| Total Power | 0.73 | 0.45 |
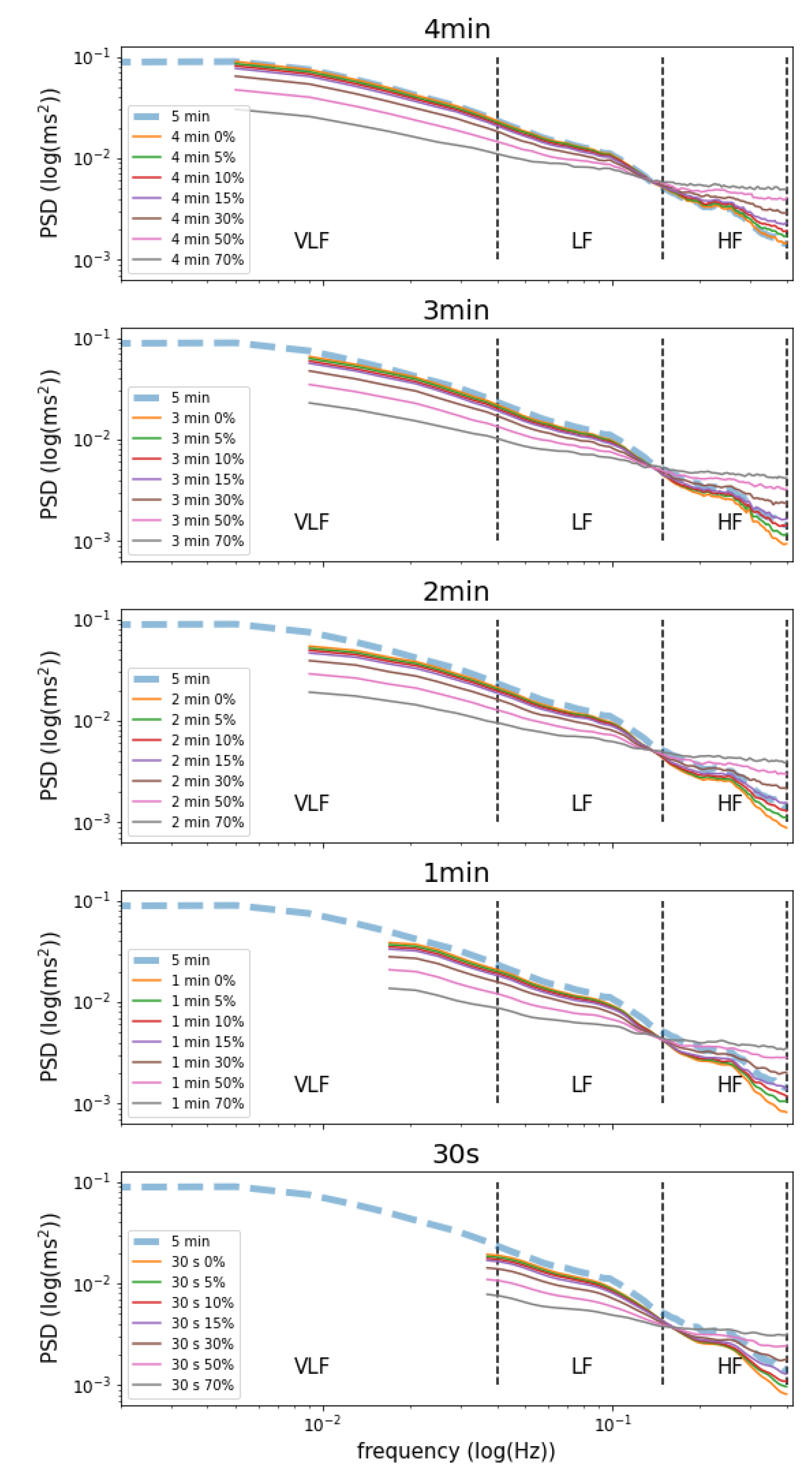
| Time Window | Missing Values | VLF | LF | HF | LF/HF | Total Power |
|---|---|---|---|---|---|---|
| 5 min | — | 1891.44 ± 46.10 | 1208.11 ± 30.84 | 689.49 ± 24.84 | 3.31 ± 0.04 | 3907.50 ± 86.95 |
| 4 min | 0% | 1533.88 ± 37.96 ** | 1208.11 ± 30.84 ** | 689.48 ± 24.84 ** | 3.31 ± 0.04 ** | 3549.94 ± 80.76 ** |
| 5% | 1462.12 ± 35.36 ** | 1183.27 ± 31.12 ** | 736.11 ± 25.53 ** | 2.72 ± 0.03 | 3496.31 ± 80.57 ** | |
| 10% | 1397.1 ± 34.58 ** | 1147.97 ± 30.16 ** | 782.07 ± 25.4 ** | 2.31 ± 0.02 | 3438.69 ± 79.8 ** | |
| 15% | 1326.12 ± 33.39 ** | 1112.15 ± 24.97 ** | 828 ± 24.78 ** | 2.02 ± 0.02 | 3374.25 ± 71.88 ** | |
| 30% | 1128.62 ± 27.88 * | 1027.24 ± 17.10 ** | 954.92 ± 22.97 * | 1.44 ± 0.01 | 3208.93 ± 59.54 ** | |
| 50% | 853.10 ± 22.20 * | 917.61 ± 19.01 ** | 1123.03 ± 30.88 * | 0.99 ± 0.01 | 2976.95 ± 66.27 ** | |
| 70% | 567.70 ± 15.31 | 819.50 ± 27.86 ** | 1313.26 ± 32.97 * | 0.69 ± 0.004 | 2778.61 ± 73.01 ** | |
| 3 min | 0% | 1127.11 ± 25.10 * | 1110.41 ± 19.26 ** | 547.88 ± 25.18 ** | 3.41 ± 0.04 | 2897.76 ± 58.20 ** |
| 5% | 1079.4 ± 24.31 * | 1083.32 ± 18.99 ** | 591.89 ± 26.46 ** | 2.81 ± 0.03 | 2861.37 ± 59.38 * | |
| 10% | 1023.22 ± 22.57 * | 1047.71 ± 15.22 ** | 625.92 ± 17.50 ** | 2.4 ± 0.02 | 2799.93 ± 46.28 * | |
| 15% | 975.01 ± 21.49 * | 1020.62 ± 16.45 ** | 666.64 ± 18.70 ** | 2.09 ± 0.02 | 2761.14 ± 48.79 * | |
| 30% | 827.45 ± 18.66 * | 937.27 ± 13.08 ** | 785.34 ± 13.43 ** | 1.48 ± 0.01 | 2640.77 ± 40.29 * | |
