Plant Leaf Position Estimation with Computer Vision
Abstract
:1. Introduction
2. Materials and Methods
2.1. Robotic Platform Description
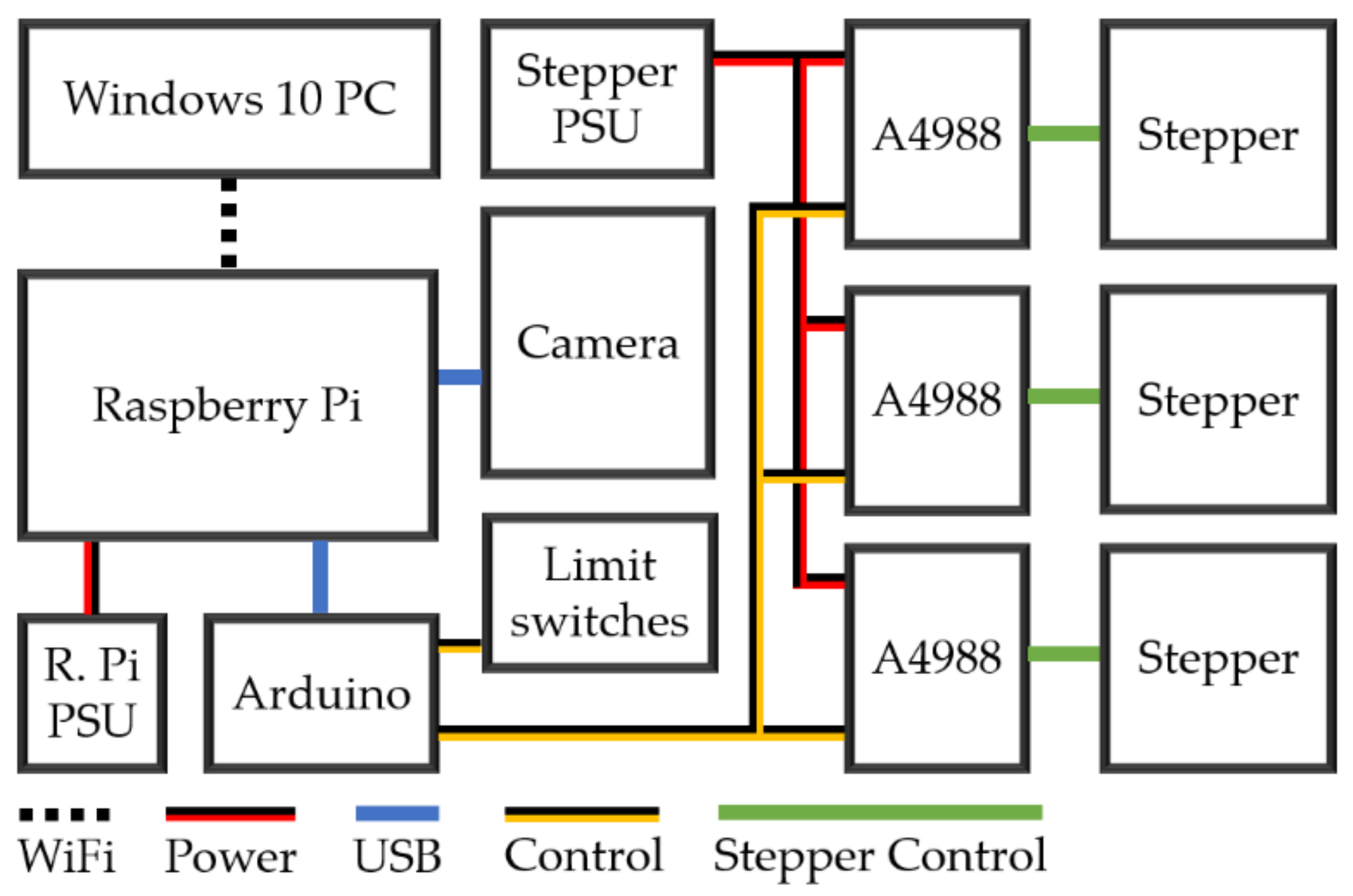
2.2. Electrical Build Description
