3.1. Dataset and Experimental Setups
To evaluate the performance of our proposed method and compare it with previous studies, we conducted experiments using a public dataset, namely the in vivo GI endoscopic dataset [
30]. We called this dataset Hamlyn-GI for convenience, as this dataset is collected and provided by the Hamlyn Center for Robotic Surgery [
30]. This dataset was originally collected for tracking and retargeting of GI endoscopic pathological sites using Olympus narrow-band imaging and Pentax i-Scan endoscope devices [
30]. Specifically, this dataset contains 10 video sequences of GI endoscopic scans. Each video is saved in the format of successive still images. In the study by Ye et al. [
30], the authors first manually defined a pathological site (a small polyp or suspected region) at the beginning of the endoscopic image sequence. Then, they tracked and retargeted this region for the remaining sequence of images. The information regarding the selected region and ground-truth tracked-retargeted region is provided for each video sequence in an annotation file. Because these regions are carefully set by experts and the polyp region or possible polyp regions are focused on, we used the information in the annotation file as an indicator of the existence of pathological sites in the still images. In the case of a still image containing a pathological site region, an approximate location of the pathological site is provided in the annotation file; otherwise, a negative value is provided. Based on the provided information in the annotation file, we preclassified the still images into two categories: With and without the existence of the pathological site in the stomach; that is, if the annotation of a still image is provided, then the still image is considered to contain the pathological site and assigned to the “with pathological site” class; otherwise, the still image is assigned to the “without pathological site” class. In
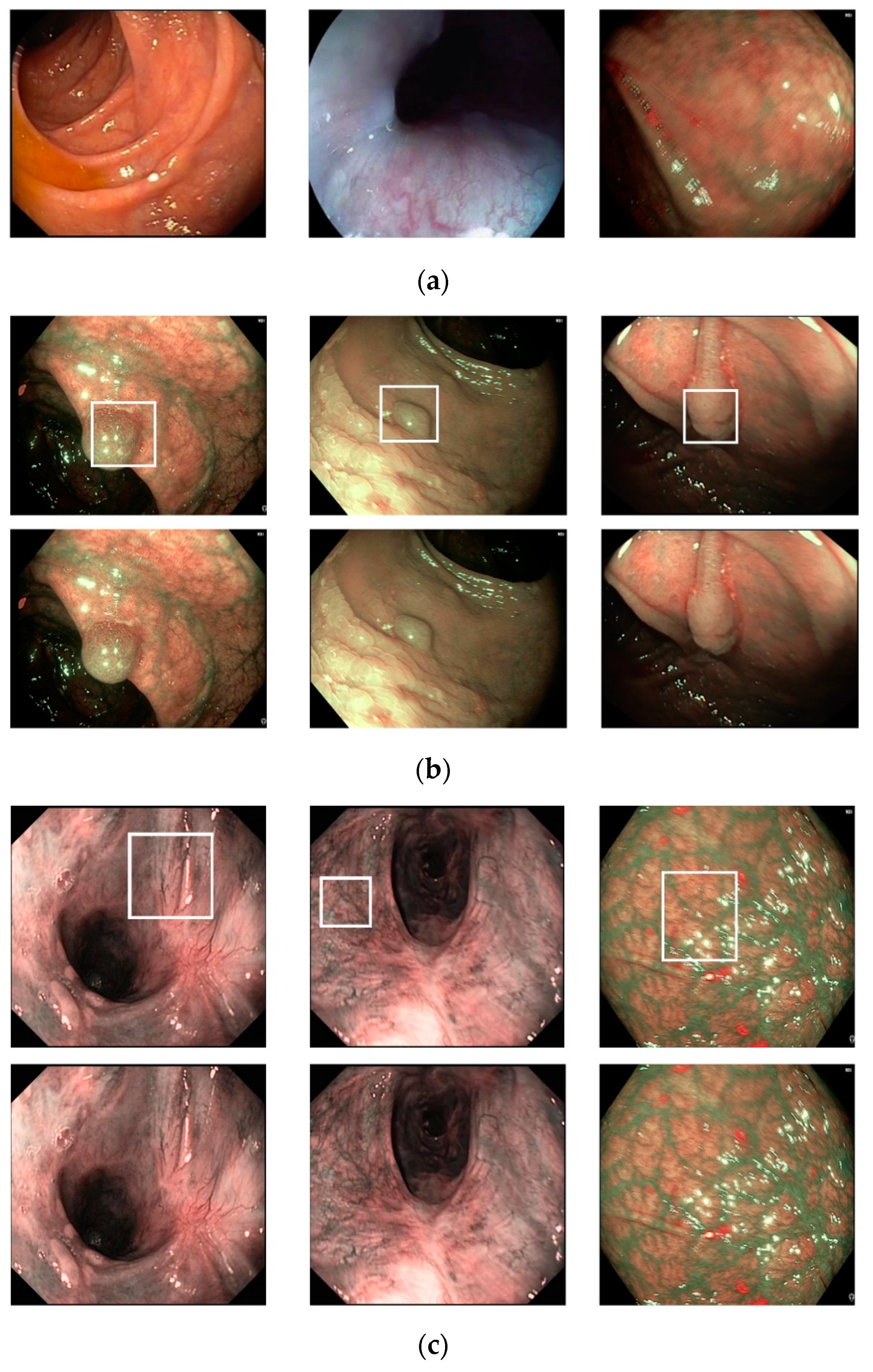
Figure 6, we show certain examples of images from the Hamlyn-GI dataset.
Figure 6a shows example images without the existence of pathological sites, whereas
