Wide Field Spectral Imaging with Shifted Excitation Raman Difference Spectroscopy Using the Nod and Shuffle Technique
Abstract
1. Introduction
2. Materials and Methods
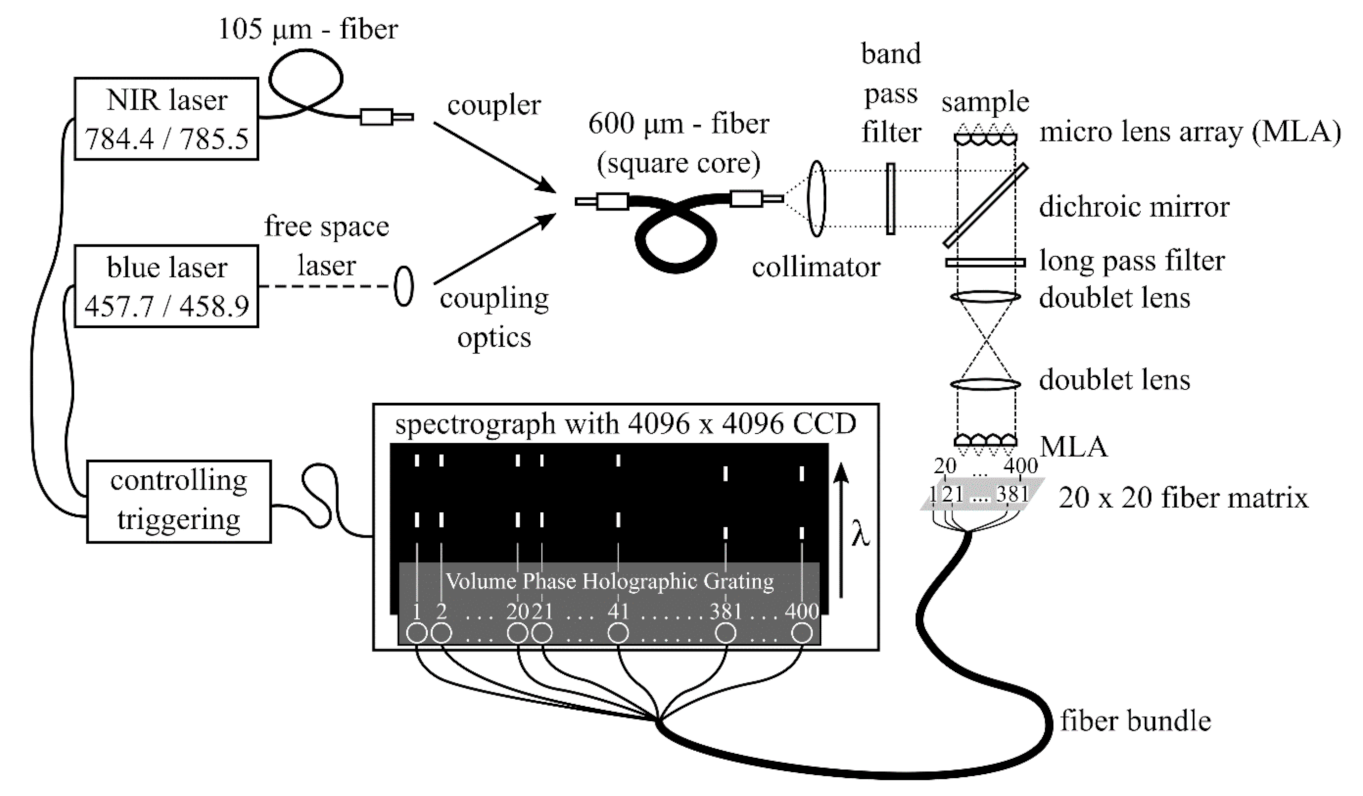
2.1. Wide Field SERDS Setup
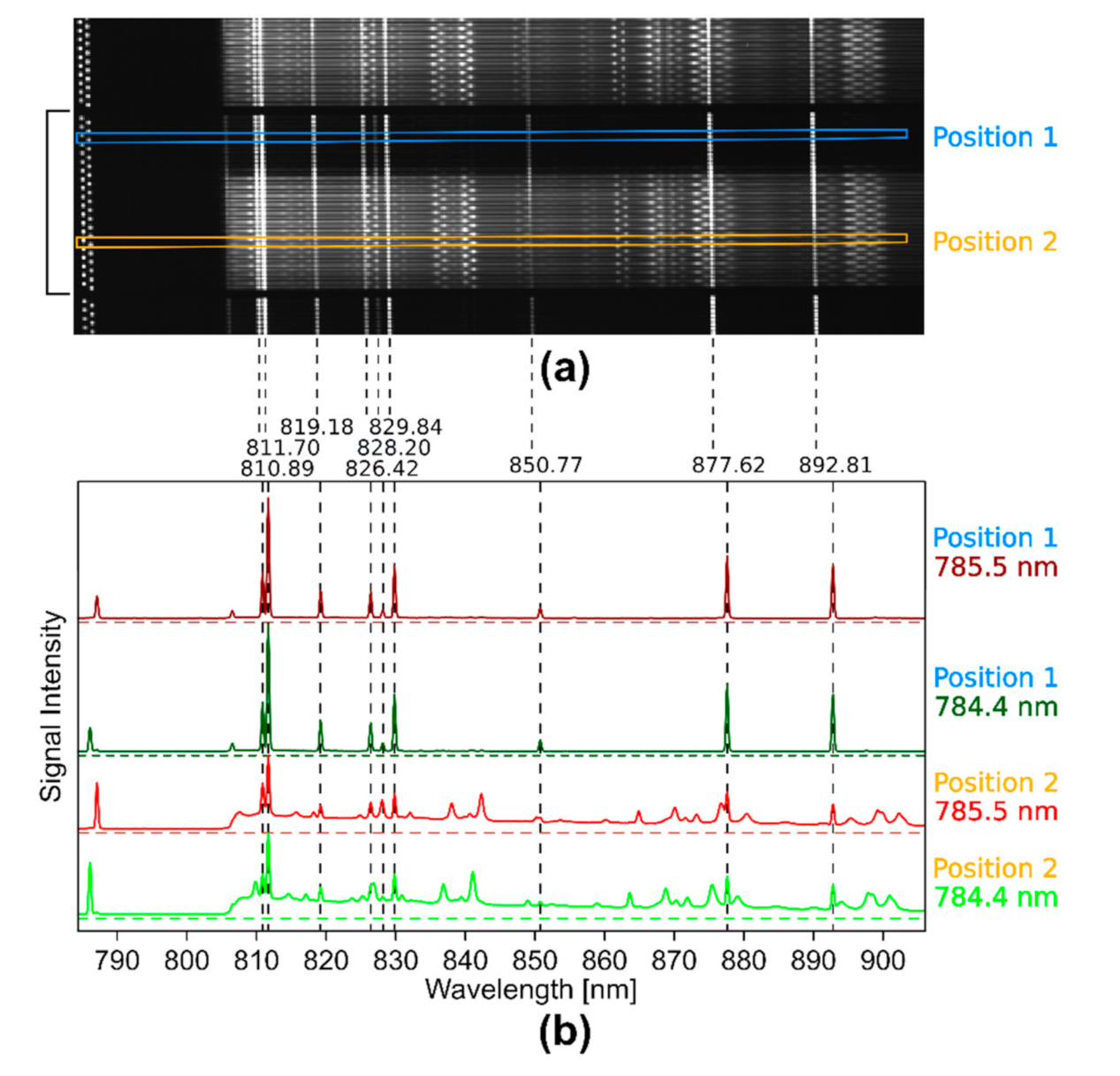
- A fiber-coupled near infrared (NIR) laser source LS 2-VBG, Ushio, Tokyo, Japan (formerly PD-LD, Pennington, USA) was used. The laser emits at 784.43 and 785.48 nm with a maximum laser power of 400 mW. This 1.05 nm shift (17 cm−1) in excitation wavelength was the largest possible shift without a high loss of laser intensity due to the filters used in the setup. This small shift in wavenumbers was used for the investigation of two pharmaceutical pills, which show Raman spectra with narrow band profiles.
