Portable Sleep Apnea Syndrome Screening and Event Detection Using Long Short-Term Memory Recurrent Neural Network
Abstract
1. Introduction
- This study is the first work to apply two triaxial accelerometers, a single-lead ECG and a finger oximeter for portable sleep apnea syndrome screening. The clinic experiments were performed by recording overnight signals of suspected patients under the approval of Institutional Review Board of hospital. Most works designed new sensing devices without clinic experiments or developed new detection algorithms by using public database.
- This work proposes a complete systematic framework of sensing devices and algorithms to detect and identify OSA, central sleep apnea (CSA) and hypopnea (HYP) events. This system can not only evaluate AHI values but also provide reliable event-level classification results of various sleep apnea events. Except our previous work [23] based on piezo-electronic bands, most works can only detect OSA events and evaluate AHI only.
2. Material and Methods
2.1. Material
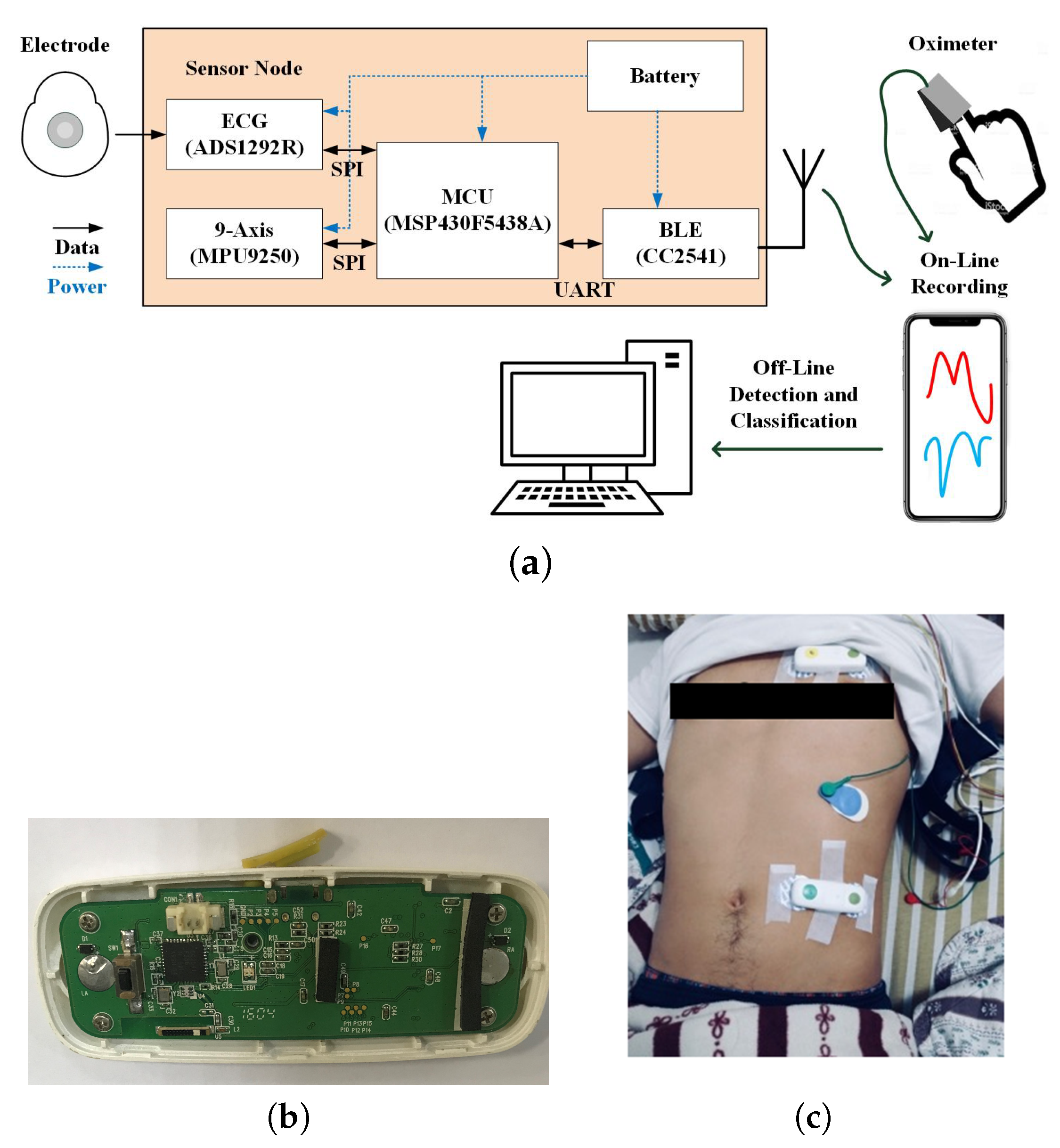
2.2. Integrated Sensing System
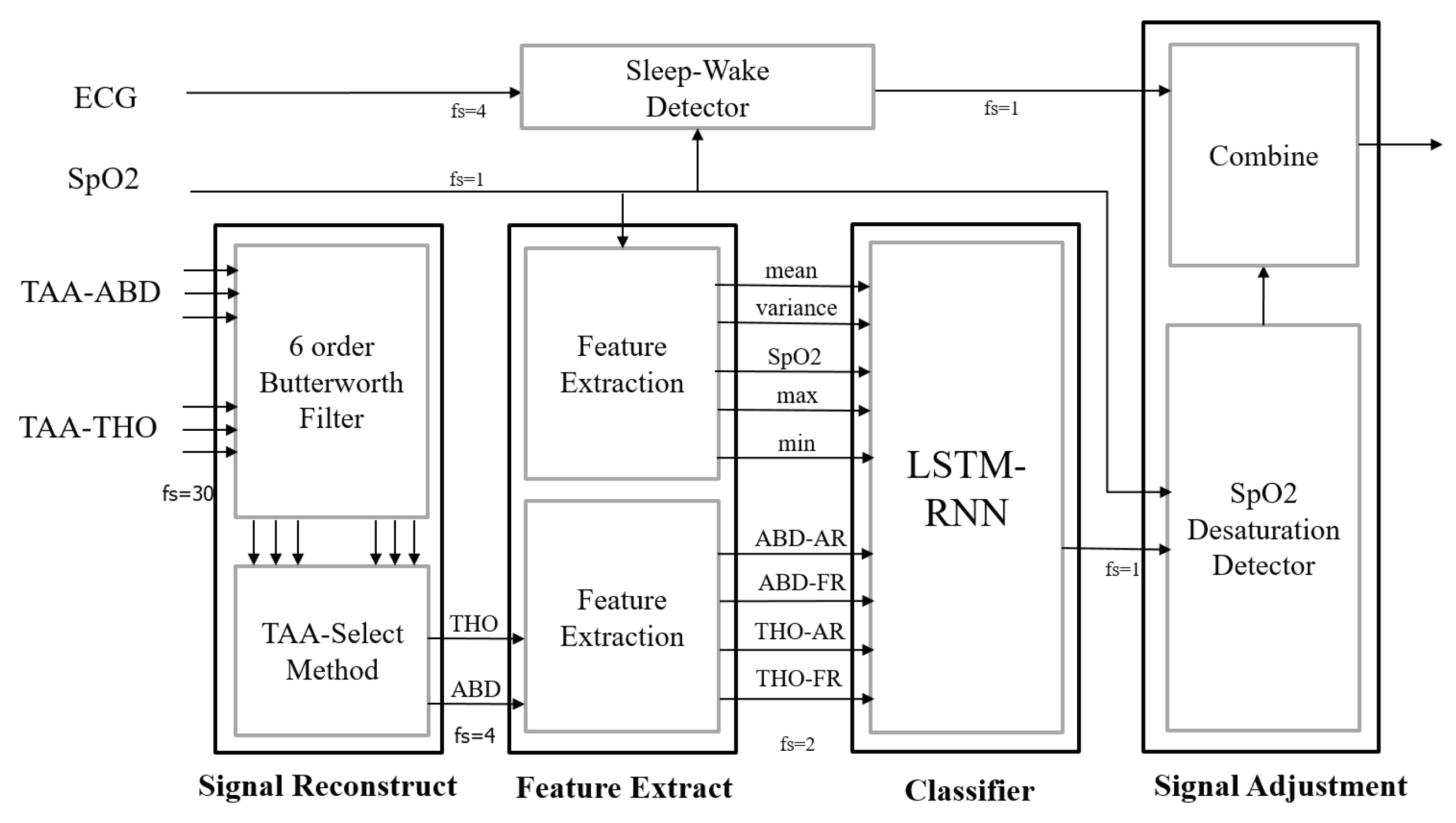
2.3. Signal Preprocessing
2.4. Feature Extraction
2.4.1. Features of the THO and ABD signals
2.4.2. Features of SpO Signal
2.5. Neutral Network Model, Event Classification and AHI Evaluation
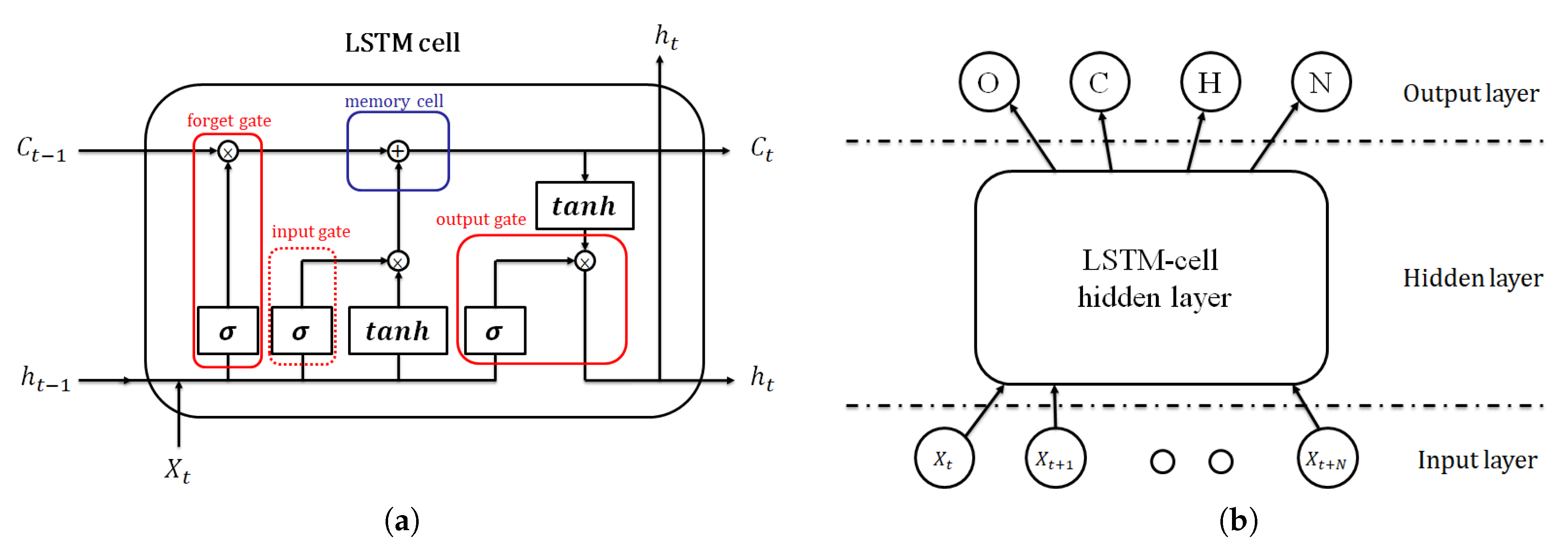
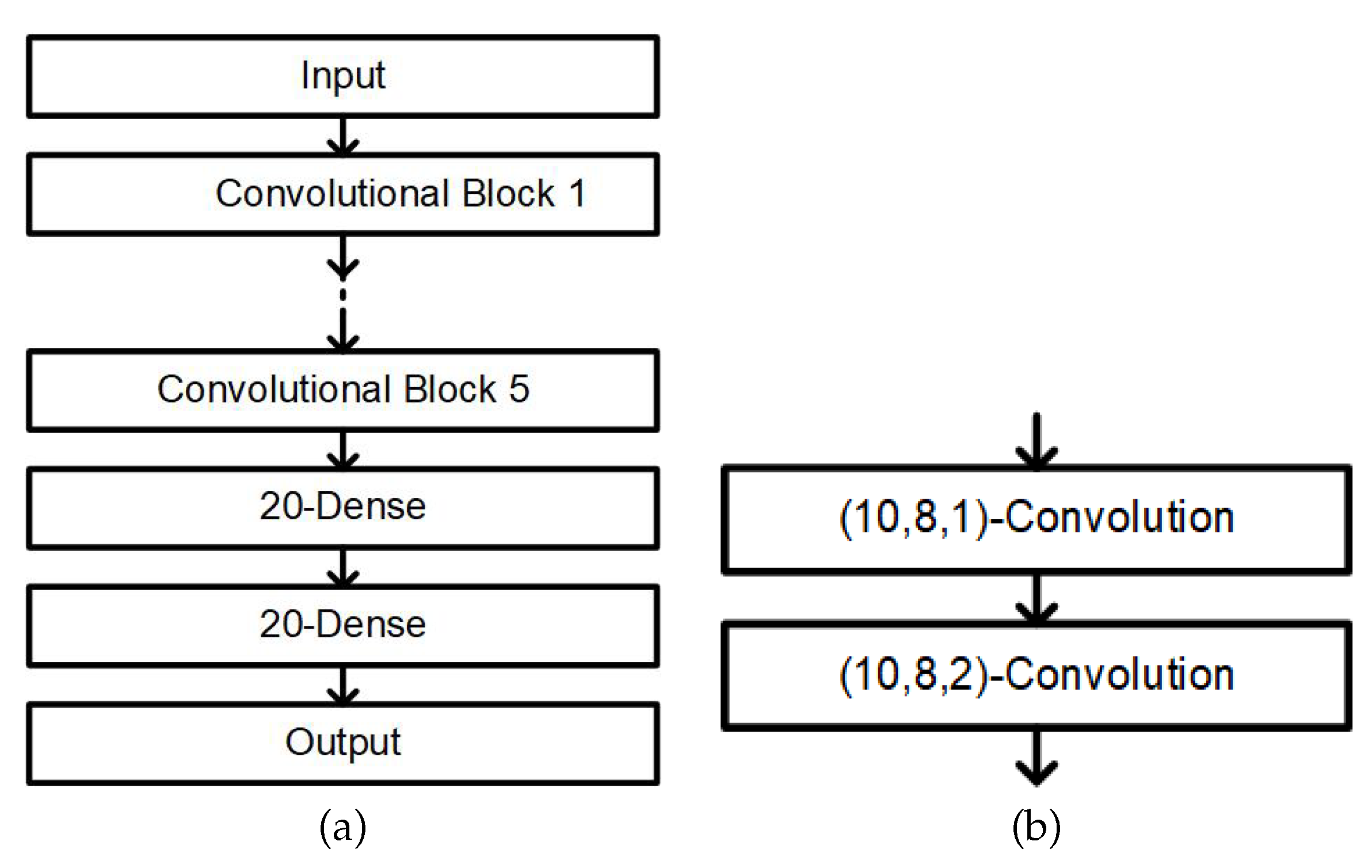
2.5.1. Neural Network Model Classifier