Figure 6b shows images containing pathological sites, which are marked with white bounding boxes. From
Figure 6b, it can be observed that the bounding boxes are approximately provided by the author of the dataset. Therefore, they do not fit the correct location of pathological site regions. Consequently, it is difficult to perform a detection to solve this problem. Instead, we classified the input still images of this dataset into two classes: With and without the existence of pathological sites. In
Table 3, we list the detailed statistical information regarding the Hamlyn-GI dataset. In total, the Hamlyn-GI dataset contains 7894 images.
To measure the performance of the proposed method, we performed two-fold cross-validation. For this purpose, we divided the Hamlyn-GI dataset into two separate parts, namely training and testing datasets. In the first fold, we assigned images of the first five video sequences (video files 1–5 in
Table 3) as the training dataset and the images of the remaining five video sequences (video files 6–10 in
Table 3) as the testing dataset. In the second fold, we exchanged the training and testing datasets of the first fold, i.e., training dataset contains images of the last five video sequences (video files 6–10 in
Table 3) and the testing dataset contains images of the first five video sequences (video files 1–5 in
Table 3). This division method ensures that the images of the same person (identity) only exist in either the training or testing dataset. Finally, the overall performance of the dataset with a two-fold cross-validation approach is measured by calculating the average (weighted by the number of testing images) of the two folds. In
Table 4, we list a detailed description of the training and testing datasets in our experiments. In this table, “With PS” indicates the existence of a pathological site condition; further, “Without PS” indicates the absence of the existence of a pathological site condition. Although it is possible to use other cross-validation methods such as three-fold, five-fold, or leave-one-out approaches, we decided to use a two-fold approach in our experiments to save the processing time of the experiments.