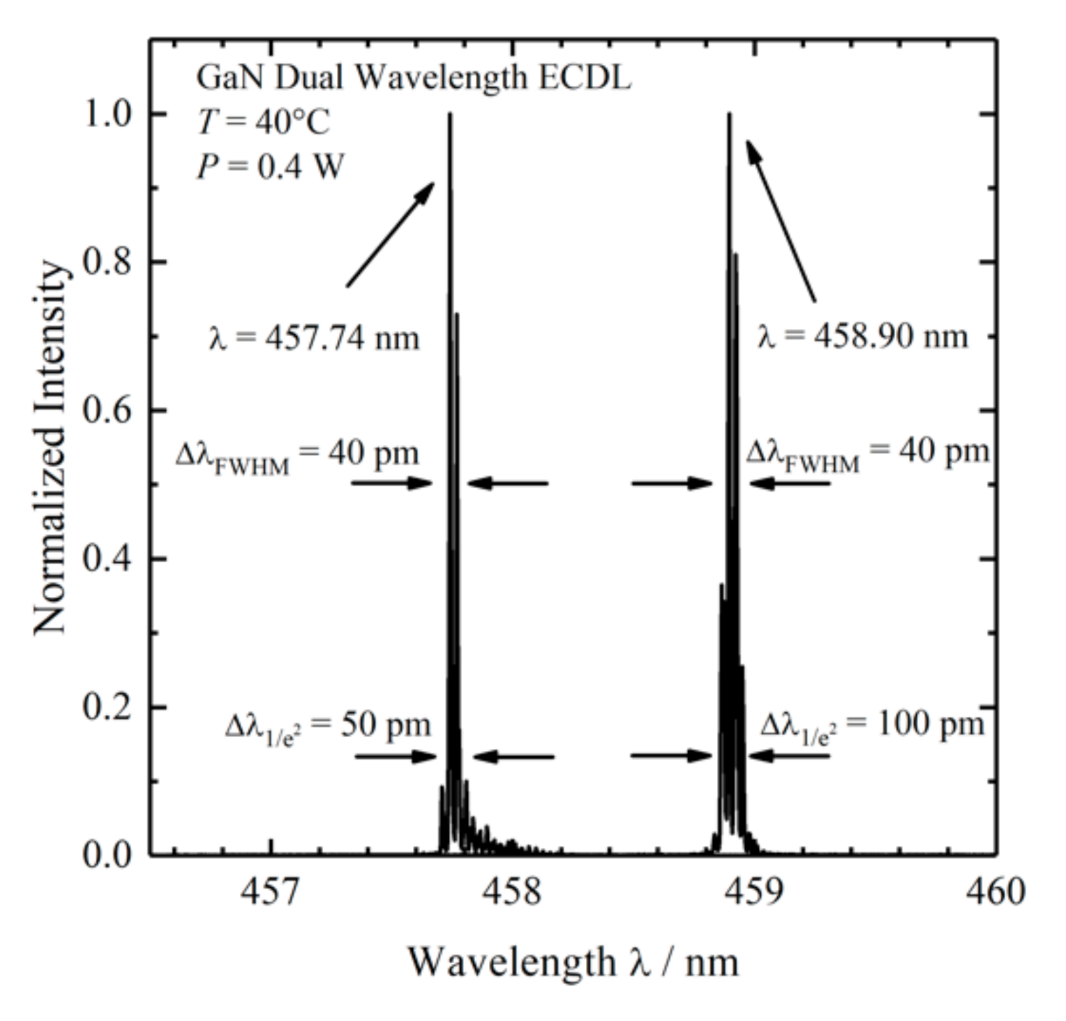
- A custom-made laser source based on two external wavelength stabilized blue laser diodes with spatially overlapping laser emissions at 457.74 and 458.90 nm in a conduction cooled package mount with a 25 × 25 mm2 footprint was designed and realized [51]. The diode lasers are wavelength stabilized at both target wavelengths with volume Bragg gratings with spectral bandwidths of <0.2 nm (<10 cm−1) and diffraction efficiencies of 15%. The spectral distance of 1.16 nm (55 cm−1) is selected for SERDS with respect to the spectral resolution of the spectrometer with 0.22 nm (≈10.4 cm−1) and for the investigation of biological tissue samples, since these samples have broader Raman band structures. For the spatial overlap of both laser emissions, using a high-reflection coated prism as well as a polarizing beam splitter, one laser is rotated by 90°. During the measurements, the laser was mounted on a heatsink (hsa-series, Ostech, Berlin, Germany), which was set to an operating temperature of 40 °C. For the individual operation of both laser diodes, two laser drivers were used (ds-series, Ostech, Berlin, Germany). The optical output power at both wavelengths was 0.4 W at 0.5 A. Figure 1 shows the corresponding emission spectra.
