An Analysis Review of Detection Coronavirus Disease 2019 (COVID-19) Based on Biosensor Application
Abstract
1. Introduction and Overview of Coronaviruses
1.1. SARS-CoV-2 Structure and Genome
1.2. Background Virus Detection
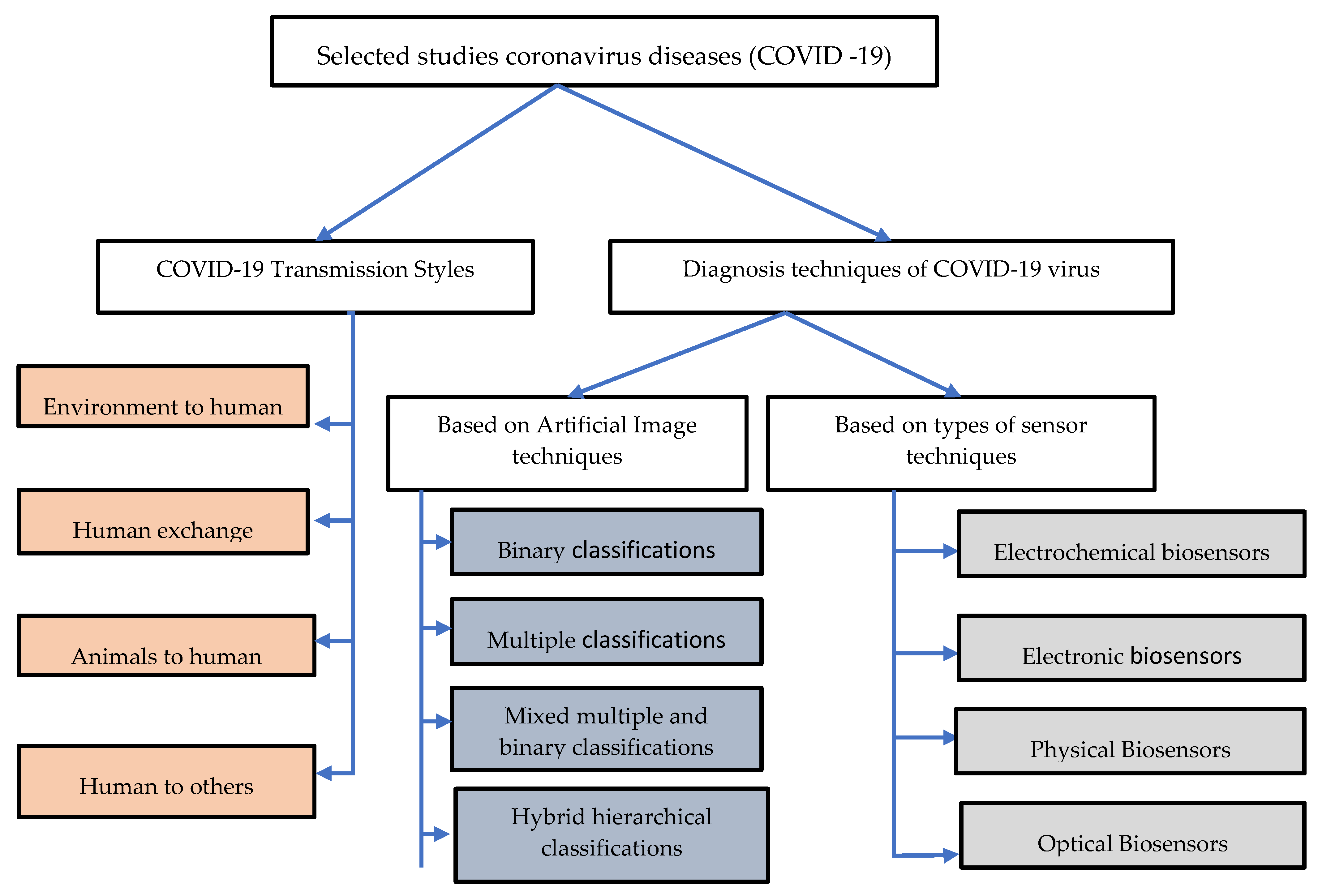
2. Taxonomy of Literature Research of Coronavirus Diseases (COVID-19)
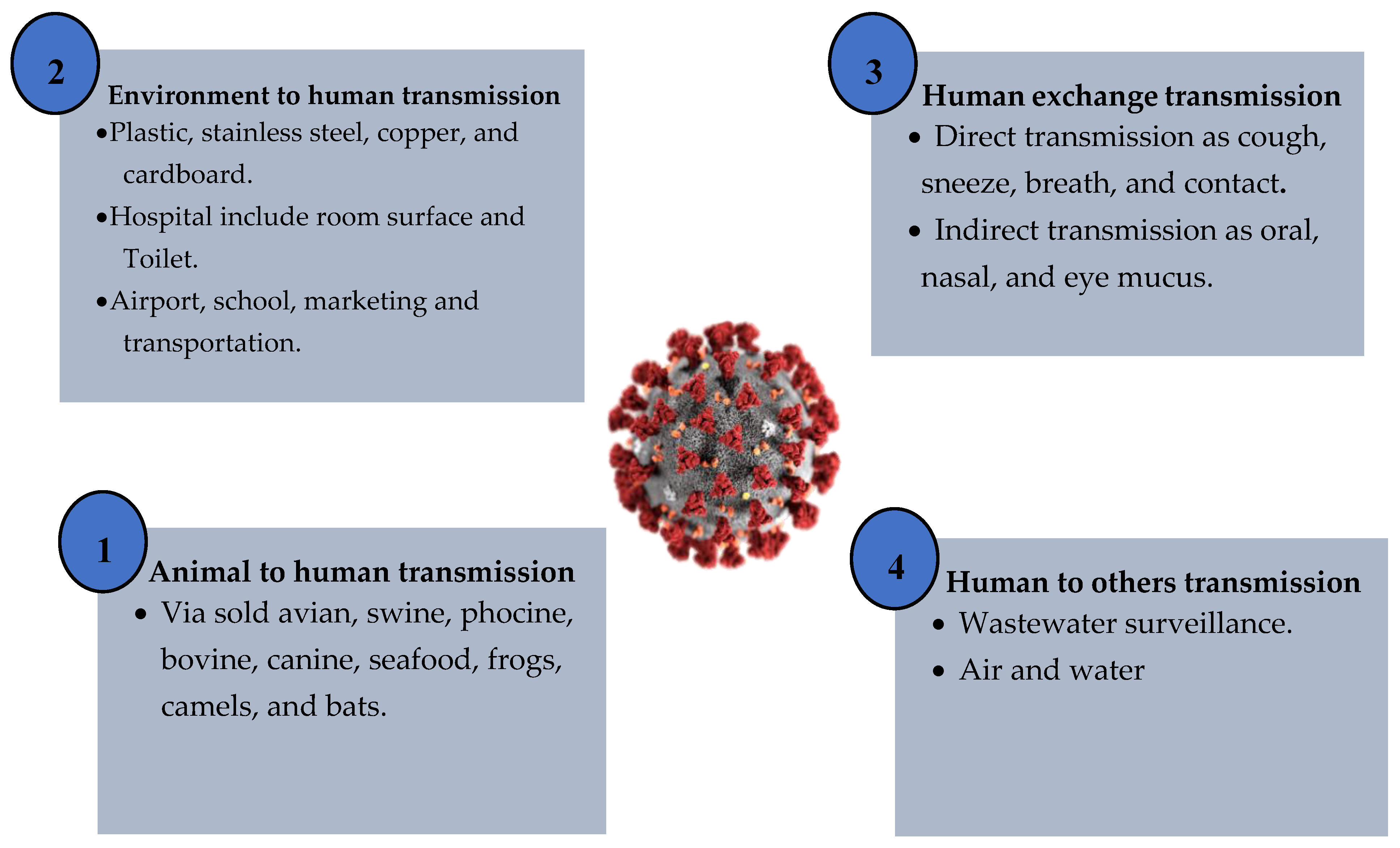
3. COVID-19 Transmission Styles
3.1. Environment to Human
3.2. Human Exchange
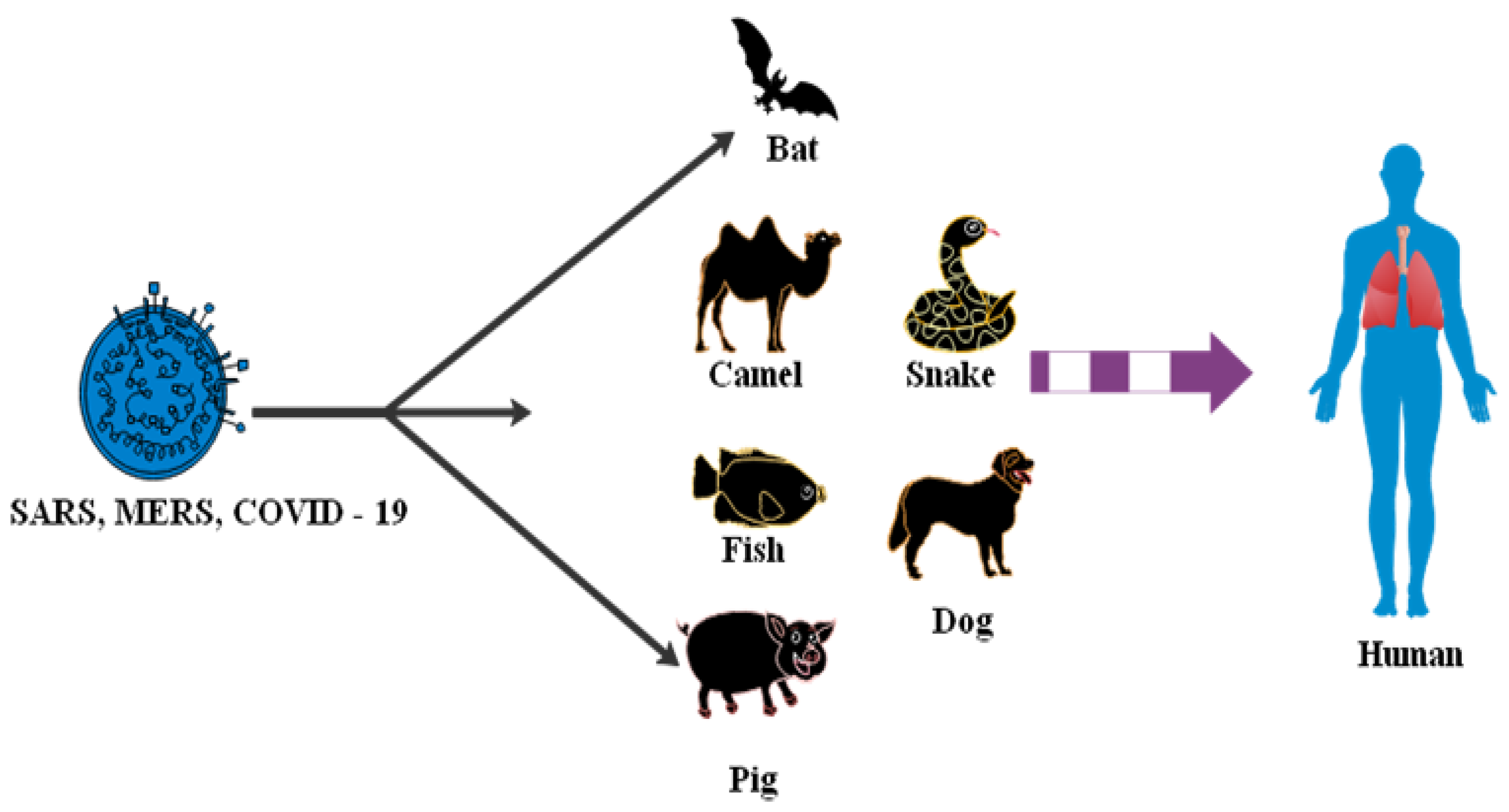
3.3. Animals to Human
3.4. Human to Others
Analysis Outcome of COVID-19 Transmission
4. Diagnosis Techniques of the COVID-19 Virus
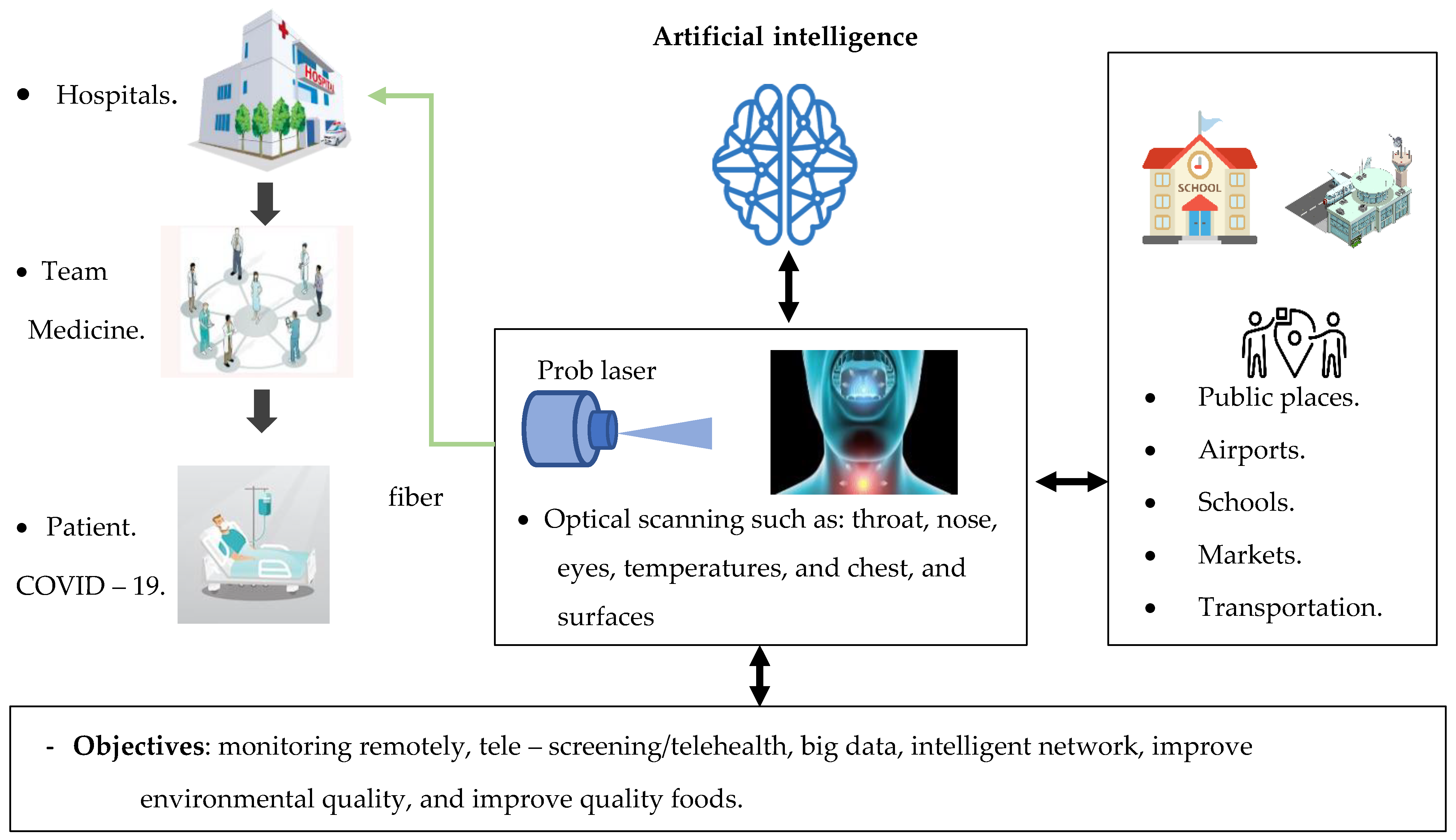