3.2. Training
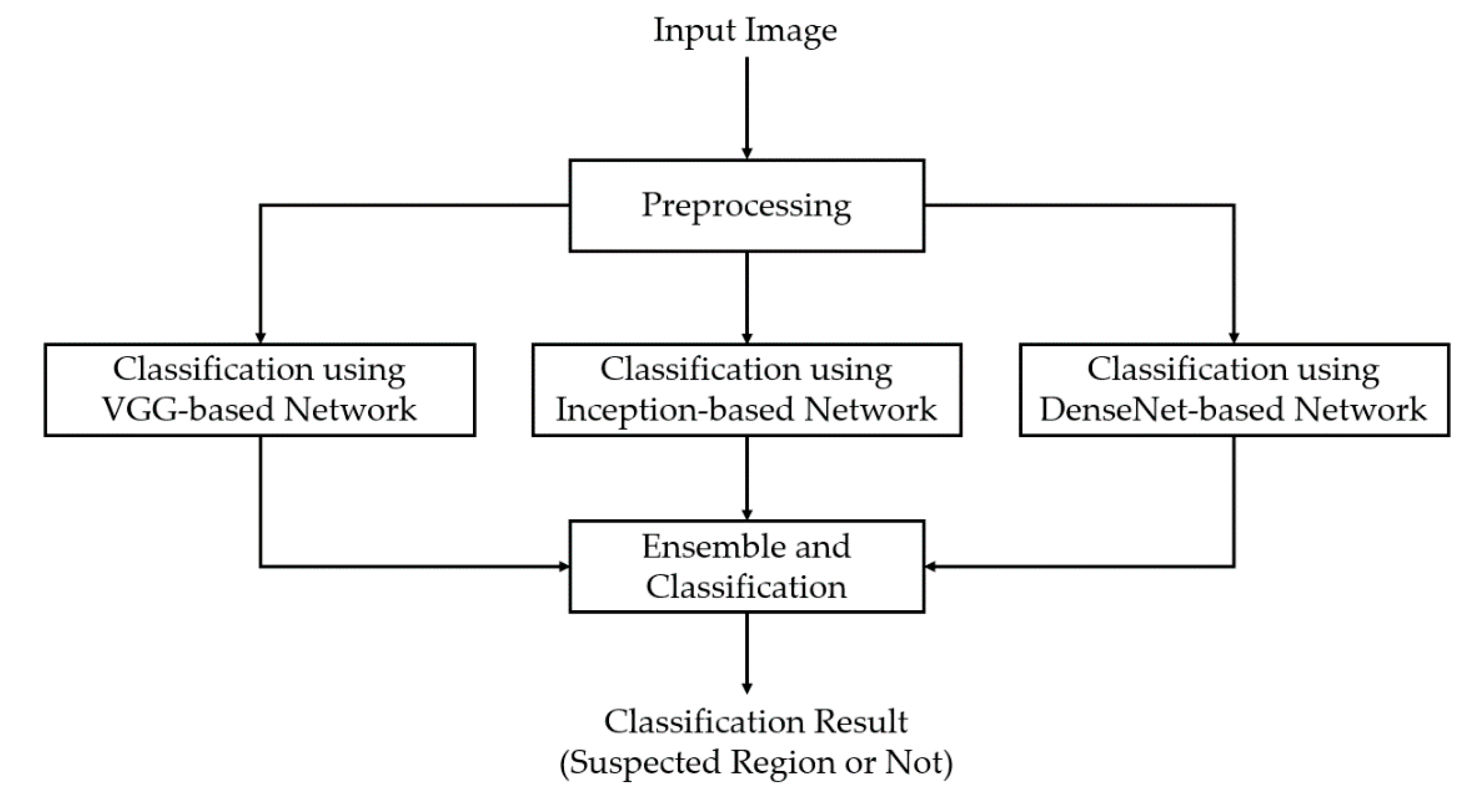
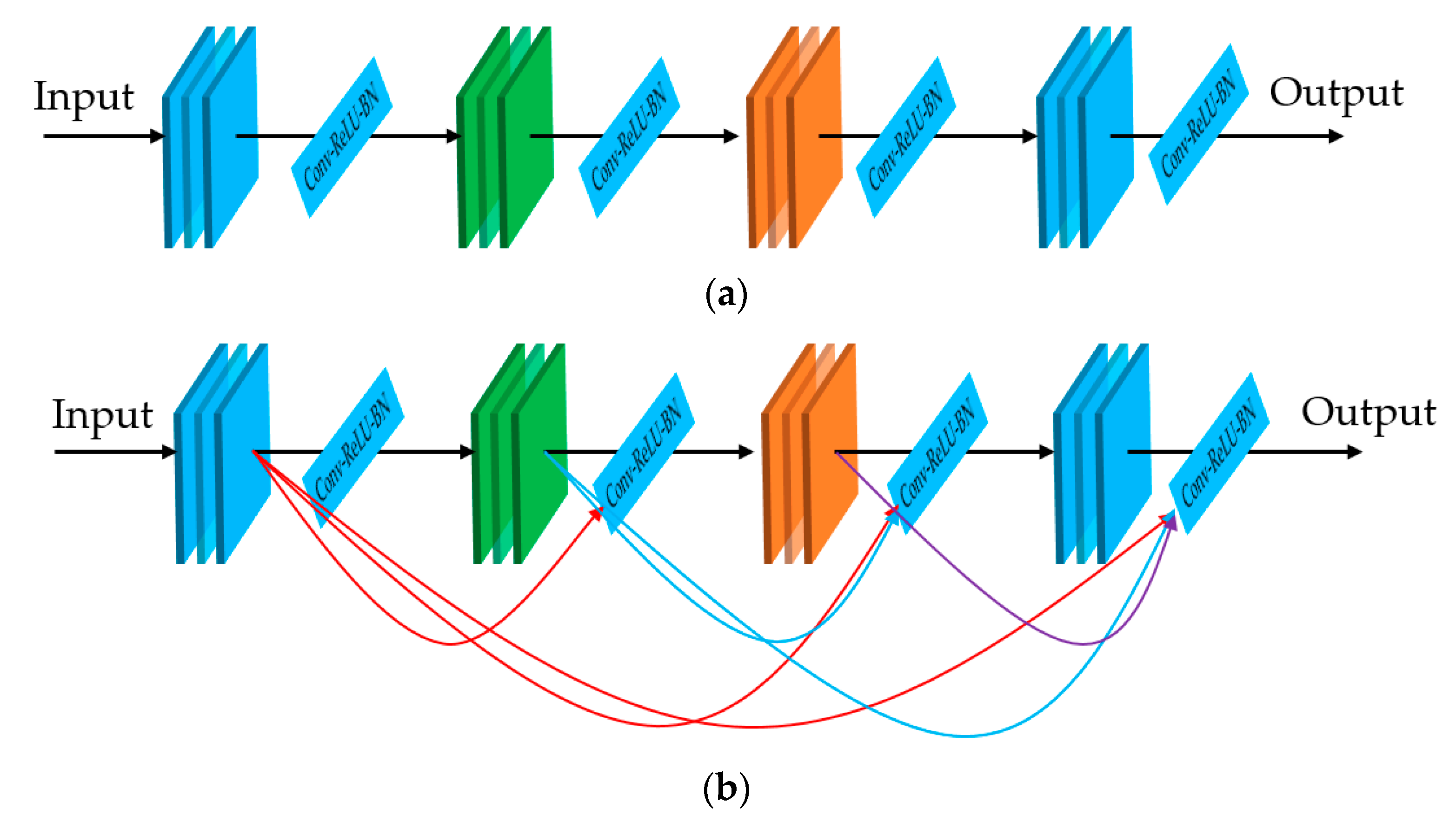
In our first experiment, we performed a training process to train the three deep learning-based models which are illustrated in
Figure 1 and
Table 2. For this experiment, we programmed our network using Python programming language with the help of the Tensorflow library [
39] for the implementation of deep learning-based models. A detailed description of the parameters is listed in
Table 5. For the training method, we used the adaptive moment estimation (Adam) optimizer with an initial learning rate of 0.0001, and we trained each model with 30 epochs. As the epoch increases, the network parameters become finer; therefore, we continuously reduced the learning rate after every epoch. In addition, a batch size of 32 was used in our experiment.