2.5.2. Oxygen Desaturation Detection
2.5.3. Sleep–Wake Classification
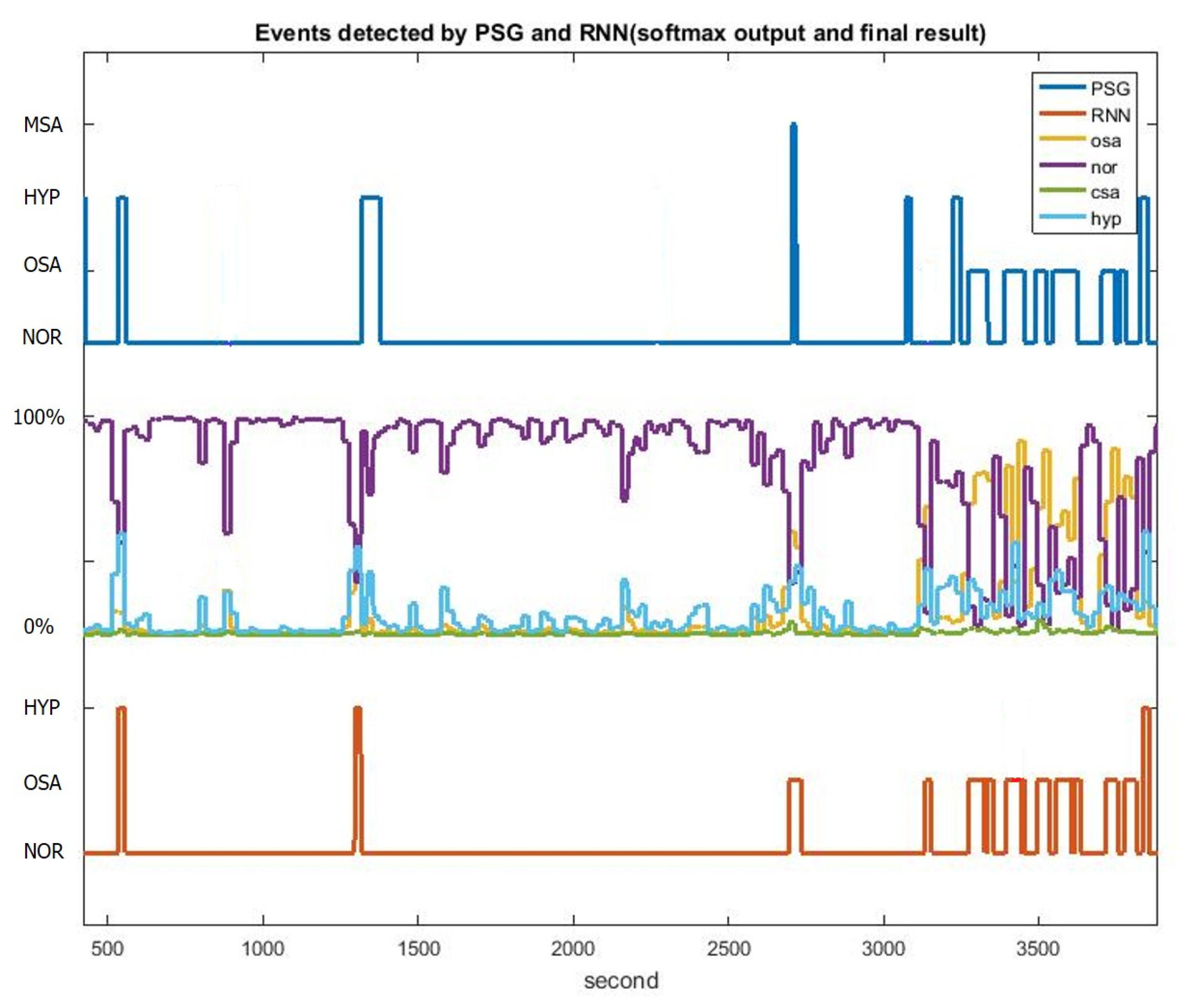
3. Results
LSTM-RNN with Oxygen Desaturation and Sleep–Wake Detection
4. Discussion
Limitation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Peppard, P.E.; Young, T.; Barnet, J.H.; Palta, M.; Hagen, E.W.; Hla, K.M. Increased prevalence of sleep-disordered breathing in adults. Am. J. Epidemiol. 2013, 177, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Kapur, V.K.; Auckley, D.H.; Chowdhuri, S.; Kuhlmann, D.C.; Mehra, R.; Ramar, K.; Harrod, C.G. Clinical Practice Guideline for Diagnostic Testing for Adult Obstructive Sleep Apnea: An American Academy of Sleep Medicine Clinical Practice Guideline. J. Clin. Sleep Med. 2017, 15, 479–504. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.B. The AASM Manual for the Scoring of Sleep and Associated Events. Available online: http://aasm.org/clinical-resources/scoring-manual/ (accessed on 25 October 2020).