2.2. Nod and Shuffle
- Readout noise is only applied once to the complete set of data.
- Readout time only factors into the complete measurement time once. As for a large CCD chip, the readout time can take up to a minute or more, the total measurement time is significantly increased, when reading out the CCD chip after every single measurement. In our case, a sample was measured with 200 accumulations for each wavelength before reading out the CCD once. If the CCD was read out after each acquisition, readout times alone would have taken 400 min instead of 1 min with nod and shuffle.
- The technique allows measurements with very short acquisition times, which were accumulated with multiple iterations over a long time period using a wide field imaging setup. The accumulation of single measurements with very short acquisition times is favorable in the presence of signal fluctuations, e.g., photobleaching and fluctuating high-intensity background light (see Section 3.2.1).
- Spectra measured at the same spatial position with different wavelengths are recorded on the same pixels of the CCD chip. Therefore, Raman images of both excitation wavelengths have the same artifacts originating from the sensitivity and noise variations of the CCD pixels.
2.3. Samples and Experimental Parameters
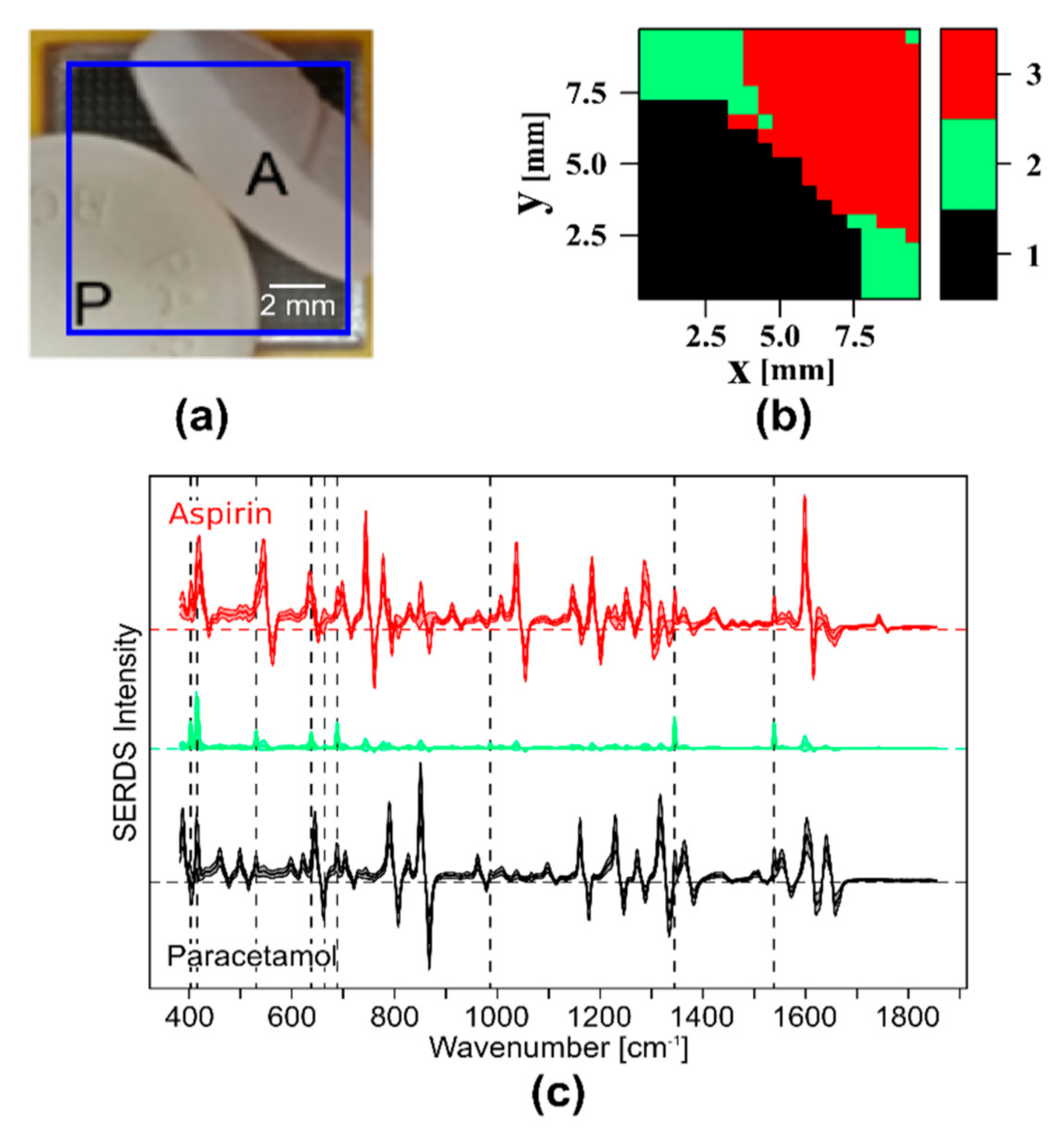
- Sample 1: Paracetamol tablet and a piece of an aspirin tablet were arranged on the large field of view probe head. Both samples show intense Raman bands.