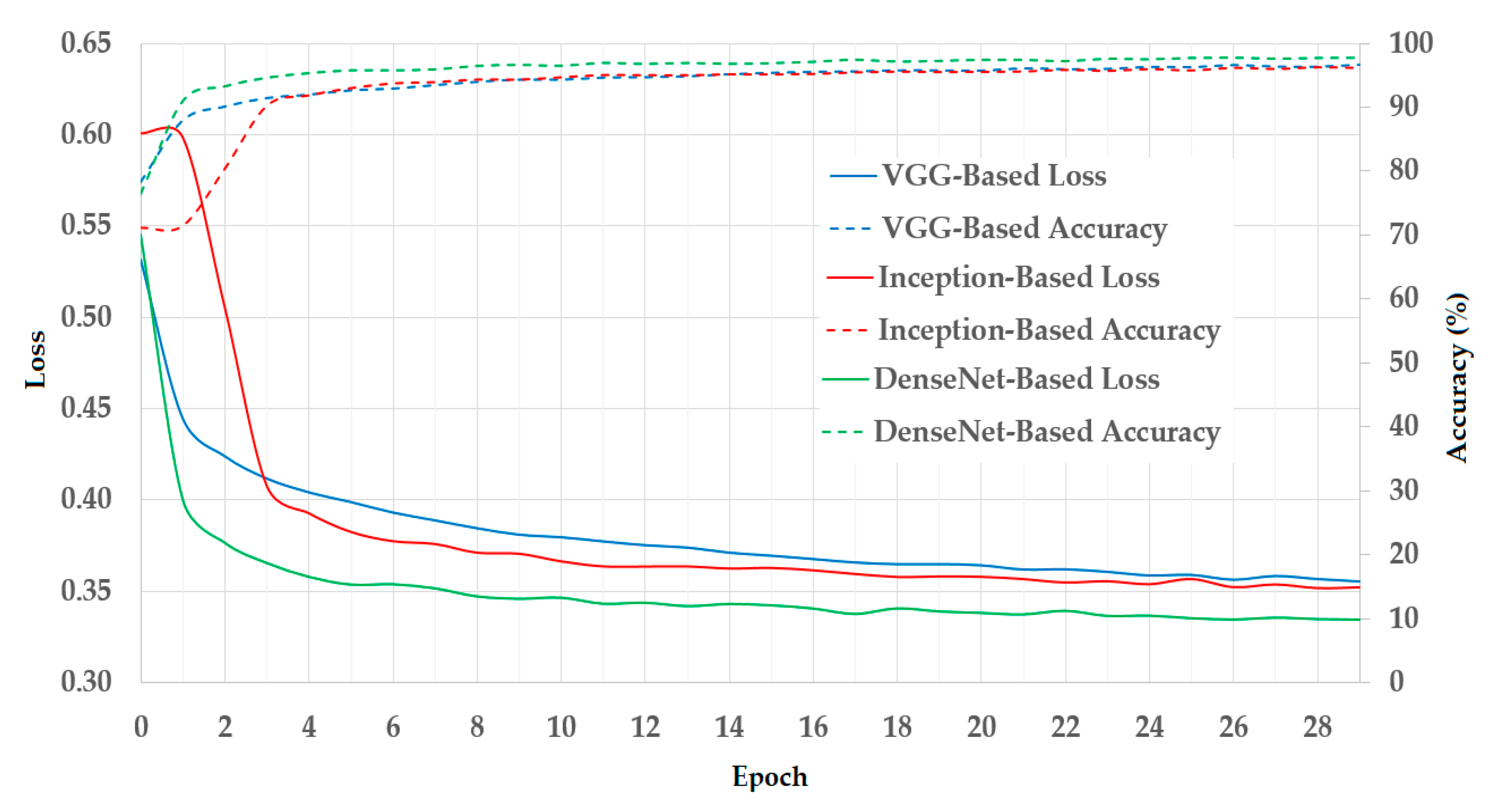
In
Figure 7, we illustrated the results of the training process using the training datasets. As mentioned in
Section 3.1, we used a two-fold cross-validation procedure in our experiments. Therefore, we calculated the average result of the two folds and presented it in
Figure 7. In this figure, we show the curves of the loss and training accuracy of the training procedure for all three CNN models. As shown in these curves, the losses continuously decrease while the training accuracies increase with the increase in the training epoch. Thus, we can consider that the training procedures were successful in our experiments.