4.1. Based on Artificial Intelligence Techniques
4.1.1. Binary Classifications
4.1.2. Multiple Classifications
4.1.3. Mixed Multiple and Binary Classifications
4.1.4. Hybrid Hierarchical
Analysis Outcome of Diagnostic COVID-19 Based on Artificial Intelligence Techniques
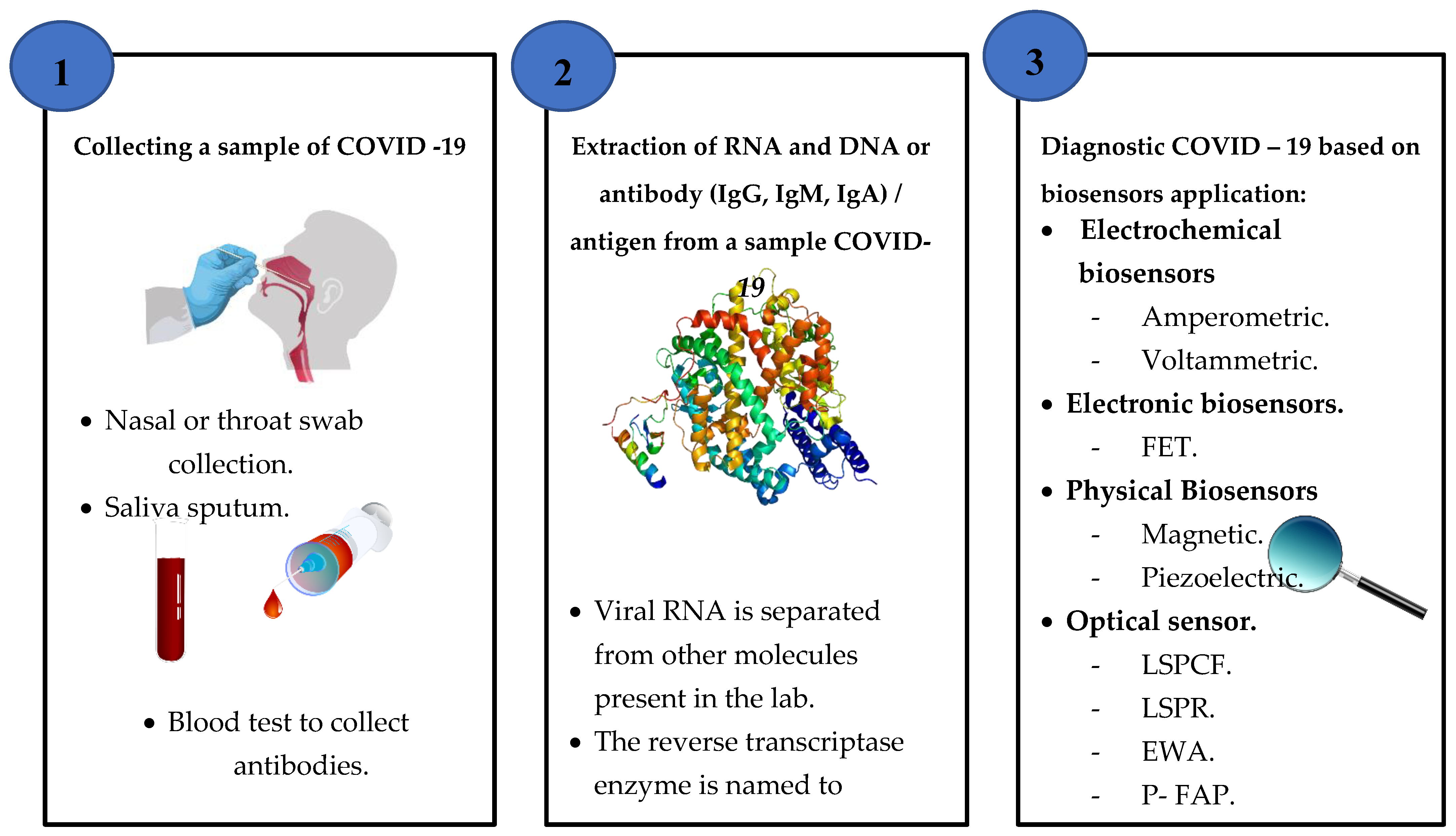
4.2. Based on Biosensors Application
4.2.1. Electrochemical Biosensors
4.2.2. Electronic Biosensors
4.2.3. Physical Biosensors
4.2.4. Optical Biosensors
Analysis Outcome of the Diagnosis of COVID-19 Based on Biosensor Application
- Direct virus identification. A perfect virus is detected only by biosensors or more generally by cultured cell techniques [125].
- Viral recognition of RNA/DNA using RT-PCR and PCR principles, whether attributed with fluorescence in regular nuclear acid platforms or using advanced methods including LSPR, SPR, QCM and other sensor techniques [45].
- Detection of an antibody or antigen. Bioassays use absorption coefficient monitors and many optical and electronic biomedical sensors that basically calculate molecular kinetics. High-resolution scanning probe microscopy with a 1000-fold resolution less than the optical wavelength range in the size of a fraction of a nanometer is used for the surface properties of viruses [126].