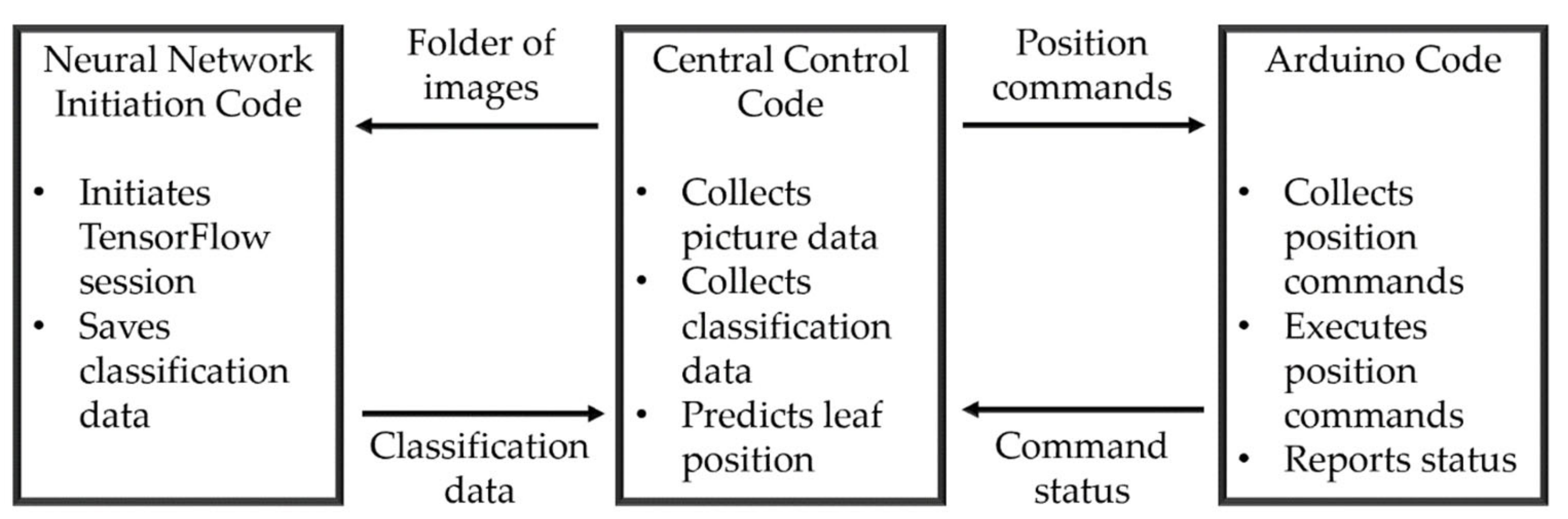
2.3. Software Build Description
2.3.1. Neural