3.4. Comparisons with the State-of-the-Art Methods and Processing Time
Based on the experimental results presented in the above sections, we compared the classification performance of our proposed method with those of previous studies. As explained in
Section 1, most of the previous studies used the conventional CNN (training a single shallow network or extracting image features from a pretrained network) for classification purposes. As discussed in
Section 2.3 and
Section 3.3, our proposed method applies the ensemble learning technique to three individual deep CNNs, i.e., VGG-, inception-, and DenseNet-based networks. Therefore, we can consider that the performances of previous studies correspond to the case of using each individual network. Based on this assumption, we conducted a comparison experiment and its results are listed in
Table 9. From the table, it can be observed that by using the individual CNNs, we obtained the overall classification accuracies of 68.912%, 66.505% and 52.609% using the VGG-, inception-, and DenseNet-based networks, respectively. The best accuracy of 68.912% was obtained using the VGG-based method. Using our proposed method, we obtained the best accuracy of 70.775% with the AVERAGE combination rule, which is higher than the accuracy produced by the use of an individual network. Moreover, our proposed method with the VOTING combination rule also produced an accuracy of 70.369%, which is also higher than the best value of 68.912% of the individual network. From these experimental results, we can conclude that our proposed method outperforms previous studies in the pathological site classification system. However, as the best accuracy is still low (70.775%), the classification system still requires enhancements in our future works.
In the final experiment, we measured the processing time of our proposed method for pathological site classification problems. For this purpose, we created a deep CNN program in Python programming language using the Tensorflow library [
39]. To run the program, we used a desktop computer with an Intel Core i7-6700 central processing unit with a working clock of 3.4 GHz and 64 GB of RAM memory. To increase the speed of the deep learning networks, we used a graphical processing unit, namely GeForce Titan X, to run the inference of the three deep learning models [
40]. The detailed experimental results are listed in
Table 10. We also performed the comparisons of the processing time between the proposed method and the previous studies. As shown in
Table 10, it takes about 37.646 ms, 67.472 ms and 65.901 ms to classify an input image using VGG-based [
11,
25], Inception-based [
26], and DenseNet-based [
31] network, respectively. Using our proposed method, it takes 179.440 ms to classify a single input image, and our proposed method can work at a speed of 5.6 frames per second. From these experiment results, we can find that our proposed method takes longer processing time than previous studies. However, it is acceptable in medical image processing applications where the accuracy, but not processing time, is the primary requirement.
3.5. Analysis and Discussion
As shown in our experimental result, the average rule outperforms the max and voting rule in enhancing classification performance. This is caused by the fact that the average rule uses the prediction scores of individual CNN models directly, whereas the max rule is performed based on only the maximum classification score of one CNN model among several CNN models, and the voting rule is performed based on majority decisions of CNN models. As a result, the combined classification score by average rule contains more detailed classification information from all CNN models than the other two combination rules.