5. Enhancement Design of Sensor Output Performance
5.1. Sensitivity
5.2. Signal Noise Ratio
5.3. Full Width at Half Maximum/Minimum (FWHM) and Q-Factor
5.4. Figure of Merit (FOM)
5.5. Detection Limitation
6. Monitoring and Diagnosing the COVID-19 Virus in the Environment
7. Challenges of the Techniques for Detecting COVID-19
7.1. Collection and Transport of Samples
7.2. AI Image Techniques
7.3. Biosensors
8. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| Abbreviation | Description |
| COVID-19 | Coronavirus disease |
| MERS-CoV | Middle East Respiratory Syndrome |
| SARS-CoV-2 | Coronavirus Severe Acute Respiratory Syndrome 2 |
| SARS-CoV | Coronavirus Severe Acute Respiratory Syndrome |
| 2019-nCoV | 2019 novel coronavirus |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
| LFR | Low-Frequency Raman |
| UFOP | U-bent Fiber Optic Probe |
| P-FAB | Plasmonic Fiber-Optic Absorbance Biosensor |
| FET | Field Effect Transistor |
| Zr QDs | Chiral Zirconium Quantum Dots |
| PPM | Parts Per Million |
| QCM | Quartz Crystal Microbalance |
| LFA | Lateral Flow Assays |
References
- Dong, E.; Du, H.; Gardner, L. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Lancet Inf. Dis. 2020, 19, 533–534. [Google Scholar] [CrossRef]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nat. Cell Biol. 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Mitra, A.; Deutsch, B.; Ignatovich, F.; Dykes, C.; Novotny, L. Nano-optofluidic detection of single viruses and nanoparticles. ACS Nano 2010, 4, 1305–1312. [Google Scholar] [CrossRef]
- Wei, Q.; Qi, H.; Luo, W.; Tseng, D.; Ki, S.J.; Wan, Z.; Göröcs, Z.; Bentolila, L.A.; Wu, T.-T.; Sun, R.; et al. Fluorescent imaging of single nanoparticles and viruses on a smart phone. ACS Nano 2013, 7, 9147–9155. [Google Scholar] [CrossRef]
- Caucheteur, C.; Guo, T.; Albert, J. Review of plasmonic fiber optic biochemical sensors: Improving the limit of detection. Anal. Bioanal. Chem. 2015, 407, 3883–3897. [Google Scholar] [CrossRef]
- Shpacovitch, V.; Temchura, V.; Matrosovich, M.; Hamacher, J.; Skolnik, J.; Libuschewski, P.; Siedhoff, D.; Weichert, F.; Marwedel, P.; Müller, H.; et al. Application of surface plasmon resonance imaging technique for the detection of single spherical biological submicrometer particles. Anal. Biochem. 2015, 486, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.T.; Zhang, M.; Zhuo, Y.; Kwon, L.; Race, C. Recent advances in biosensing with photonic crystal surfaces: A Review. IEEE Sens. J. 2015, 16, 3349–3366. [Google Scholar] [CrossRef] [PubMed]
- Hamed, A.M. Image processing of corona virus using interferometry. Opt. Photon J. 2016, 6, 75–86. [Google Scholar] [CrossRef]
- Sakamoto, H.; Minpou, Y.; Sawai, T.; Enami, Y.; Suye, S.-I. A Novel optical biosensing system using Mach–Zehnder-type optical waveguide for influenza virus detection. Appl. Biochem. Biotechnol. 2016, 178, 687–694. [Google Scholar] [CrossRef]
- Zhao, X.; Tsao, Y.-C.; Lee, F.-J.; Tsai, W.-H.; Wang, C.-H.; Chuang, T.-L.; Wu, M.-S.; Lin, C.-W. Optical fiber sensor based on surface plasmon resonance for rapid detection of avian influenza virus subtype H6: Initial studies. J. Virol. Methods 2016, 233, 15–22. [Google Scholar] [CrossRef]
- Vinet, L.; Zhedanov, A. A ‘missing’ family of classical orthogonal polynomials. J. Phys. A: Math. Theor. 2011, 44, 86–91. [Google Scholar] [CrossRef]
- Rahman, M.S.; Anower, M.S.; Rahman, M.K.; Hasan, M.R.; Hossain, M.B.; Haque, M.I. Modeling of a highly sensitive MoS2-Graphene hybrid based fiber optic SPR biosensor for sensing DNA hybridization. Optik 2017, 140, 989–997. [Google Scholar] [CrossRef]
- Wang, L.-J.; Chang, Y.-C.; Sun, R.; Li, L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens. Bioelectron. 2017, 87, 686–692. [Google Scholar] [CrossRef]
- Takemura, K.; Adegoke, O.; Takahashi, N.; Kato, T.; Li, T.-C.; Kitamoto, N.; Tanaka, T.; Suzuki, T.; Park, E.Y. Versatility of a localized surface plasmon resonance-based gold nanoparticle-alloyed quantum dot nanobiosensor for immunofluorescence detection of viruses. Biosens. Bioelectron. 2017, 89, 998–1005. [Google Scholar] [CrossRef]
- Lenssen, J.E.; Toma, A.; Seebold, A.; Shpacovitch, V.; Libuschewski, P.; Weichert, F.; Chen, J.-J.; Hergenröder, R. Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks. In Biosignals, Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies, Madeira, Portugal, 19–21 January 2018; Springer: Berlin, Germany, 2018; Volume 4, pp. 36–47. [Google Scholar] [CrossRef]
- Siedhoff, D.; Strauch, M.; Shpacovitch, V.; Merhof, D. Unsupervised data analysis for virus detection with a surface plasmon resonance sensor. In Proceedings of the 2017 Seventh International Conference on Image Processing Theory, Tools and Applications (IPTA), St. Petersburg, FL, USA, 10–13 December 2017; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Wang, B.-T.; Wang, Q. Sensitivity-Enhanced Optical Fiber Biosensor Based on Coupling Effect Between SPR and LSPR. IEEE Sens. J. 2018, 18, 8303–8310. [Google Scholar] [CrossRef]
- Ma, R.; Oulton, R.F. Applications of nanolasers. Nat. Nanotechnol. 2019, 14, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.A.; Markov, P.; Cartwright, A.P.; Choudhury, M.H.; Afzal, F.O.; Cao, T.; Halimi, S.I.; Retterer, S.T.; Kravchenko, I.I.; Weiss, S.M. Photonic crystal nanobeam biosensors based on porous silicon. Opt. Express 2019, 27, 9536–9549. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, J.; Kim, J.H.; Lee, S.; Kang, M. Sensitive Detection of Multiple Fluoresence Probes based on Surface-enhanced Raman Scattering (SERS) for MERS-CoV. In Proceedings of the 2019 IEEE 14th International Conference on Nano/Micro Engineered and Molecular Systems (NEMS), Bangkok, Thailand, 11–14 April 2019; pp. 498–501. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 1. [Google Scholar] [CrossRef]
- Alqudah, A.M. Covid-2019 Detection using X-Ray Images and Artificial Intelligence Hybrid Systems. In Biomedical Signal and Image Analysis and Project; Biomedical Signal and Image Analysis and Machine Learning Lab: Boca Raton, FL, USA, 2019. [Google Scholar]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus Disease (COVID-19) based on Deep Features and Support Vector Machine. Int. J. Math. Eng. Manag. Sci. 2020, 5, 643–651. [Google Scholar] [CrossRef]