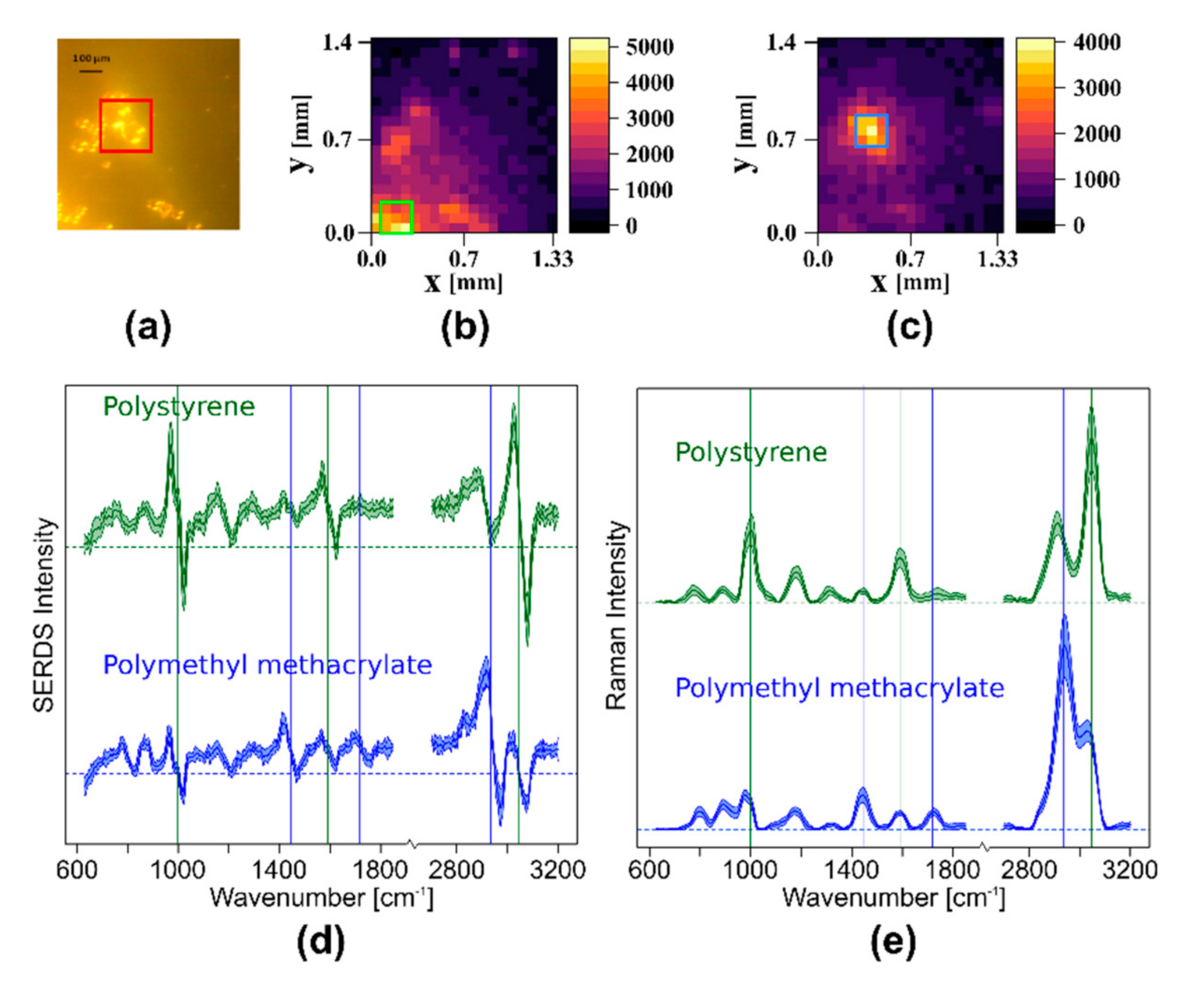
- Sample 2: Polystyrene (PS) beads (50 μm diameter) and polymethyl methacrylate (PMMA) beads (120 μm diameter) were mixed and applied on a CaF2 window. Both samples show intense Raman bands and have diameters to demonstrate the lateral resolution.
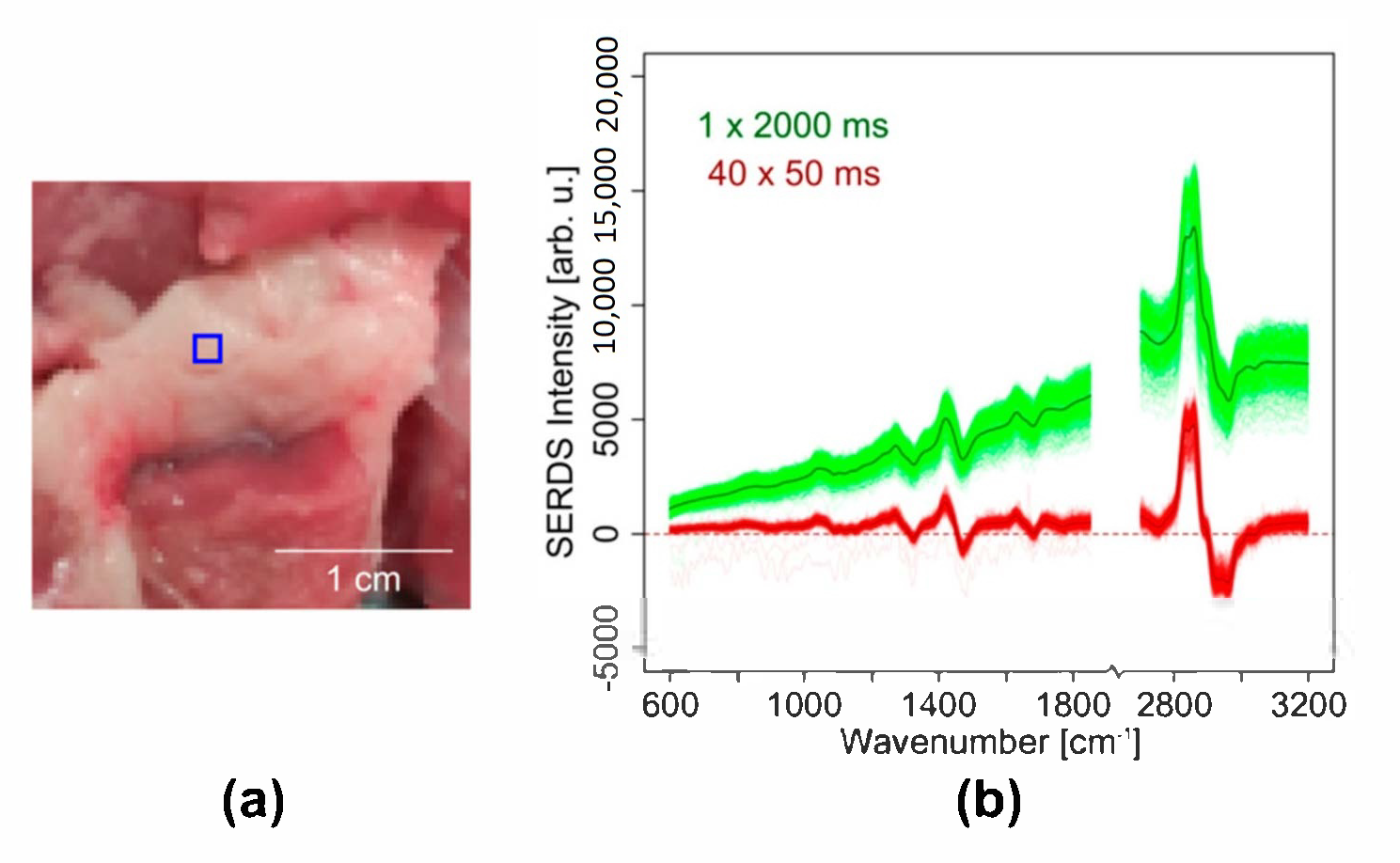
- Sample 3: The lipid-rich part of a pork chop fresh from a butcher was placed on a CaF2 window. Pork tissue served as model for biological material with less intense Raman bands and elevated background.