| 50% | 625.49 ± 13.81 | 822.96 ± 12.18 * | 960.21 ± 17.47 * | 1.01 ± 0.01 | 2484.46 ± 40.24 * | |
| 70% | 429.55 ± 9.17 | 719.21 ± 13.36 * | 1137.89 ± 20.49 * | 0.70 ± 0.004 | 2350.27 ± 41.60 * | |
| 2 min | 0% | 1035.59 ± 24.53 * | 1082.92 ± 18.27 ** | 518.80 ± 16.84 ** | 3.42 ± 0.04 | 2743.70 ± 48.45 * |
| 5% | 989.72 ± 23.62 * | 1054.75 ± 17.79 ** | 554.49 ± 18.46 ** | 2.86 ± 0.03 | 2702.02 ± 49.17 * | |
| 10% | 946.56 ± 22.56 * | 1028.3 ± 17.47 ** | 592.71 ± 16.89 ** | 2.47 ± 0.03 | 2667.16 ± 47.16 * | |
| 15% | 893.91 ± 21.12 * | 997.58 ± 16.83 ** | 639.02 ± 19.08 ** | 2.14 ± 0.02 | 2626.85 ± 48.55 * | |
| 30% | 753.04 ± 17.71 | 904.07 ± 14.64 ** | 743.62 ± 14.54 ** | 1.51 ± 0.01 | 2486.35 ± 41.52 * | |
| 50% | 572.83 ± 13.52 | 786.10 ± 12.41 * | 895.60 ± 15.57 * | 1.02 ± 0.01 | 2326.40 ± 38.32 * | |
| 70% | 394.79 ± 9.66 | 682.95 ± 14.42 * | 1067.63 ± 28.46 * | 0.71 ± 0.005 | 2204.56 ± 50.53 * | |
| 1 min | 0% | 616.19 ± 16.25 | 1060.77 ± 17.50 ** | 491.83 ± 10.75 ** | 3.46 ± 0.04 | 2272.02 ± 38.02 * |
| 5% | 588.58 ± 15.48 | 1033.71 ± 16.93 ** | 527.44 ± 11.94 ** | 2.93 ± 0.03 | 2249.53 ± 38.05 * | |
| 10% | 559.61 ± 14.65 | 999.09 ± 16.35 ** | 557.29 ± 9.69 ** | 2.51 ± 0.03 | 2212.12 ± 35.94 * | |
| 15% | 534.03 ± 13.78 | 964.84 ± 15.65 ** | 589.81 ± 10.8 ** | 2.21 ± 0.02 | 2181.41 ± 35.56 * | |
| 30% | 453.45 ± 11.82 | 880.46 ± 14.18 * | 693.12 ± 12.15 ** | 1.57 ± 0.02 | 2110.12 ± 34.48 * | |
| 50% | 337.15 ± 8.56 | 741.32 ± 12.09 * | 831.16 ± 14.37 ** | 1.05 ± 0.01 | 1976.68 ± 32.74 * | |