- Pevernagie, D.A.; Gnidovec–Strazisar, B.; Grote, L.; Heinzer, R.; McNicholas, W.T.; Penzel, T.; Randerath, W.; Schiza, S.; Verbraecken, J.; Arnardottir, E.S. On the rise and fall of the apnea–hypopnea index: A historical review and critical appraisal. J. Sleep Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Corral-Peñafiel, J.; Pepin, J.-L.; Barbe, F. Ambulatory monitoring in the diagnosis and management of obstructive sleep apnoea syndrome. Eur. Respir. Rev. 2013, 22, 312–324. [Google Scholar] [CrossRef]
- Collop, N.A.; Anderson, W.M.; Boehlecke, B.; Claman, D.; Goldberg, R.; Gottlieb, D.J.; Hudgel, D.; Sateia, M.; Schwab, R. Clinical guidelines for the use of unattended portable monitors in the diagnosis of obstructive sleep apnea in adult patients. J. Clin. Sleep Med. 2007, 3, 737–747. [Google Scholar]
- Li, K.; Pan, W.; Li, Y.; Jiang, Q.; Liu, G. A method to detect sleep apnea based on deep neural network and hidden Markov model using single-lead ECG signal. Neurocomputing 2018, 294, 94–101. [Google Scholar] [CrossRef]
- Jung, D.W.; Hwang, S.H.; Yoon, H.N.; Lee, Y.J.G.; Jeong, D.U.; Park, K.S. Nocturnal Awakening and Sleep Efficiency Estimation Using Unobtrusively Measured Ballistocardiogram. IEEE Trans. Biomed. Eng. 2014, 61, 131–1138. [Google Scholar] [CrossRef] [PubMed]
- Almazaydeh, L.; Faezipour, M.; Elleithy, K. A neural network system for detection of obstructive sleep apnea through SpO2 signal features. Int. J. Adv. Comput. Sci. Appl. 2012, 3, 7–11. [Google Scholar] [CrossRef]
- Avci, C.; Akba, A. Sleep apnea classification based on respiration signals by using ensemble methods. Bio-Med. Mater. Eng. 2015, 26, S1703–S1710. [Google Scholar] [CrossRef]
- Jane, R.; Fiz, J.A.; Solà-Soler; Mesquita, J.; Morera, J. Snoring Analysis for the Screening of Sleep Apnea Hypopnea Syndrome with a Single-Channel Device Developed using Polysomnographic and Snoring Databases. In Proceedings of the 33rd Annual International Conference of the IEEE Engineering in Medicine an Biology Society (EMBC), Boston, MA, USA, 30 August–3 September 2011; pp. 8331–8333. [Google Scholar]
- Eiseman, N.A.; Westover, M.B.; Mietus, J.E.; Thomas, R.J.; Bianchi, M.T. Classification algorithms for predicting sleepiness and sleep apnea severity. J. Sleep Res. 2012, 21, 101–112. [Google Scholar] [CrossRef]
- De Chazal, P.; Sadr, N. Sleep apnoea classification using heart rate variability, ECG derived respiration and cardiopulmonary coupling parameters. In Proceedings of the 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3203–3206. [Google Scholar]
- Rodrigues, P.P.; Santos, D.F.; Leite, L. Obstructive sleep apnoea diagnosis: The Bayesian network model revisited. In Proceedings of the 28th International Symposium on Computer-Based Medical Systems, Sao Carlos, Brazil, 22–25 June 2015; pp. 115–120. [Google Scholar]
- Dey, D.; Chaudhuri, S.; Munshi, S. Obstructive sleep apnoea detection using convolutional neural network based deep learning framework. Biomed. Eng. Lett. 2018, 8, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Quiceno-Manrique, A.; Alonso-Hernandez, J.; Travieso-Gonzalez, C.; Ferrer-Ballester, M.; Castellanos-Dominguez, G. Detection of obstructive sleep apnea in ECG recordings using time-frequency distributions and dynamic features. In Proceedings of the 31st IEEE Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Minneapolis, MN, USA, 3–6 September 2009; pp. 5559–5562. [Google Scholar]
- Mendez, M.; Ruini, D.; Villantieri, O.; Matteucci, M.; Penzel, T.; Bianchi, A. Detection of sleep apnea from surface ECG based on features extracted by an autoregressive model. In Proceedings of the 29th IEEE International Conference on Engineering in Medicine and Biology Society (EMBS), Lyon, France, 22–26 August 2007; pp. 6105–6108. [Google Scholar]