Network Training and Initiating
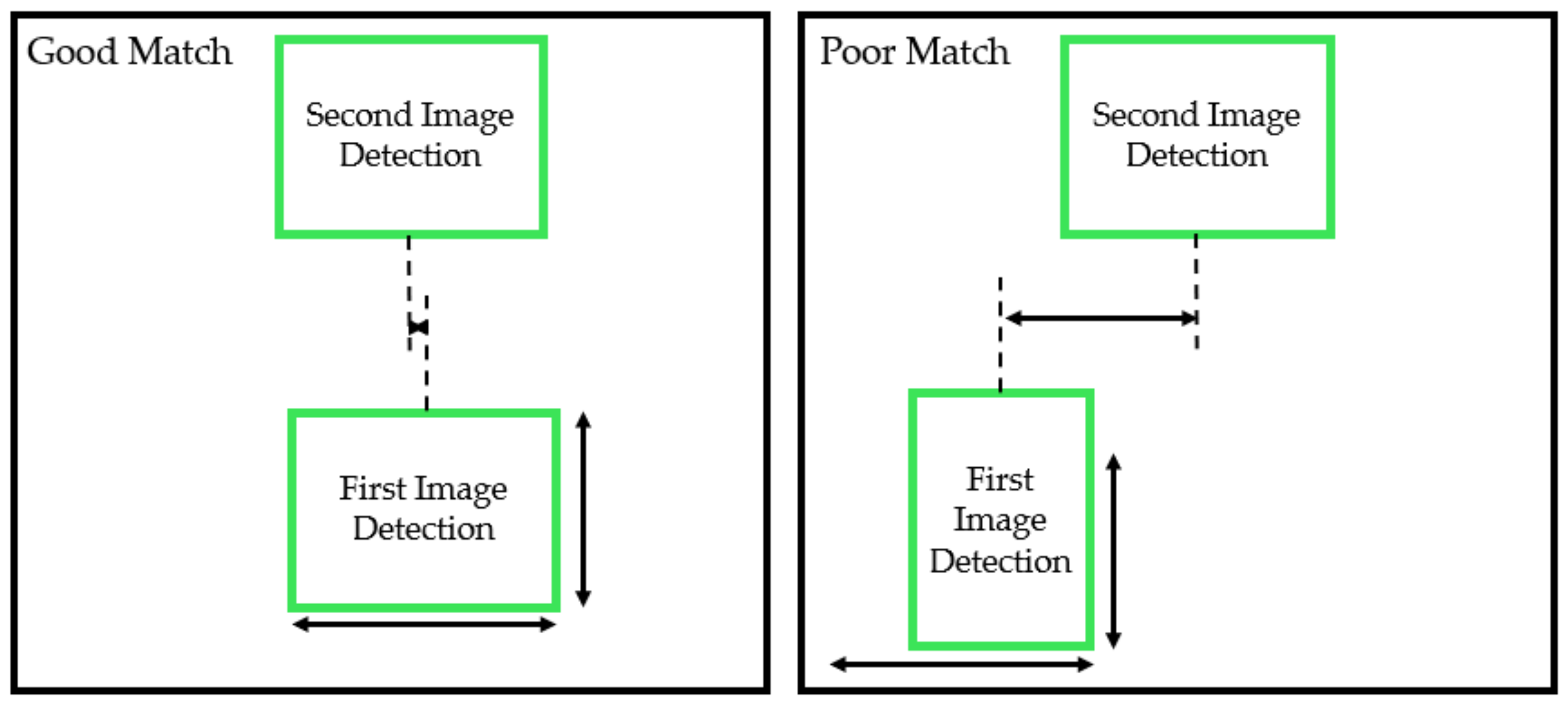
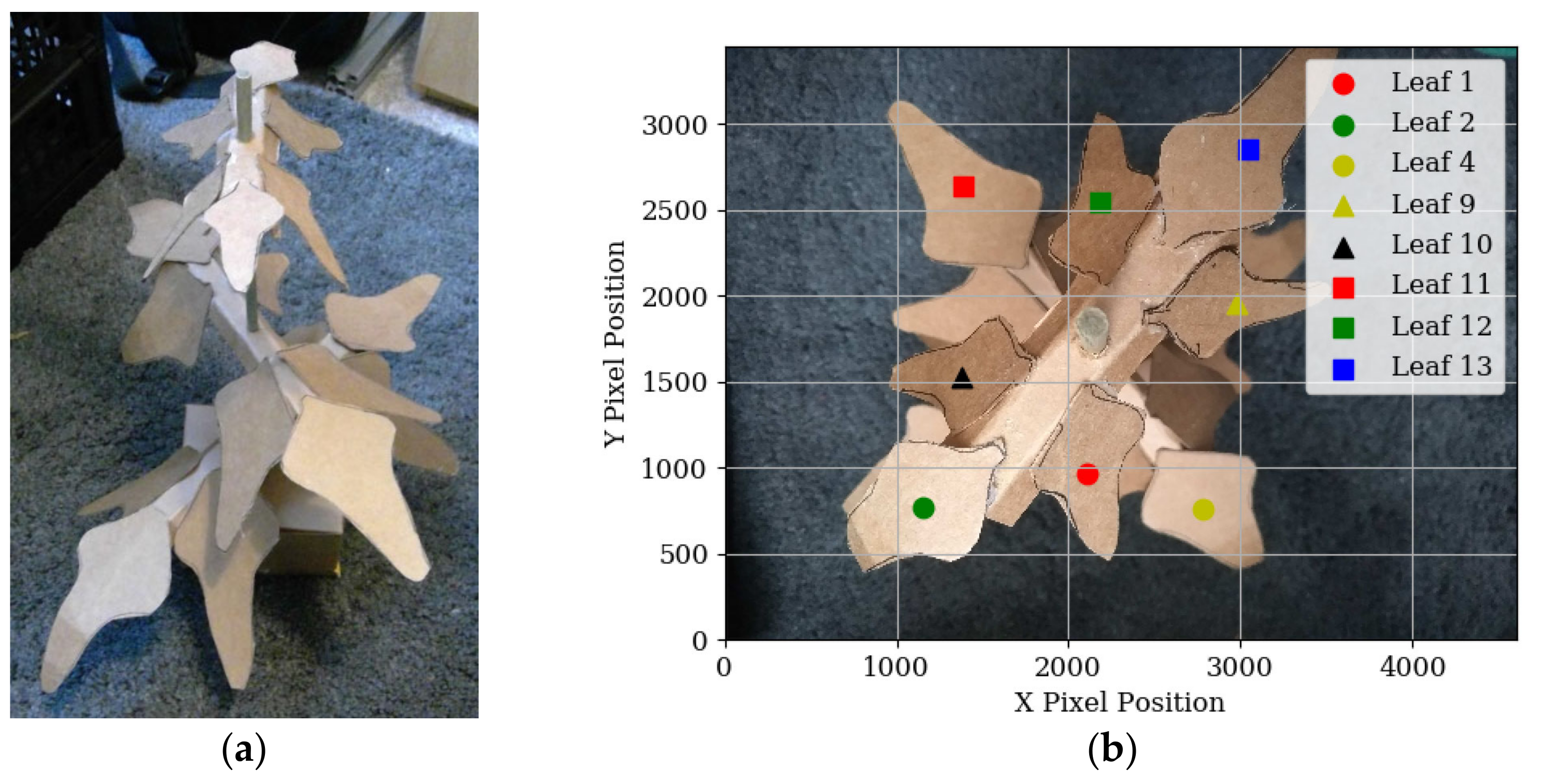