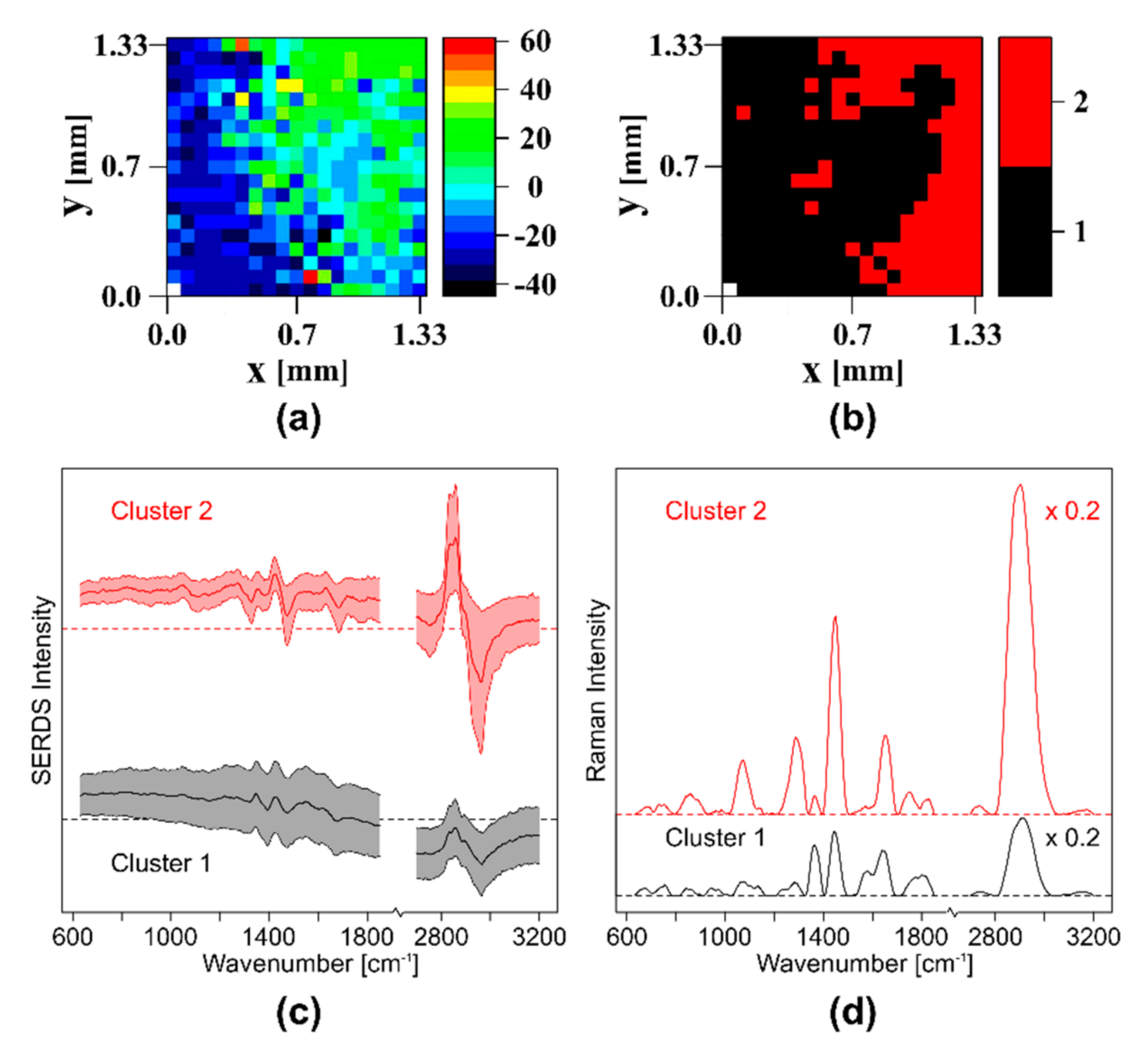
- Sample 4: The transition region from meat to fat of a pork chop fresh from a butcher was placed on a CaF2 window.
2.4. Data Handling
3. Results and Discussion
3.1. Wide-Field SERDS Imaging Using Nod and Shuffle and the Large Field of View (1 cm2) Probe Head
3.1.1. Illustration of the Raw Wide-Field Raman Images on the CCD Chip Using Nod and Shuffle
3.1.2. Filtering Out Room Light by SERDS Imaging
3.2. Wide-Field SERDS Imaging Using Nod and Shuffle, the Smaller Field of View Probe Head (0.02 cm2), and the Rapidly Shifting Dual-Wavelength Blue Diode Laser Source
3.2.1. Wide Field SERDS Imaging with Nod and Shuffle to Differentiate Different Polymer Beads
3.2.2. Photobleaching Compensation in Wide Field SERDS Imaging by Very Fast Nod and Shuffle
- Two Raman images were measured with a 50 ms acquisition time and 40 accumulations for each excitation wavelength before reading out the CCD chip once. Thus, the overall acquisition time was 2 s for each excitation wavelength.
- Two Raman images were measured with a 2 s acquisition time and one accumulation for each excitation wavelength before reading out the CCD chip once. Thus, the total acquisition time was also 2 s for each excitation wavelength.