We used a public dataset, namely Hamlyn-GI, which showed the large visual differences among the collected images [
30]. To measure the performance of the proposed method, we first divided the entire Hamlyn-GI dataset into training and testing sets. The division is done by ensuring that the training and testing datasets are different, i.e., images of the same patient only exist in either training or testing datasets as open world evaluation. As a result, these training and testing set are independent from the other as shown in
Figure 6 (for example, the left-bottom and middle-bottom images are the samples of training data whereas the others are those of testing one for the 1st fold validation). We used the training dataset to train the classification models, and testing dataset to measure the performance of the trained models. Therefore, the measured performance is the optimal generalization because the testing set is independent from the training set.
To demonstrate the efficiency of our proposed method, we show certain examples of the classification results in
Figure 9. As shown in this figure, images without a pathological site are accurately classified as negative cases (
Figure 9a), while images with pathological sites (small polyp regions appear in
Figure 9b, complex vascular structure regions in
Figure 9c) are classified as positive cases. However, our proposed method also produces certain incorrect classification results, as shown in
Figure 10. In
Figure 10a, we show certain sample images that were classified as positive cases (images with pathological sites) even if they did not contain pathological sites. The errors were caused either by the fact that the endoscopic images contain complex vascular structures (the left-most image in
Figure 10a) or owing to imperfect input images (middle and right-most images in
Figure 10a). In
Figure 10b,c, we show certain example images in which our classification method incorrectly classified. As we can observe from these figures, certain text boxes in the input images can cause classification errors (right-most image in
Figure 10c). In addition, the input images contain small or blurred polyp regions, resulting in classification errors, which can be solved by super-resolution reconstruction or deblurring in our future works.
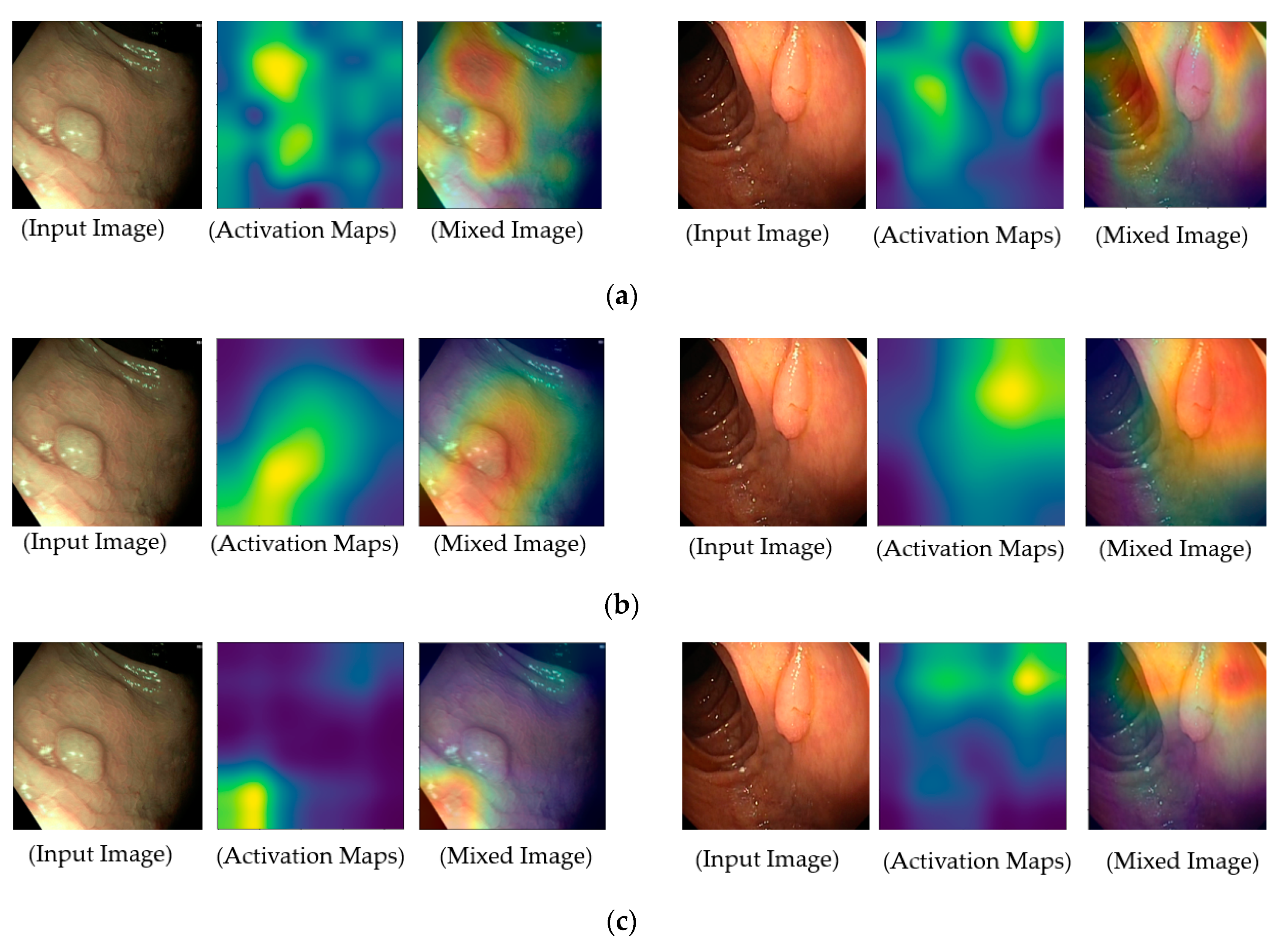
To provide a deep look inside the actual operation of deep learning-based models for classification problems in our study, we measured the regions of focus in the input images on which the classification models are used for accomplishing their functionalities, and the results are illustrated in
Figure 11. For this purpose, we obtained the class activation maps using the gradient-weighted class activation mapping (grad-CAM) method [