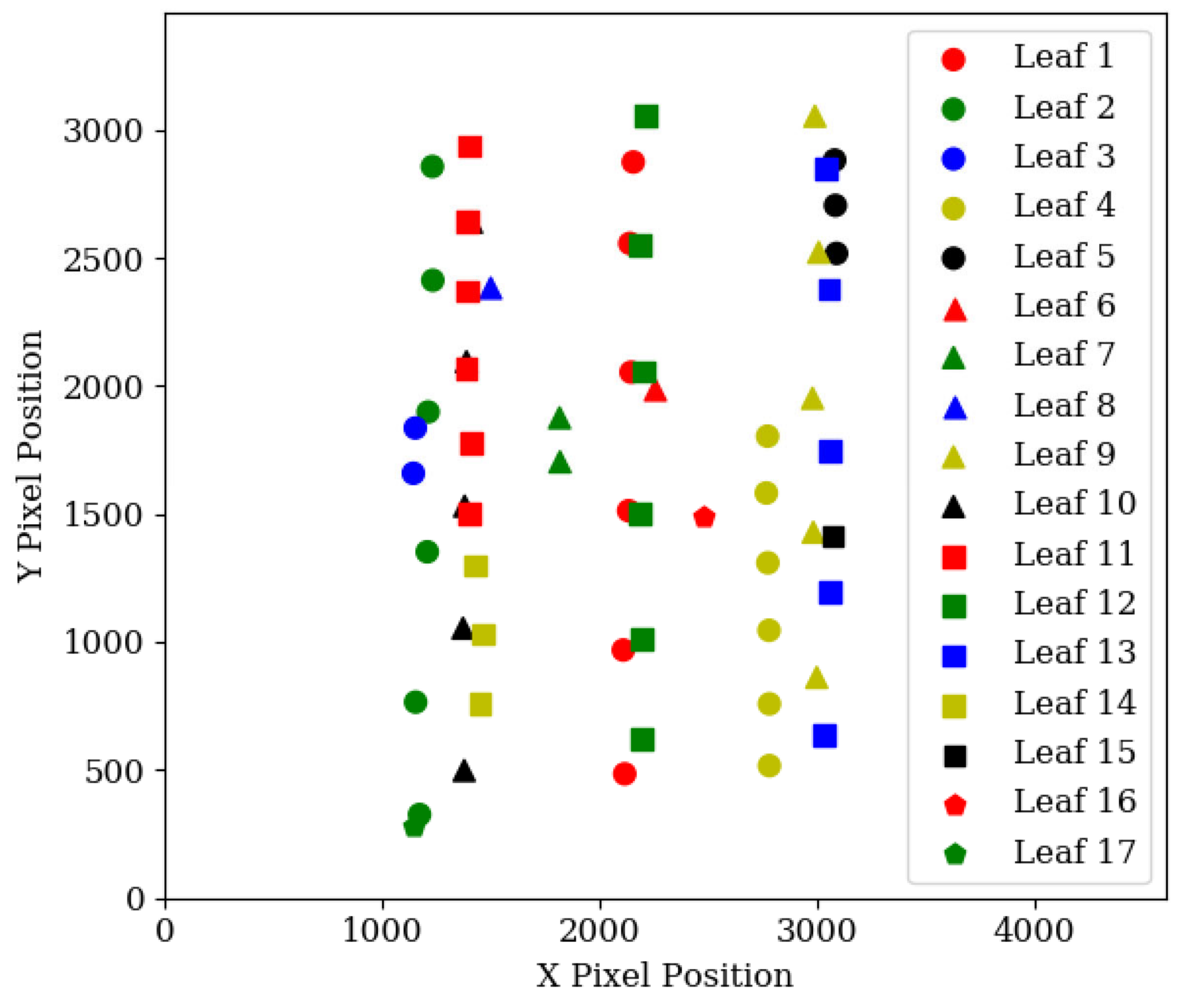
2.3.2. Leaf Detection Grouping