3.2.3. Wide Field SERDS Imaging of Different Tissues in Pork Meat Using Nod and Shuffle
4. Conclusions
5. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Krafft, C.; Schie, I.W.; Meyer, T.; Schmitt, M.; Popp, J. Developments in spontaneous and coherent Raman scattering microscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1819–1849. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.J.E.; Shore, A.; Stone, N. Raman spectroscopy for rapid intra-operative margin analysis of surgically excised tumour specimens. Analyst 2019, 144, 6479–6496. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.X.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Schmitt, M.; Schie, I.W.; Cialla-May, D.; Matthaeus, C.; Bocklitz, T.; Matthäus, C.; Bocklitz, T.; Popp, J. Label-free molecular imaging of biological cells and tissues by linear and non-linear Raman spectroscopic approaches. Angew. Chem. Int. Ed. 2017, 4392–4430. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef]
- Monici, M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [CrossRef]
- Cordero, E.; Korinth, F.; Stiebing, C.; Krafft, C.; Schie, I.; Popp, J. Evaluation of Shifted Excitation Raman Difference Spectroscopy and Comparison to Computational Background Correction Methods Applied to Biochemical Raman Spectra. Sensors 2017, 17, 1724. [Google Scholar] [CrossRef]
- Wei, D.; Chen, S.; Liu, Q. Review of Fluorescence Suppression Techniques in Raman Spectroscopy. Appl. Spectrosc. Rev. 2015, 50, 387–406. [Google Scholar] [CrossRef]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Martens, H.; Stark, E. Extended multiplicative signal correction and spectral interference subtraction: New preprocessing methods for near infrared spectroscopy. J. Pharm. Biomed. Anal. 1991, 9, 625–635. [Google Scholar] [CrossRef]
- Pirzer, M.; Sawatzki, J. Method and Device for Correcting a Spectrum. U.S. Patent Application No. 2006/0211562 A1, 21 September 2006. [Google Scholar]
- Kneen, M.A.; Annegarn, H.J. Algorithm for fitting XRF, SEM and PIXE X-ray spectra backgrounds. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 1996, 109–110, 209–213. [Google Scholar] [CrossRef]
- Morháč, M. An algorithm for determination of peak regions and baseline elimination in spectroscopic data. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2009, 600, 478–487. [Google Scholar] [CrossRef]
- Morháč, M.; Matoušek, V. Peak clipping algorithms for background estimation in spectroscopic data. Appl. Spectrosc. 2008, 62, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Appl. Spectrosc. 2003, 57, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Bergholt, M.S.; Zheng, W.; Lin, K.; Wang, J.; Xu, H.; Ren, J.L.; Ho, K.Y.; Teh, M.; Yeoh, K.G.; Huang, Z. Characterizing variability of in vivo Raman spectroscopic properties of different anatomical sites of normal colorectal tissue towards cancer diagnosis at colonoscopy. Anal. Chem. 2015, 87, 960–966. [Google Scholar] [CrossRef]
- Desroches, J.; Jermyn, M.; Pinto, M.; Picot, F.; Tremblay, M.A.; Obaid, S.; Marple, E.; Urmey, K.; Trudel, D.; Soulez, G.; et al. A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Cordero, E.; Rüger, J.; Marti, D.; Mondol, A.S.; Hasselager, T.; Mogensen, K.; Hermann, G.G.; Popp, J.; Schie, I.W. Bladder tissue characterization using probe-based Raman spectroscopy: Evaluation of tissue heterogeneity and influence on the model prediction. J. Biophotonics 2020, 13, 1–15. [Google Scholar] [CrossRef]
- Ariese, F.; Meuzelaar, H.; Kerssens, M.M.; Buijs, J.B.; Gooijer, C. Picosecond Raman spectroscopy with a fast intensified CCD camera for depth analysis of diffusely scattering media. Analyst 2009, 134, 1192–1197. [Google Scholar] [CrossRef]
- Kögler, M.; Heilala, B. Time-gated Raman spectroscopy—A review. Meas. Sci. Technol. 2020, 32, 1–17. [Google Scholar] [CrossRef]
- De Luca, A.; Dholakia, K.; Mazilu, M. Modulated Raman Spectroscopy for Enhanced Cancer Diagnosis at the Cellular Level. Sensors 2015, 15, 13680–13704. [Google Scholar] [CrossRef]
- Dochow, S.; Bergner, N.; Krafft, C.; Clement, J.; Mazilu, M.; Praveen, B.B.; Ashok, P.C.; Marchington, R.; Dholakia, K.; Popp, J. Classification of Raman spectra of single cells with autofluorescence suppression by wavelength modulated excitation. Anal. Methods 2013, 5, 4608–4614. [Google Scholar] [CrossRef]
- Craig, D.; Mazilu, M.; Dholakia, K. Quantitative detection of pharmaceuticals using a combination of paper microfluidics and wavelength modulated Raman spectroscopy. PLoS ONE 2015, 10, e0123334. [Google Scholar] [CrossRef] [PubMed]
- Wirth, M.J.; Chou, S.H. Comparison of Time and Frequency Domain Methods for Rejecting Fluorescence from Raman Spectra. Anal. Chem. 1988, 60, 1882–1886. [Google Scholar] [CrossRef]
- Shreve, A.P.; Cherepy, N.J.; Mathies, R.A. Effective Rejection of Fluorescence Interference in Raman Spectroscopy Using a Shifted Excitation Difference Technique. Appl. Spectrosc. 1992, 46, 707–711. [Google Scholar] [CrossRef]
- Korinth, F.; Mondol, A.S.; Stiebing, C.; Schie, I.W.; Krafft, C.; Popp, J. New methodology to process shifted excitation Raman difference spectroscopy data: A case study of pollen classification. Sci. Rep. 2020, 10, 11215. [Google Scholar] [CrossRef]
- Gebrekidan, M.T.; Erber, R.; Hartmann, A.; Fasching, P.A.; Emons, J.; Beckmann, M.W.; Braeuer, A. Breast Tumor Analysis Using Shifted-Excitation Raman Difference Spectroscopy (SERDS). Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef]
- Martins, M.A.d.S.; Ribeiro, D.G.; Pereira dos Santos, E.A.; Martin, A.A.; Fontes, A.; da Silva Martinho, H. Shifted-excitation Raman difference spectroscopy for in vitro and in vivo biological samples analysis. Biomed. Opt. Express 2010, 1, 617–626. [Google Scholar] [CrossRef]
- Noack, K.; Eskofier, B.; Kiefer, J.; Dilk, C.; Bilow, G.; Schirmer, M.; Buchholz, R.; Leipertz, A. Combined shifted-excitation Raman difference spectroscopy and support vector regression for monitoring the algal production of complex polysaccharides. Analyst 2013, 138, 5639–5646. [Google Scholar] [CrossRef]
- Gebrekidan, M.T.; Knipfer, C.; Stelzle, F.; Popp, J.; Will, S.; Braeuer, A. A shifted-excitation Raman difference spectroscopy (SERDS) evaluation strategy for the efficient isolation of Raman spectra from extreme fluorescence interference. J. Raman Spectrosc. 2016, 47, 198–209. [Google Scholar] [CrossRef]