- Liu, S.-L.; Wang, Z.-G.; Xie, H.-Y.; Liu, A.-A.; Lamb, D.C.; Pang, D.-W. Single-Virus Tracking: From Imaging Methodologies to Virological Applications. Chem. Rev. 2020, 120, 1936–1979. [Google Scholar] [CrossRef]
- Xu, B.; Xing, Y.; Peng, J.; Zheng, Z.; Tang, W.; Sun, Y.; Xu, C.; Peng, F. Chest CT for detecting COVID-19: A systematic review and meta-analysis of diagnostic accuracy. Eur. Radiol. 2020, 30, 5720–5727. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Majeed, T.; Rashid, R.; Ali, D.; Asaad, A. Problems of Deploying CNN Transfer Learning to Detect COVID-19 from Chest X-rays. medRxiv 2020. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Loey, M.; Smarandache, F.; Khalifa, N.E.M. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- Asif, S.; Wenhui, Y.; Jin, H.; Tao, Y.; Jinhai, S. Classification of COVID-19 from Chest X-ray images using Deep Convolutional Neural Networks. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses—Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef] [PubMed]
- Nag, P.; Sadani, K.; Mukherji, S. Optical Fiber Sensors for Rapid Screening of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 5, 233–236. [Google Scholar] [CrossRef]
- Bhalla, N.; Pan, Y.; Yang, Z.; Payam, A.F. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14, 7783–7807. [Google Scholar] [CrossRef] [PubMed]
- Murugan, D.; Bhatia, H.; Sai, V.; Satija, J. P-FAB: A Fiber-Optic Biosensor Device for Rapid Detection of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 5, 211–215. [Google Scholar] [CrossRef]
- Naudé, W. Artificial Intelligence against COVID-19: An Early Review; No. 13110; IZA—Institute of Labor Economics: Bonn, Germany, 2020. [Google Scholar]
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv 2020, 1–19. [Google Scholar] [CrossRef]
- Jacobi, L.; Damle, V.H.; Rajeswaran, B.; Tischler, Y.R. Low-Frequency Raman Spectroscopy as a Diagnostic Tool for COVID-19 and other Coronaviruses. R. Soc. Open Sci. 2020, 7, 1–28. [Google Scholar]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Lee, H.-C.; Diao, K.-Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Hemdan, E.E.-D.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Janczuk-Richter, M.; Gromadzka, B.; Richter, Ł.; Panasiuk, M.; Zimmer, K.; Mikulic, P.; Bock, W.J.; Maćkowski, S.; Smietana, M.; Niedziolka-Jonsson, J. Immunosensor Based on Long-Period Fiber Gratings for Detection of Viruses Causing Gastroenteritis. Sensors 2020, 20, 813. [Google Scholar] [CrossRef]
- Horve, P.F.; Lloyd, S.; Mhuireach, G.A.; Dietz, L.; Fretz, M.; MacCrone, G.; Wymelenberg, K.V.D.; Ishaq, S.L. Building upon current knowledge and techniques of indoor microbiology to construct the next era of theory into microorganisms, health, and the built environment. J. Expo. Sci. Environ. Epidemiol. 2019, 30, 219–235. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Lloyd-Smith, J.O.; De Wit, E.; Munster, V.J.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Sutjipto, S.; Lee, P.H.; Son, T.T.; Young, B.E.; et al. Detection of Air and Surface Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Hospital Rooms of Infected Patients. medRxiv 2020, 125. [Google Scholar] [CrossRef]
- Razzini, K.; Castrica, M.; Menchetti, L.; Maggi, L.; Negroni, L.; Orfeo, N.V.; Pizzoccheri, A.; Stocco, M.; Muttini, S.; Balzaretti, C.M. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci. Total Environ. 2020, 742, 140540. [Google Scholar] [CrossRef]
- Otter, J.; Donskey, C.J.; Yezli, S.; Douthwaite, S.T.; Goldenberg, S.; Weber, D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250. [Google Scholar] [CrossRef]
- Xu, H.; Yan, C.; Fu, Q.; Xiao, K.; Yu, Y.; Han, D.; Wang, W.; Cheng, J. Possible environmental effects on the spread of COVID-19 in China. Sci. Total Environ. 2020, 731, 139211. [Google Scholar] [CrossRef]
- Dietz, L.; Horve, P.F.; Coil, D.A.; Fretz, M.; Eisen, J.A.; Van Den Wymelenberg, K. 2019 Novel Coronavirus (COVID-19) Pandemic: Built Environment Considerations to Reduce Transmission. mSystems 2020, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods - A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yan, X. Hospital indoor air quality monitoring for the detection ofSARS- CoV-2 (COVID-19) virus. J. Mater. Sci. Technol. 2020, 43, 175–188. [Google Scholar] [CrossRef]
- Ding, Z.; Qian, H.; Xu, B.; Huang, Y.; Miao, T.; Yen, H.-L.; Xiao, S.; Cui, L.; Wu, X.; Shao, W.; et al. Toilets dominate environmental detection of SARS-CoV-2 virus in a hospital. medRxiv 2020. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Tang, Y.-W. Laboratory diagnosis of emerging human coronavirus infections—The state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Perez-Campos-Mayoral, L.; Hernández-Huerta, M.T.; Mayoral-Andrade, G.; Mayoral, E.P.-C.; Pérez-Campos, E. A letter to the editor on World Health Organization declares global emergency: A review of the 2019 novel Coronavirus (COVID-19). Int. J. Surg. 2020, 79, 163–164. [Google Scholar] [CrossRef]
- Willcox, M.D.; Walsh, K.; Od, J.J.N.; Morgan, P.B.; Jones, L.W. The ocular surface, coronaviruses and COVID-19. Clin. Exp. Optom. 2020, 103, 418–424. [Google Scholar] [CrossRef]
- Ho, D.; Low, R.; Tong, L.; Gupta, V.; Veeraraghavan, A.; Agrawal, R. COVID-19 and the Ocular Surface: A Review of Transmission and Manifestations. Ocul. Immunol. Inflamm. 2020, 28, 726–734. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Li, Y.; Cheng, L.; Zhou, X.; Ren, B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.; Zaloga, D.J.; Friderici, C.S. COVID-19 Personal Protective Equipment (PPE) for the emergency physician. Vis. J. Emerg. Med. 2020, 19, 100740. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Kabir, E.; Azzouz, A.; Raza, N.; Bhardwaj, S.K.; Tsang, D.C.W.; Tabatabaei, M.; Kukkar, D. Recent Advances in Monitoring, Sampling, and Sensing Techniques for Bioaerosols in the Atmosphere. ACS Sens. 2020, 5, 1254–1267. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Cohen, J. Wuhan seafood market may not be source of novel virus spreading globally. Science 2020, 10. [Google Scholar] [CrossRef]
- Letko, M.C.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Genet. 2020, 18, 461–471. [Google Scholar] [CrossRef]