| 70% | 226.16 ± 5.78 | 623.91 ± 11.57 * | 964.02 ± 17.28 * | 0.71 ± 0.005 | 1867.88 ± 33.62 | |
| 30 s | 0% | — | 1043.22 ± 19.46 ** | 497.03 ± 10.04 ** | 3.47 ± 0.05 | 1634.33 ± 27.33 |
| 5% | — | 1009.19 ± 19.01 ** | 518.67 ± 10.16 ** | 3.01 ± 0.04 | 1618.65 ± 27.22 | |
| 10% | — | 973.86 ± 18.21 ** | 536.36 ± 9.59 ** | 2.64 ± 0.04 | 1597.54 ± 26.45 | |
| 15% | — | 932.54 ± 17.52 ** | 571.69 ± 10.78 ** | 2.29 ± 0.03 | 1588.48 ± 26.87 | |
| 30% | — | 820.17 ± 15.69 * | 628.70 ± 11.05 ** | 1.65 ± 0.02 | 1521.87 ± 26.04 | |
| 50% | — | 671.04 ± 13.42 * | 720.85 ± 13.05 ** | 1.08 ± 0.01 | 1451.23 ± 25.83 | |
| 70% | — | 539.98 ± 11.27 | 839.28 ± 16.60 * | 0.72 ± 0.01 | 1425.51 ± 27.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, A.; Pedreschi, D.; Clifton, D.A.; Morelli, D. Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts. Sensors 2020, 20, 7122. https://doi.org/10.3390/s20247122
Rossi A, Pedreschi D, Clifton DA, Morelli D. Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts. Sensors. 2020; 20(24):7122. https://doi.org/10.3390/s20247122
Chicago/Turabian StyleRossi, Alessio, Dino Pedreschi, David A. Clifton, and Davide Morelli. 2020. "Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts" Sensors 20, no. 24: 7122. https://doi.org/10.3390/s20247122
APA StyleRossi, A., Pedreschi, D., Clifton, D. A., & Morelli, D. (2020). Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts. Sensors, 20(24), 7122. https://doi.org/10.3390/s20247122