- Sowoidnich, K.; Kronfeldt, H.-D. In-Situ Species Authentication of Frozen-Thawed Meat and Meat Juice Using Shifted Excitation Raman Difference Spectroscopy. In Biophotonics: Photonic Solutions for Better Health Care VI, Proceedings of the SPIE Photonics Europe, Strasbourg, France, 22–26 April 2018; Popp, J., Tuchin, V.V., Pavone, F.S., Eds.; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; p. 106850. [Google Scholar]
- Sowoidnich, K.; Kronfeldt, H.-D. Fluorescence Rejection by Shifted Excitation Raman Difference Spectroscopy at Multiple Wavelengths for the Investigation of Biological Samples. ISRN Spectrosc. 2012, 2012, 256326 . [Google Scholar] [CrossRef]
- Knipfer, C.; Motz, J.; Adler, W.; Brunner, K.; Gebrekidan, M.T.; Hankel, R.; Agaimy, A.; Will, S.; Braeuer, A.; Neukam, F.W.; et al. Raman difference spectroscopy: A non-invasive method for identification of oral squamous cell carcinoma. Biomed. Opt. Express 2014, 5, 3252–3265. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Strycker, B.D.; Commer, B.; Wang, K.; Shaw, B.D.; Scully, M.O.; Sokolov, A.V. Molecular origin of the Raman signal from Aspergillus nidulans conidia and observation of fluorescence vibrational structure at room temperature. Sci. Rep. 2020, 10, 5428. [Google Scholar] [CrossRef] [PubMed]
- Schmälzlin, E.; Moralejo, B.; Rutowska, M.; Monreal-Ibero, A.; Sandin, C.; Tarcea, N.; Popp, J.; Roth, M.M. Raman imaging with a fiber-coupled multichannel spectrograph. Sensors 2014, 14, 21968–21980. [Google Scholar] [CrossRef] [PubMed]
- Schmälzlin, E.; Moralejo, B.; Bodenmüller, D.; Darvin, M.E.; Thiede, G.; Roth, M.M. Ultrafast imaging Raman spectroscopy of large-area samples without stepwise scanning. J. Sens. Sens. Syst. 2016, 5, 261–271. [Google Scholar] [CrossRef]
- Stewart, S.; Priore, R.J.; Nelson, M.P.; Treado, P.J. Raman Imaging. Annu. Rev. Anal. Chem. 2012, 5, 337–360. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.; Kielhorn, M.; Schmitt, M.; Popp, J.; Heintzmann, R. Light sheet Raman micro-spectroscopy. Optica 2016, 3, 452–457. [Google Scholar] [CrossRef]
- Sinjab, F.; Liao, Z.; Notingher, I. Applications of Spatial Light Modulators in Raman Spectroscopy. Appl. Spectrosc. 2019, 73, 727–746. [Google Scholar] [CrossRef]
- Allington-Smith, J. Basic principles of integral field spectroscopy. New Astron. Rev. 2006, 50, 244–251. [Google Scholar] [CrossRef]
- Bacon, R.; Monnet, G. Recent Trends in Integral Field Spectroscopy. In Optical 3D-Spectroscopy for Astronomy; Wiley-VCH: Weinheim, Germany, 2017; pp. 115–128. [Google Scholar]
- Glazebrook, K.; Bland-Hawthorn, J. Microslit Nod-Shuffle Spectroscopy: A Technique for Achieving Very High Densities of Spectra. Publ. Astron. Soc. Pac. 2002, 113, 197–214. [Google Scholar] [CrossRef][Green Version]
- Roth, M.M.; Cardiel, N.; Cenarro, J.; Schönberner, D.; Steffen, M. Nod & Shuffle 3D Spectroscopy. In Scientific Detectors for Astronomy 2005; Beletic, J.E., Beletic, J.W., Amico, P., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 99–108. [Google Scholar]
- Roth, M.M.; Fechner, T.; Wolter, D.; Kelz, A.; Becker, T. Ultra-Deep Optical Spectroscopy with PMAS. Using the Nod-and-Shuffle Technique. Exp. Astron. 2002, 14, 99–105. [Google Scholar] [CrossRef]
- Sowoidnich, K.; Maiwald, M.; Sumpf, B.; Towrie, M.; Matousek, P. Charge-Shifting Optical Lock-In Detection with Shifted Excitation Raman Difference Spectroscopy for the Analysis of Fluorescent Heterogeneous Samples. In Proceedings of the Biomedical Vibrational Spectroscopy 2020: Advances in Research and Industry, San Francisco, CA, USA, 1–6 February 2020; p. 112360K. [Google Scholar]
- Sowoidnich, K.; Towrie, M.; Maiwald, M.; Sumpf, B.; Matousek, P. Shifted Excitation Raman Difference Spectroscopy with Charge-Shifting Charge-Coupled Device (CCD) Lock-In Detection. Appl. Spectrosc. 2019, 73, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Heming, R.; Herzog, H.; Deckert, V. Optical CCD lock-in device for Raman difference spectroscopy. DGaO Proc. 2008, 109, 33. [Google Scholar]
- Schmälzlin, E.; Urrutia, T.; Korinth, F.; Stiebing, C.; Krafft, C.; Popp, J.; Roth, M.M. Bildgebende Differenz-Raman-Spektroskopie mit “Nod and Shuffle“-Technik. In Proceedings of the 14th Dresdner Sensor-Symposium, Dresden, Germany, 2–4 December 2019; pp. 96–101. [Google Scholar] [CrossRef]
- Moralejo, B.; Roth, M.M.; Godefroy, P.; Fechner, T.; Bauer, S.M.; Schmälzlin, E.; Kelz, A.; Haynes, R. The Potsdam MRS Spectrograph: Heritage of MUSE and the Impact of Cross-Innovation in the Process of Technology Transfer. In Proceedings of the SPIE Astronomical Telescopes + Instrumentation, Edinburgh, UK, 26 June–1 July 2016; p. 991222. [Google Scholar]
- Moralejo, B.; Schmälzlin, E.; Bodenmüller, D.; Fechner, T.; Roth, M.M. Improving the frame rates of Raman image sequences recorded with integral field spectroscopy using windowing and binning methods. J. Raman Spectrosc. 2018, 49, 372–375. [Google Scholar] [CrossRef]
- Sumpf, B.; Müller, A.; Maiwald, M. Tailored Diode Lasers: Enabling Raman Spectroscopy in the Presence of Disturbing Fluorescence and Background Light. In Proceedings of the SPIE BiOS, San Francisco, CA, USA, 2–7 February 2019; p. 1089411. [Google Scholar]
- Schmälzlin, E.; Moralejo, B.; Gersonde, I.; Schleusener, J.; Darvin, M.E.; Thiede, G.; Roth, M.M. Nonscanning large-area Raman imaging for ex vivo/in vivo skin cancer discrimination. J. Biomed. Opt. 2018, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kelz, A.; Verheijen, M.A.W.; Roth, M.M.; Bauer, S.M.; Becker, T.; Paschke, J.; Popow, E.; Sánchez, S.F.; Laux, U. PMAS: The Potsdam Multi-Aperture Spectrophotometer. II. The Wide Integral Field Unit PPak. Publ. Astron. Soc. Pac. 2006, 118, 129–145. [Google Scholar] [CrossRef]
- Sandin, C.; Becker, T.; Roth, M.M.; Gerssen, J.; Monreal-Ibero, A.; Böhm, P.; Weilbacher, P. P3D: A general data-reduction tool for fiber-fed integral-field spectrographs. Astron. Astrophys. 2010, 515, A35. [Google Scholar] [CrossRef]
- Sandin, C.; Becker, T.; Roth, M.M.; Gerssen, J.; Monreal-Ibero, A.; Böhm, P.; Weilbacher, P. p3d: General Data-Reduction Tool for Fiber-Fed Integral-Field Spectrographs; Verison 2.7; Astrophysics Source Code Library: Houghton, MI, USA, 2020; Available online: http://ascl.net/1205.002 (accessed on 20 November 2020).