- Hu, B.; Ge, X.; Wang, L.-F.; Shi, Z. Bat origin of human coronaviruses. Virol. J. 2015, 12, 1–10. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Lapolla, P.; Lee, R.; Mingoli, A. Wastewater as a red flag in COVID-19 spread. Public Health 2020, 185, 26. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Silverman, A.I.; Boehm, A.B. Systematic Review and Meta-Analysis of the Persistence and Disinfection of Human Coronaviruses and Their Viral Surrogates in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 544–553. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Lim, X.F.; Lim, A.S.; Sutjipto, S.; Lee, P.H.; et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020, 11, 2800. [Google Scholar] [CrossRef] [PubMed]
- Kronbichler, A.; Effenberger, M.; Eisenhut, M.; Lee, K.H.; Shinc, J.I. Seven recommendations to rescue the patients and reduce the mortality from COVID-19 infection: An immunological point of view. Autoimmun. Rev. 2020, 19, 102570. [Google Scholar] [CrossRef]
- Fagone, P.; Ciurleo, R.; Lombardo, S.D.; Iacobello, C.; Palermo, C.I.; Shoenfeld, Y.; Bendtzen, K.; Bramanti, P.; Nicoletti, F. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev. 2020, 19, 102571. [Google Scholar] [CrossRef]
- Del Papa, N.; Sambataro, G.; Minniti, A.; Pignataro, F.; Caporali, R. Novel COronaVirus Disease 2019 (COVID-19) epidemic: What are the risks for systemic sclerosis patients? Autoimmun. Rev. 2020, 19, 102558. [Google Scholar] [CrossRef]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323, 1610. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, R.; Zhao, J.; Molina, M.J. Understanding transmission and intervention for the COVID-19 pandemic in the United States. Sci. Total Environ. 2020, 748, 141560. [Google Scholar] [CrossRef]
- El Zowalaty, M.E.; Järhult, J.D. From SARS to COVID-19: A previously unknown SARS-related coronavirus (SARS-CoV-2) of pandemic potential infecting humans—Call for a One Health approach. One Health 2020, 9, 100124. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Albahri, O.; Zaidan, A.; Zaidan, B.; Abdulkareem, K.H.; Al-Qaysi, Z.; Alamoodi, A.; Aleesa, A.; Chyad, M.; Alesa, R.; Kem, L.; et al. Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. J. Infect. Public Health 2020, 13, 1381–1396. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, D.; Dong, J.; Wang, N.; Huang, H.; Xu, H.; Xia, C. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J. Radiol. 2020, 21, 505–508. [Google Scholar] [CrossRef]
- Tuncer, T.; Dogan, S.; Ozyurt, F. An automated Residual Exemplar Local Binary Pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image. Chemom. Intell. Lab. Syst. 2020, 203, 104054. [Google Scholar] [CrossRef]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef] [PubMed]
- Wallis, L.A. COVID-19 Severity Scoring Tool for low resourced settings. Afr. J. Emerg. Med. 2020. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020, 121, 103805. [Google Scholar] [CrossRef]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable Deep Learning for Pulmonary Disease and Coronavirus COVID-19 Detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Laghi, A. Cautions about radiologic diagnosis of COVID-19 infection driven by artificial intelligence. Lancet Digit. Health 2020, 2, e225. [Google Scholar] [CrossRef]
- McCall, B. COVID-19 and artificial intelligence: Protecting health-care workers and curbing the spread. Lancet Digit. Health 2020, 2, e166–e167. [Google Scholar] [CrossRef]
- Pereira, R.M.; Bertolini, D.; Teixeira, L.O.; Silla, C.N.; Costa, Y.M. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020, 194, 105532. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Basset, M.; Mohamed, R.; Elhoseny, M.; Chakrabortty, R.K.; Ryan, M.J. A Hybrid COVID-19 Detection Model Using an Improved Marine Predators Algorithm and a Ranking-Based Diversity Reduction Strategy. IEEE Access 2020, 8, 79521–79540. [Google Scholar] [CrossRef]
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Goode, J.A.; Rushworth, J.V.H.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2014, 31, 6267–6276. [Google Scholar] [CrossRef]
- Justino, C.I.; Rocha-Santos, T.A.; Duarte, A.C. Review of analytical figures of merit of sensors and biosensors in clinical applications. TrAC Trends Anal. Chem. 2010, 29, 1172–1183. [Google Scholar] [CrossRef]
- Cetin, A.E.; Coskun, A.F.; Galarreta, B.C.; Huang, M.; Herman, D.; Ozcan, A.; Altug, H. Handheld high-throughput plasmonic biosensor using computational on-chip imaging. Light Sci. Appl. 2014, 3, e122. [Google Scholar] [CrossRef]
- Mimee, M.; Nadeau, P.; Hayward, A.; Carim, S.; Flanagan, S.; Jerger, L.; Collins, J.; McDonnell, S.; Swartwout, R.; Citorik, R.J.; et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 2018, 360, 915–918. [Google Scholar] [CrossRef]
- Alonso-Lomillo, M.; Domínguez-Renedo, O.; Martínez, M.A. Screen-printed biosensors in microbiology; a review. Talanta 2010, 82, 1629–1636. [Google Scholar] [CrossRef]
- Kim, J.; Campbell, A.S.; De Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]
- Melnychuk, N.; Egloff, S.; Runser, A.; Reisch, A.; Klymchenko, A.S. Light-Harvesting Nanoparticle Probes for FRET-Based Detection of Oligonucleotides with Single-Molecule Sensitivity. Angew. Chem. 2020, 132, 6878–6885. [Google Scholar] [CrossRef]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv 2020, 1–20. [Google Scholar] [CrossRef]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Hasan, M.; Hossain, S.; Ahommed, M.; Daizy, M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: State of the art. Biosens. Bioelectron. 2020, 166, 112431. [Google Scholar] [CrossRef]
- Russo, L.; Bueno, J.L.; Bergua, J.F.; Costantini, M.; Giannetto, M.; Puntes, V.; De La Escosura-Muñiz, A.; Merkoçi, A. Low-Cost Strategy for the Development of a Rapid Electrochemical Assay for Bacteria Detection Based on AuAg Nanoshells. ACS Omega 2018, 3, 18849–18856. [Google Scholar] [CrossRef]
- Dudok de Wit, C. Medische informatie voor verpleegkundigen. Autologe bloedtransfusie. Tijdschr. Voor Ziekenverpl. 1987, 41, 655–657. [Google Scholar]
- Rauf, S.; Mishra, G.K.; Azhar, J.; Mishra, R.K.; Goud, K.Y.; Nawaz, M.A.H.; Marty, J.-L.; Hayat, A. Carboxylic group riched graphene oxide based disposable electrochemical immunosensor for cancer biomarker detection. Anal. Biochem. 2018, 545, 13–19. [Google Scholar] [CrossRef]