- Novak, D.; Mucha, K.; Al-Ani, T. Long short-term memory for apnea detection based on heart rate variability. In Proceedings of the 30th IEEE Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Vancouver, BC, Canada, 20–25 August 2008; pp. 5234–5237. [Google Scholar]
- Ng, A.; Chung, J.; Gohel, M.; Yu, W.; Fan, K.; Wong, T. Evaluation of the performance of using mean absolute amplitude analysis of thoracic and abdominal signals for immediate indication of sleep apnoea events. J. Clin. Nursing 2008, 17, 2360–2366. [Google Scholar] [CrossRef] [PubMed]
- Garde, A.; Dekhordi, P.; Ansermino, J.M.; Dumont, G.A. Identifying individual sleep apnea/hypoapnea epochs using smartphone-based pulse oximetry. In Proceedings of the 38th IEEE Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Orlando, FL, USA, 16–20 August 2016; pp. 3195–3198. [Google Scholar]
- Mendonca, F.; Mostafa, S.S.; Ravelo-Garcia, A.G.; Morgado-Dias, F.; Penzel, T. A Review of Obstructive Sleep Apnea Detection Approaches. IEEE J. Biomed. Health Inf. 2019, 23, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Collop, N.A.; Tracy, S.L.; Kapur, V.; Mehra, R.; Kuhlmann, D.; Fleishman, S.A.; Ojile, J.M. Obstructive sleep apnea devices for out-of-center (OOC) testing: Technology evaluation. J. Clin. Sleep Med. 2011, 7, 531–548. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Wu, H.T.; Hsu, C.A.; Huang, P.C.; Huang, Y.H.; Lo, Y.L. Sleep apnea detection based on thoracic and abdominal movement signals of wearable piezo-electric bands. IEEE J. Biomed. Health Inf. 2017, 21, 1533–1545. [Google Scholar] [CrossRef]
- Daubechies, I.; Lu, J.; Wu, H.-T. Synchrosqueezed Wavelet Transforms: An empirical mode decomposition-like tool. Appl. Comput. Harmon. Anal. 2011, 30, 243–261. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Cheng, M.-Y.; Wu, H.-T. Nonparametric and adaptive modeling of dynamic seasonality and trend with heteroscedastic and dependent errors. J. R. Stat. Soc. Ser. B Stat. Methodol. 2014, 76, 651–782. [Google Scholar] [CrossRef]
- Wu, H.-T. Instantaneous frequency and wave shape function (I). Appl. Comput. Harmon. Anal. 2013, 35, 181–199. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Auli, M.; Galley, M.; Quirk, C.; Zweig, G. Joint language and translation modeling with recurrent neural networks. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Seattle, WA, USA, 18–21 October 2013; pp. 1044–1054. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the Advances in Neural Information Processing Systems (NIPS), Montreal, QC, Canada, 8–13 December 2014; pp. 3104–3112. [Google Scholar]
- Vinyals, O.; Toshev, A.; Bengio, S.; Erhan, D. Show and tell: A neural image caption generator. In Proceedings of the IEEE Conference on Computer Version and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3156–3164. [Google Scholar]
- Lipton, Z.C.; Kale, D.C.; Elkan, C.; Wetzel, R. Learning to Diagnose with LSTM Recurrent Neural Networks. Available online: https://arxiv.org/abs/1511.03677 (accessed on 25 October 2020).
- Snyder, F.; Hobson, J.A.; Morrison, D.F.; Goldfrank, F. Changes in respiration, heart rate, and systolic blood pressure in human sleep. Eur. J. Appl. Physiol. 1964, 19, 417–422. [Google Scholar] [CrossRef]
- Malik, J.; Lo, Y.L.; Wu, H.T. Sleep-wake classification via quantifying heart rate variability by convolutional neural network. Physiol. Meas. 2018, 39, 085004. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.T.; Wu, J.C.; Huang, P.C.; Lin, T.Y.; Wang, T.; Huang, Y.H.; Lo, Y.L. Phenotype-based and self-learning inter-individual sleep apnea screening with a level IV-like monitoring system. Front. Physiol. 2018, 9, 723. [Google Scholar] [CrossRef]