41]. Grad-CAM is a popular method that explains the working of a deep CNN. In the activation maps of these figures, the brighter regions indicate the regions that are focused on in the feature maps, which our system uses for classification purposes. As shown in
Figure 11, our deep learning models focus on pathological sites (possible polyps) or complex vascular structures in classifying input images into negative or positive classes. By providing class activation maps to medical doctors, our system can provide a reasonable explanation about why an input image is classified as positive data, and it can demonstrate the functionality of explainable artificial intelligence. In addition, it is a significantly important characteristic of a high-performance classification system in CAD applications.
In our experiments, we use only three sub-models for ensemble algorithm because of two reasons. First, these models (VGG-based, Inception-based, and DenseNet-based) are different in network architecture. Therefore, each sub-model has its own advantages and disadvantages that can be compensated by the other sub-models. Second, although we can use more sub-models to perform ensemble algorithm, the complexity and processing time of the proposed method are increased, which can cause the difficulty in training and deployment of model. Because of these reasons, we only used three sub-models in our experiments. However, it is possible to use more sub-models to possibly enhance the system performance. In that case, our work can be seen as a specific example to demonstrate the enhancement possibility of ensemble algorithm in pathological site classification problem. When the number of sub-models increases, it not only increases the complexity and computation of system, but also represents noises to system that caused by error cases of every individual system.
For a demonstration purpose, we additionally performed experiments with more than three sub-models, i.e., ensemble method with four sub-models. For this purpose, we used an additional CNN classifier based on residual network [
23]. The experimental results are given in
Table 11. From these experimental results, we see that we obtained a highest classification accuracy of 69.686% using the average rule that is little worse than the accuracy of 70.775% obtained using three sub-models mentioned in
Section 3.3. The reason is that when the number of models increases, it also represents noises to the ensemble system caused by error cases of the new model. In addition, the residual network also uses the short-cut connection in the similar way as the DenseNet-based network. Therefore, the residual network shares similar characteristics with DenseNet-based network. Because of these reasons, the performance of ensemble system based on four sub-models is little reduced compared to the case of using three sub-models.