- Zhu, J.; Gan, H.; Wu, J.; Ju, H. Molecular Machine Powered Surface Programmatic Chain Reaction for Highly Sensitive Electrochemical Detection of Protein. Anal. Chem. 2018, 90, 5503–5508. [Google Scholar] [CrossRef]
- Kaisti, M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef]
- Zuo, B.; Li, S.; Guo, Z.; Zhang, J.; Chen, C. Piezoelectric Immunosensor for SARS-Associated Coronavirus in Sputum. Anal. Chem. 2004, 76, 3536–3540. [Google Scholar] [CrossRef]
- Pietschmann, J.; Voepel, N.; Spiegel, H.; Krause, H.-J.; Schröper, F. Brief Communication: Magnetic Immuno-Detection of SARS-CoV-2 specific Antibodies. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wu, K.; Saha, R.; Su, D.; Krishna, V.D.; Liu, J.; Cheeran, M.C.-J.; Wang, J.-P. Magnetic Immunoassays: A Review of Virus and Pathogen Detection Before and Amidst the Coronavirus Disease-19 (COVID-19). arXiv 2020, arXiv:2007.04809. [Google Scholar]
- Yu, X.; Chen, F.; Wang, R.; Li, Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E. coli O157:H7 using a QCM sensor. J. Biotechnol. 2018, 266, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Lord, H.L.; Zhan, W.; Pawliszyn, J. Fundamentals and applications of needle trap devices. Anal. Chim. Acta 2010, 677, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Amstutz, P.; Tang, C.-M.; Hang, J.; Zhu, P.; Zhang, Y.; Shelton, D.R.; Karns, J.S. Integrating Waveguide Biosensor. Breast Cancer 2009, 503, 389–401. [Google Scholar] [CrossRef]
- Socorro-Leránoz, A.; Santano, D.; Del Villar, I.; Matias, I. Trends in the design of wavelength-based optical fibre biosensors (2008–2018). Biosens. Bioelectron. X 2019, 1, 100015. [Google Scholar] [CrossRef]
- Huang, J.C.; Chang, Y.F.; Chen, K.H.; Su, L.C.; Lee, C.W.; Chen, C.C.; Chen, Y.M.; Chou, C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009, 25, 320–325. [Google Scholar] [CrossRef]
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, G.F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J.; et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nat. Cell Biol. 2013, 500, 227–231. [Google Scholar] [CrossRef]
- Roh, C.; Jo, S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011, 86, 1475–1479. [Google Scholar] [CrossRef]
- Weng, X.; Neethirajan, S. Immunosensor Based on Antibody-Functionalized MoS2 for Rapid Detection of Avian Coronavirus on Cotton Thread. IEEE Sens. J. 2018, 18, 4358–4363. [Google Scholar] [CrossRef]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef]
- Saylan, Y.; Erdem, Ö.; Ünal, S.; Denizli, A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yan, X.; Cao, W.; Wang, C.; Feng, J.; Duan, J.; Xie, S. Probing the structure of the SARS coronavirus using scanning electron microscopy. Antivir. Ther. 2004, 9, 287–289. [Google Scholar] [PubMed]
- Papageorgiou, N.; Lichière, J.; Baklouti, A.; Ferron, F.; Sévajol, M.; Canard, B.; Coutard, B. Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Chiavaioli, F.; Gouveia, C.; Jorge, P.; Baldini, F. Towards a Uniform Metrological Assessment of Grating-Based Optical Fiber Sensors: From Refractometers to Biosensors. Biosensors 2017, 7, 23. [Google Scholar] [CrossRef]
- Wang, C.T.; Jiang, B.; Zhou, Y.W.; Jiang, T.W.; Liu, J.H.; Zhu, G.D.; Cai, W.B. Exploiting the surface-enhanced IR absorption effect in the photothermally induced resonance AFM-IR technique toward nanoscale chemical analysis. Anal. Chem. 2019, 91, 10541–10548. [Google Scholar] [CrossRef]
- Volz, S. Thermal Nanosystems and Nanomaterials; Springer: Berlin, Germany, 2009; Volume 118. [Google Scholar]
- Saah, A.J.; Hoover, D.R. Sensitivity’ and ‘specificity’ revisited: The signification of these terms in analytical and diagnostic language. Ann. Dermatol. Venereol. 1998, 125, 291–294. [Google Scholar]
- Förster, R.; Weidlich, S.; Nissen, M.; Wieduwilt, T.; Kobelke, J.; Goldfain, A.M.; Chiang, T.K.; Garmann, R.F.; Manoharan, V.N.; Lahini, Y.; et al. Tracking and Analyzing the Brownian Motion of Nano-objects Inside Hollow Core Fibers. ACS Sens. 2020, 5, 879–886. [Google Scholar] [CrossRef]
- Deckert, V.; Deckert-Gaudig, T.; Cialla-May, D.; Popp, J.; Zell, R.; Deinhard-Emmer, S.; Sokolov, A.V.; Yi, Z.; Scully, M.O. Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS. Proc. Natl. Acad. Sci. USA 2020, 117, 13169–27824. [Google Scholar] [CrossRef] [PubMed]
- Burkhartsmeyer, J.; Wang, Y.; Wong, K.S.; Gordon, R. Optical Trapping, Sizing, and Probing Acoustic Modes of a Small Virus. Appl. Sci. 2020, 10, 394. [Google Scholar] [CrossRef]
- Hales, J.E.; Matmon, G.; Dalby, P.A.; Ward, J.M.; Aeppli, G. Virus lasers for biological detection. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kukushkin, V.I.; Ivanov, N.M.; Novoseltseva, A.A.; Gambaryan, A.S.; Yaminsky, I.V.; Kopylov, A.M.; Zavyalova, E.G. Highly sensitive detection of influenza virus with SERS aptasensor. PLoS ONE 2019, 14, e0216247. [Google Scholar] [CrossRef] [PubMed]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Chua, G.T.; Kwok, K.L.; Wong, J.S.; Au, D.C.Y.; Lam, Y.Y.; Wong, W.H.; Ho, M.H.; Chan, G.C.; Chui, C.S.; et al. False-positive SARS-CoV-2 serology in 3 children with Kawasaki disease. Diagn. Microbiol. Infect. Dis. 2020, 98, 115141. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhi, S.; Chen, M.; Su, X.; Kang, L.; Li, C.; Su, X.; Zhang, S.; Ge, S.; Li, W. Heat inactivation decreases the qualitative real-time RT-PCR detection rates of clinical samples with high cycle threshold values in COVID-19. Diagn. Microbiol. Infect. Dis. 2020, 98, 115109. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Lin, C.; Xiang, J.; Yan, M.; Li, H.; Huang, S.; Shen, C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19). Clin. Chem. Lab. Med. 2020, 58, 1089–1094. [Google Scholar] [CrossRef]
- Naudé, W. Artificial intelligence vs COVID-19: Limitations, constraints and pitfalls. AI Soc. 2020, 35, 761–765. [Google Scholar] [CrossRef]
- Joseph, W.S. Letters to the Editor. J. Am. Podiatr. Med. Assoc. 2006, 96, 272. [Google Scholar] [CrossRef]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296, E41–E45. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.; Leung, W.S.; So, R.T.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31, 107725. [Google Scholar] [CrossRef] [PubMed]
- Weston, S.; Frieman, M.B. Respiratory Viruses. Ref. Modul. Biomed. Sci. 2018, 85–101. [Google Scholar] [CrossRef]