- Whitney, C.W.; Gottlieb, D.J.; Redline, S.; Norman, R.G.; Dodge, R.R.; Shahar, E.; Surovec, S.; Nieto, F.J. Reliability of scoring respiratory disturbance indices and sleep staging. Sleep 1998, 21, 749–757. [Google Scholar] [CrossRef]





| Severity | Gender | AHI | BMI | Age | * TST | ** SE | *** REM | **** NREM |
|---|---|---|---|---|---|---|---|---|
| (times/hr) | (kg/m) | (year) | (min) | (%) | (%) | (%) | ||
| Normal | all (9) male (6) female (3) | 1.8 ± 1.6 | 23.2 ± 3.3 | 30.0 ± 8.2 | 312.9 ± 37.2 | 84.4 ± 9.9 | 15.4 ± 5.7 | 84.6 ± 5.7 |
| Mild | all (17) male (13) female (4) | 9.4 ± 2.4 | 25.6 ± 3.7 | 49.0 ± 11.3 | 313.4 ± 28.0 | 84.3 ± 7.3 | 17.8 ± 5.8 | 82.2 ± 5.8 |
| Moderate | all (28) male (21) female (7) | 21.7 ± 4.1 | 25.4 ± 2.7 | 48.5 ± 12.9 | 311.0 ± 40.7 | 83.9 ± 11.1 | 14.9 ± 5.6 | 85.1 ± 5.6 |
| Severe | all (61) male (50) female (11) | 61.1 ± 24.0 | 30.2 ± 6.0 | 50.3 ± 12.3 | 291.0 ± 52.6 | 79.4 ± 13.8 | 11.0 ± 6.2 | 89.0 ± 6.2 |
| Level of Severity | Training Subjects | Testing Subjects | Total Subjects |
|---|---|---|---|
| Normal | 6 | 3 | 9 |
| Mild | 9 | 8 | 17 |
| Moderate | 14 | 14 | 28 |
| Severe | 30 | 31 | 61 |
| All levels | 59 | 56 | 115 |
| Time Step (N) | Precision | Sensitivity | F1 Score | AHI Difference |
|---|---|---|---|---|
| 10 s | ||||
| 15 s | ||||
| 20 s | ||||
| 25 s | ||||
| 30 s |
| LSTM-RNN Model | Sensitivity | Precision | F1 Score | AHI Difference |
|---|---|---|---|---|
| Normal | ||||
| Mild | ||||
| Moderate | ||||
| Severe | ||||
| All levels |
| Model + Oxygen Desaturation | Original SVM | Phenotype SVM [34] | Phenotype SVM+ Comoribidity [34] | LSTM-RNN |
|---|---|---|---|---|
| Precision | ||||
| Sensitivity | ||||
| F1 score | ||||
| AHI difference |
| LSTM-RNN with Desaturation and Awake Detection | Expert Label | ||||
|---|---|---|---|---|---|
| Normal | Mild | Moderate | Severe | ||
| RNN Label | Normal | 3 | 0 | 0 | 0 |
| Mild | 0 | 8 | 3 | 0 | |
| Moderate | 0 | 0 | 11 | 3 | |
| Severe | 0 | 0 | 0 | 28 | |
| Accuracy | 89.28% | ||||
| Four Events Types Classification in LSTM-RNN | Expert Label | ||||
|---|---|---|---|---|---|
| Normal | OSA | CSA | HYP | ||
| RNN Label | Normal | 37,788 | 232 | 470 | 1824 |
| OSA | 1391 | 7485 | 2132 | 1213 | |
| CSA | 21 | 35 | 1855 | 32 | |
| HYP | 556 | 1703 | 149 | 2138 | |
| Accuracy | 83.34% | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.-C.; Wu, H.-T.; Huang, P.-C.; Ma, H.-P.; Lo, Y.-L.; Huang, Y.-H. Portable Sleep Apnea Syndrome Screening and Event Detection Using Long Short-Term Memory Recurrent Neural Network. Sensors 2020, 20, 6067. https://doi.org/10.3390/s20216067
Chang H-C, Wu H-T, Huang P-C, Ma H-P, Lo Y-L, Huang Y-H. Portable Sleep Apnea Syndrome Screening and Event Detection Using Long Short-Term Memory Recurrent Neural Network. Sensors. 2020; 20(21):6067. https://doi.org/10.3390/s20216067
Chicago/Turabian StyleChang, Hung-Chi, Hau-Tieng Wu, Po-Chiun Huang, Hsi-Pin Ma, Yu-Lun Lo, and Yuan-Hao Huang. 2020. "Portable Sleep Apnea Syndrome Screening and Event Detection Using Long Short-Term Memory Recurrent Neural Network" Sensors 20, no. 21: 6067. https://doi.org/10.3390/s20216067
APA StyleChang, H.-C., Wu, H.-T., Huang, P.-C., Ma, H.-P., Lo, Y.-L., & Huang, Y.-H. (2020). Portable Sleep Apnea Syndrome Screening and Event Detection Using Long Short-Term Memory Recurrent Neural Network. Sensors, 20(21), 6067. https://doi.org/10.3390/s20216067