- p3d a General Data-Reduction Tool for Fiber-Fed Integral-Field Spectrographs. Available online: https://p3d.sourceforge.io/ (accessed on 20 November 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Beleites, C.; Sergo, V. hyperSpec: A Package to Handle Hyperspectral Data Sets in R. Available online: https://CRAN.R-project.org/package=hyperSpec (accessed on 23 November 2020).
- Harris, A. FITSio: FITS (Flexible Image Transport System) Utilities. Available online: https://rdrr.io/cran/FITSio/ (accessed on 23 November 2020).
- Beleites, C. Sofware Package for R-Ramancal: Calibration Routines for Raman Spectrometers. 2013. [Google Scholar]
- Borchers, H.W. Pracma: Practical Numerical Math Functions. Available online: https://rdrr.io/rforge/pracma/ (accessed on 23 November 2020).
- Gibb, S.; Strimmer, K. MALDIquant: A versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012, 28, 2270–2271. [Google Scholar] [CrossRef]
- Garnier, S. Viridis: Default Color Maps from “Matplotlib”. Available online: https://rdrr.io/cran/viridis/ (accessed on 23 November 2020).
- Bonifacio, A.; Beleites, C.; Sergo, V. Application of R-mode analysis to Raman maps: A different way of looking at vibrational hyperspectral data. Anal. Bioanal. Chem. 2015, 407, 1089–1095. [Google Scholar] [CrossRef]
- Arc Line Lamps on the Website of W. M. Keck Observatory. Available online: https://www2.keck.hawaii.edu/inst/lris/arc_calibrations.html (accessed on 20 November 2020).
- Horne, K. An optimal extraction algorithm for CCD spectroscopy. Publ. Astron. Soc. Pac. 1986, 98, 609. [Google Scholar] [CrossRef]
- Dochow, S.; Bergner, N.; Matthäus, C.; Praveen, B.B.; Ashok, P.C.; Mazilu, M.; Krafft, C.; Dholakia, K.; Popp, J. Etaloning, fluorescence and ambient light suppression by modulated wavelength Raman spectroscopy. Biomed. Spectrosc. Imaging 2012, 1, 383–389. [Google Scholar] [CrossRef]
- Maiwald, M.; Müller, A.; Sumpf, B.; Tränkle, G. A portable shifted excitation Raman difference spectroscopy system: Device and field demonstration. J. Raman Spectrosc. 2016, 47, 1180–1184. [Google Scholar] [CrossRef]
- Sowoidnich, K.; Towrie, M.; Matousek, P. Lock-in detection in Raman spectroscopy with charge-shifting CCD for suppression of fast varying backgrounds. J. Raman Spectrosc. 2019, 50, 983–995. [Google Scholar] [CrossRef]

| No. | Sample | Laser (λ1/λ2) in nm | Intensity in W/mm2 | Imaging Area in cm2 | Acquisition Times 1 |
|---|---|---|---|---|---|
| 1 | Aspirin/Paracetamol | NIR (784.43/785.48) | 0.004 2 | 1 | 200 × 200 ms |
| 2 | PS/PMMA | blue (457.74/458.90) | 0.1 | 0.02 | 40 × 100 ms |
| 3 | Lipid-rich pork tissue | blue (457.74/458.90) | 0.2 | 0.02 | 40 × 50 ms and 1 × 2000 ms |
| 4 | Pork tissue | blue (457.74/458.90) | 0.2 | 0.02 | 40 × 50 ms |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korinth, F.; Schmälzlin, E.; Stiebing, C.; Urrutia, T.; Micheva, G.; Sandin, C.; Müller, A.; Maiwald, M.; Sumpf, B.; Krafft, C.; et al. Wide Field Spectral Imaging with Shifted Excitation Raman Difference Spectroscopy Using the Nod and Shuffle Technique. Sensors 2020, 20, 6723. https://doi.org/10.3390/s20236723
Korinth F, Schmälzlin E, Stiebing C, Urrutia T, Micheva G, Sandin C, Müller A, Maiwald M, Sumpf B, Krafft C, et al. Wide Field Spectral Imaging with Shifted Excitation Raman Difference Spectroscopy Using the Nod and Shuffle Technique. Sensors. 2020; 20(23):6723. https://doi.org/10.3390/s20236723
Chicago/Turabian StyleKorinth, Florian, Elmar Schmälzlin, Clara Stiebing, Tanya Urrutia, Genoveva Micheva, Christer Sandin, André Müller, Martin Maiwald, Bernd Sumpf, Christoph Krafft, and et al. 2020. "Wide Field Spectral Imaging with Shifted Excitation Raman Difference Spectroscopy Using the Nod and Shuffle Technique" Sensors 20, no. 23: 6723. https://doi.org/10.3390/s20236723
APA StyleKorinth, F., Schmälzlin, E., Stiebing, C., Urrutia, T., Micheva, G., Sandin, C., Müller, A., Maiwald, M., Sumpf, B., Krafft, C., Tränkle, G., Roth, M. M., & Popp, J. (2020). Wide Field Spectral Imaging with Shifted Excitation Raman Difference Spectroscopy Using the Nod and Shuffle Technique. Sensors, 20(23), 6723. https://doi.org/10.3390/s20236723