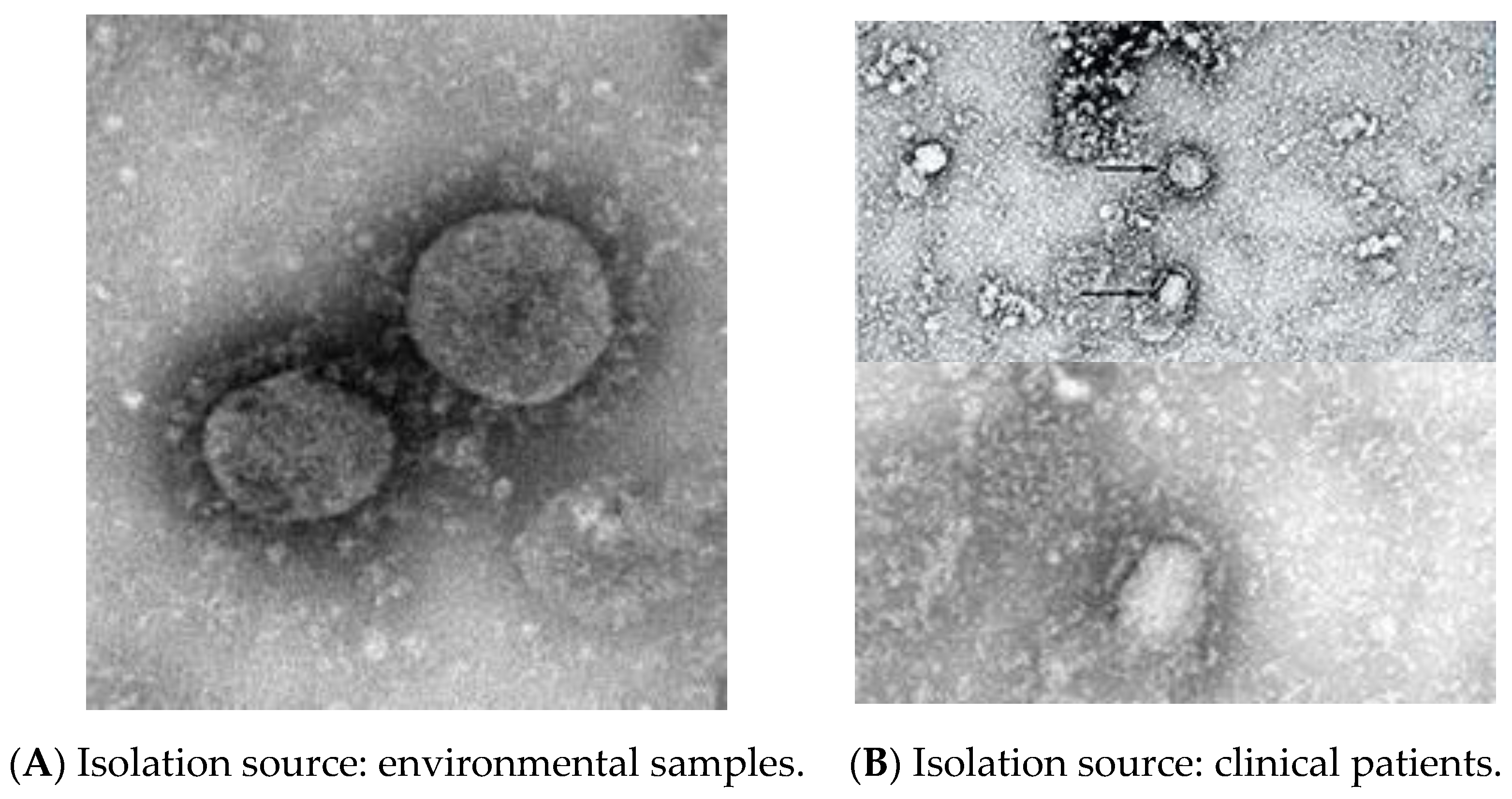


| Transmission Style | Hazard | Places | Data Published | Country | Finding | Reference |
|---|---|---|---|---|---|---|
| Environment to human | High | Plastic, stainless steel, copper and cardboard | 17 March 2020 | USA | SARS-CoV-2 | [51,56] |
| Environment to human | High | Hospital rooms | 29 May 2020 | Singapore | SARS-CoV-2 | [52,58] |
| Environment to human | High | Air and surface | 26 June 2020 | Italy | SARS-CoV-2 | [53] |
| Environment to human | High | Surface contamination | 2016 | London | SARS-CoV and MERS-CoV | [54] |
| Environment to human | High | Air, surface | 7 May 2020 | China | COVID-19 | [55] |
| Environment to human | High | Water | 28 April 2020 | Italy | SARS-CoV-2 | [57] |
| Environment to human | High | Toilet | 13 August 2020 | China | SARS-CoV-2 | [59] |
| Human exchange | High | Contacts | 24 January 2020 | China | SARS-CoV-2 | [61] |
| Human exchange | High | Ocular surface | 22 April 2020 | Australia | SARS-CoV-2 | [63] |
| Human exchange | High | Dental clinics | 19 February 2020 | China | SARS-CoV-2 | [66] |
| Animal to human | Medium | Bats | 2020 | USA | SARS-CoV-2 | [73,74] |
| Animal to human | Medium | Sold avian, swine, porcine, bovine, canine, seafood, frogs, camels | 2020 | China | SARS-CoV-2 | [71] |
| Human to others | Low | Wastewater surveillance | 18 April 2020 | Australia | SARS-CoV-2 | [76,78] |
| Type of Dataset | AI Techniques | Case Study | Efficiency of Detection (%) | Installation Data | Collection Dataset Size | AI Partition | No. of Classes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prime Data | Minor Data | Traditional Machine Learning Techniques | Deep Learning Techniques | ||||||||
| Test | Validation | Training | |||||||||
| ✕ | ✕ | ✕ | ✓ | CT scan | 96% | 19 February 2020 | 2 images | ✕ | ✕ | 13.3% | COVID-19, Flu |
| ✓ | ✕ | ✓ | ✕ | CT scan | 93.6% | 2020 | 13,500 images | ✕ | ✕ | ✕ | COVID-19 |
| ✓ | ✓ | ✓ | ✓ | CT scan | 50% | 2020 | NA | ✓ | ✕ | ✕ | COVID-19 |
| ✓ | ✓ | ✓ | ✓ | CT scan | NA | 20 February 2020 | NA | ✕ | ✕ | ✕ | COVID-19 |
| ✕ | ✓ | ✕ | ✓ | X-ray | 87.02%, 98.08% | 26 April 2020 | 127 images | ✕ | 5-fold cross | ✕ | COVID-19, no finding and pneumonia |
| ✕ | ✓ | ✕ | ✓ | X-ray | 98.03% | 21 April 2020 | 2839 images | 10% | 10% | 80% | COVID-19, normal, pneumonia |
| ✕ | ✓ | ✕ | ✓ | X-ray | 86.9% | 16 June 2020 | 430 images | 0.7 | 0.1 | 0.2 | Normal, bacterial tuberculosis, viral and COVID-19 |
| ✕ | ✓ | ✓ | ✕ | X-ray | 80% | 2020 | 8 images | ✕ | two-fold | ✕ | COVID-19 |
| ✕ | ✓ | ✓ | ✓ | X-ray | 99.27% | 2 May 2020 | 845 images | 30% | 5-fold cross | 70% | coronavirus, pneumonia and normal |
| ✓ | ✓ | ✓ | ✓ | X-ray | 87%, 98% | 2020 | 1144 images | 30% | ✕ | 70% | Normal, COVID-19, MERS SARS, Varicella, Streptococcus Pneumocystis |
| Sensor Type | Application Range | Material (nm) | Prognosis | Diagnosis | Installation Date | Target | Duration | Detection Limit | Sample Size | Detection of COVID-19 Virus | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| In Clinical | On the Surface | ||||||||||
| Optical LSPR PPT | SARS-CoV-2 | Gold | ✓ | ✓ | 8 Apr 220 | DNA | 800 s | 0.22 pM | 0.01 pM–50 Μm | ✓ | ✕ |
| FET sensor | SARS-CoV-2 | Graphene | ✓ | ✓ | 29 May 2020 | Spike protein | 4 h | 1.6 × 101 pfu/mL | 1 fg/mL | ✓ | ✕ |
| Magnetic | SARS-CoV | NA | ✓ | ✓ | 3 July 2020 | Anti-spike protein | 42 min | 2.96 ng/mL | 2 ng mL−1 | ✓ | ✕ |
| P-FABU-bent optical fiber | COVID-19 | Gold | ✓ | ✓ | 1 June 2020 | N-protein | 15 min | 10–18 M | 106 particles/mL | ✓ | ✕ |
| Electrochemical | SARS-CoV-2 | Gold | ✓ | ✓ | 11 M ay 2020 | Spike protein | 10–30 s | 90 fM | 1 μM | ✓ | ✕ |
| Electrochemical | MERS-CoV | Gold | ✓ | ✓ | 27 Feb 2019 | Spike protein | 20 min | 1.0 pg mL−1 | 10 μg mL−1 | ✓ | ✕ |
| Optical Fiber | COVID-19 | Gold | ✕ | ✓ | 11 June 2020 | IgG-type antibodies | 1 h | 100 units/mL | NA | ✓ | ✕ |
| Piezoelectric | SARS-CoV | Crystal with quartz wafer | ✕ | ✓ | 2004 | Antigen (sputum) | 1 h | 0.6 µg/mL | 0.6–4 µg/mL | ✓ | ✕ |
| Optical LSPCF | SARS-COV | Gold | ✓ | ✓ | 2009 | Nucleocapsid protein | 3 h | 1 pg/mL | ∼1 pg/mL | ✓ | ✕ |
| Optical | SARS-CoV | Quantum dots | ✓ | ✓ | 24 July 2011 | Nucleocapsid protein | 1 h | 0.1 pg/mL | 0.1 pg mL−1 | ✓ | ✕ |
| Nanocrystals optical | Coronavirus | Zirconium quantum dots | ✕ | ✓ | 29 Oct 2018 | Antibodies | 1 h | 79.15 EID/50 mL | 1000 EID/50 mL | ✓ | ✕ |
| LFA | SARS-CoV-2 | Gold | ✕ | ✓ | 21 May 2020 | IgM antibody | 15 min | NA | 10−20 μL | ✓ | ✕ |
| Challenges, Detection of COVID-19 Virus Techniques According to Critical Review Research | ||
|---|---|---|
| Virus Detection Categories | Techniques | Limitation |
| Indirect detection: DNA, RNA | Electronic sensors |
|
| Indirect detection: DNA, RNA | Magnetic sensors |
|
| Indirect detection: Spike protein | Electrochemical sensors |
|
| Indirect detection: IgM antibody, DNA, RNA | Optical sensors: LSPR, P-FAB, EWA, QCM, SPR and LSPCF |
|
| Direct detection: CT image | CT scan |
|
| Direct detection: X-ray image | CXR |
|
| All Detection Techniques of COVID-19 Viruses Based On | ||
| Viral RNA, DNA Identification |
| |
| Antibody or Antigen Identification |
| |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, B.A.; Al Mashhadany, Y.; Hafiz Mokhtar, M.H.; Dzulkefly Bin Zan, M.S.; Arsad, N. An Analysis Review of Detection Coronavirus Disease 2019 (COVID-19) Based on Biosensor Application. Sensors 2020, 20, 6764. https://doi.org/10.3390/s20236764
Taha BA, Al Mashhadany Y, Hafiz Mokhtar MH, Dzulkefly Bin Zan MS, Arsad N. An Analysis Review of Detection Coronavirus Disease 2019 (COVID-19) Based on Biosensor Application. Sensors. 2020; 20(23):6764. https://doi.org/10.3390/s20236764
Chicago/Turabian StyleTaha, Bakr Ahmed, Yousif Al Mashhadany, Mohd Hadri Hafiz Mokhtar, Mohd Saiful Dzulkefly Bin Zan, and Norhana Arsad. 2020. "An Analysis Review of Detection Coronavirus Disease 2019 (COVID-19) Based on Biosensor Application" Sensors 20, no. 23: 6764. https://doi.org/10.3390/s20236764
APA StyleTaha, B. A., Al Mashhadany, Y., Hafiz Mokhtar, M. H., Dzulkefly Bin Zan, M. S., & Arsad, N. (2020). An Analysis Review of Detection Coronavirus Disease 2019 (COVID-19) Based on Biosensor Application. Sensors, 20(23), 6764. https://doi.org/10.3390/s20236764