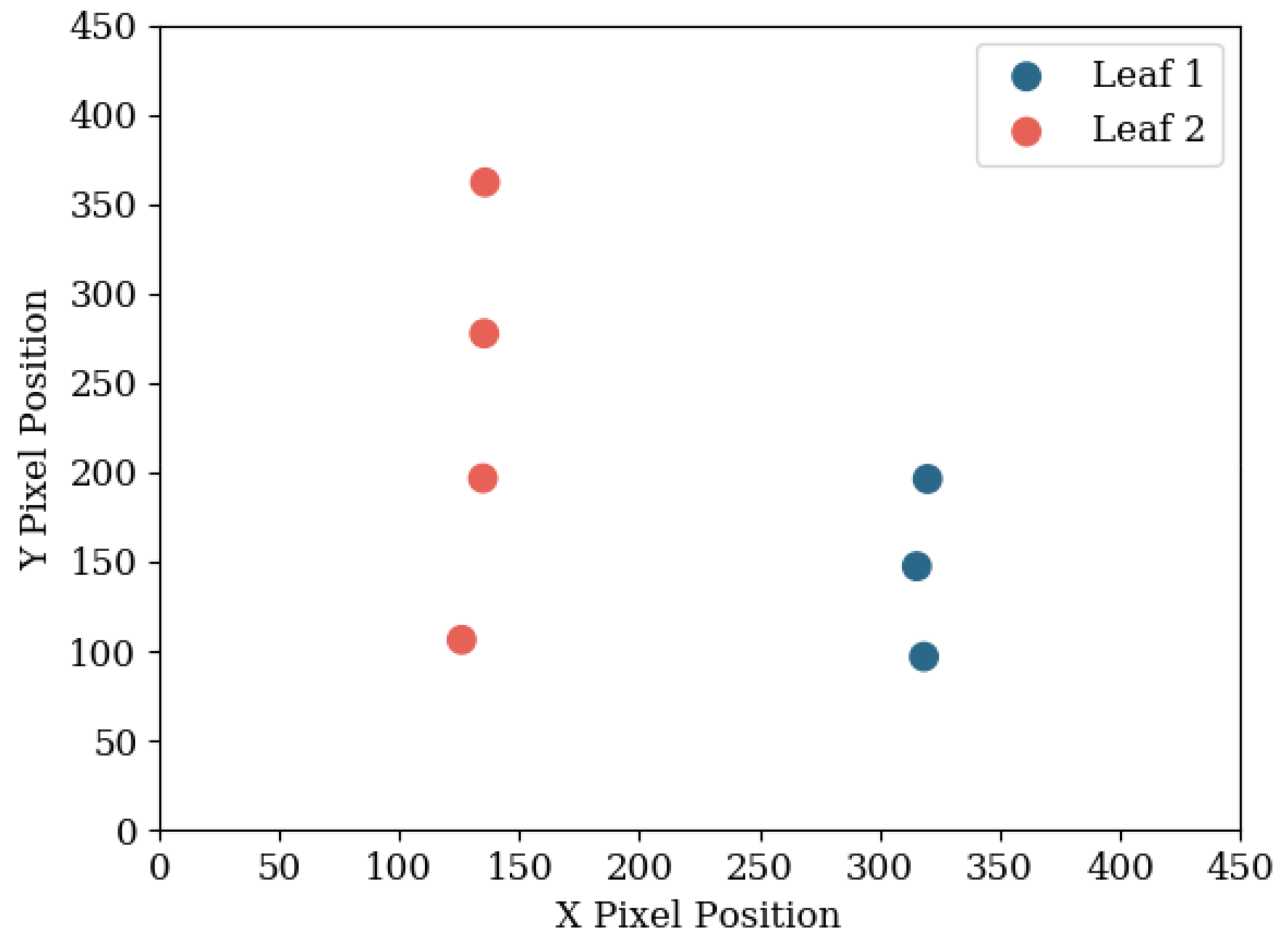
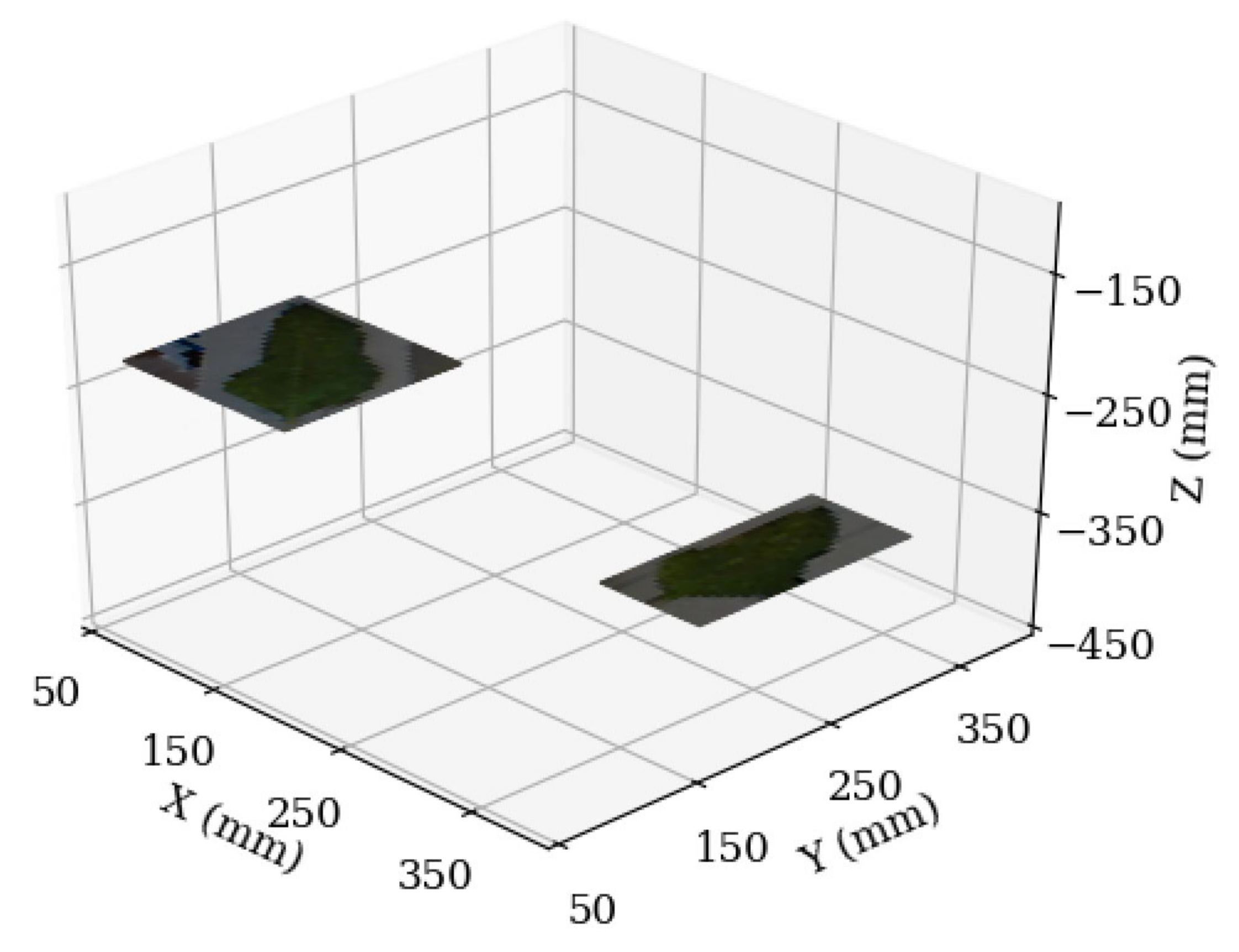
2.3.3. Depth and Position Estimation
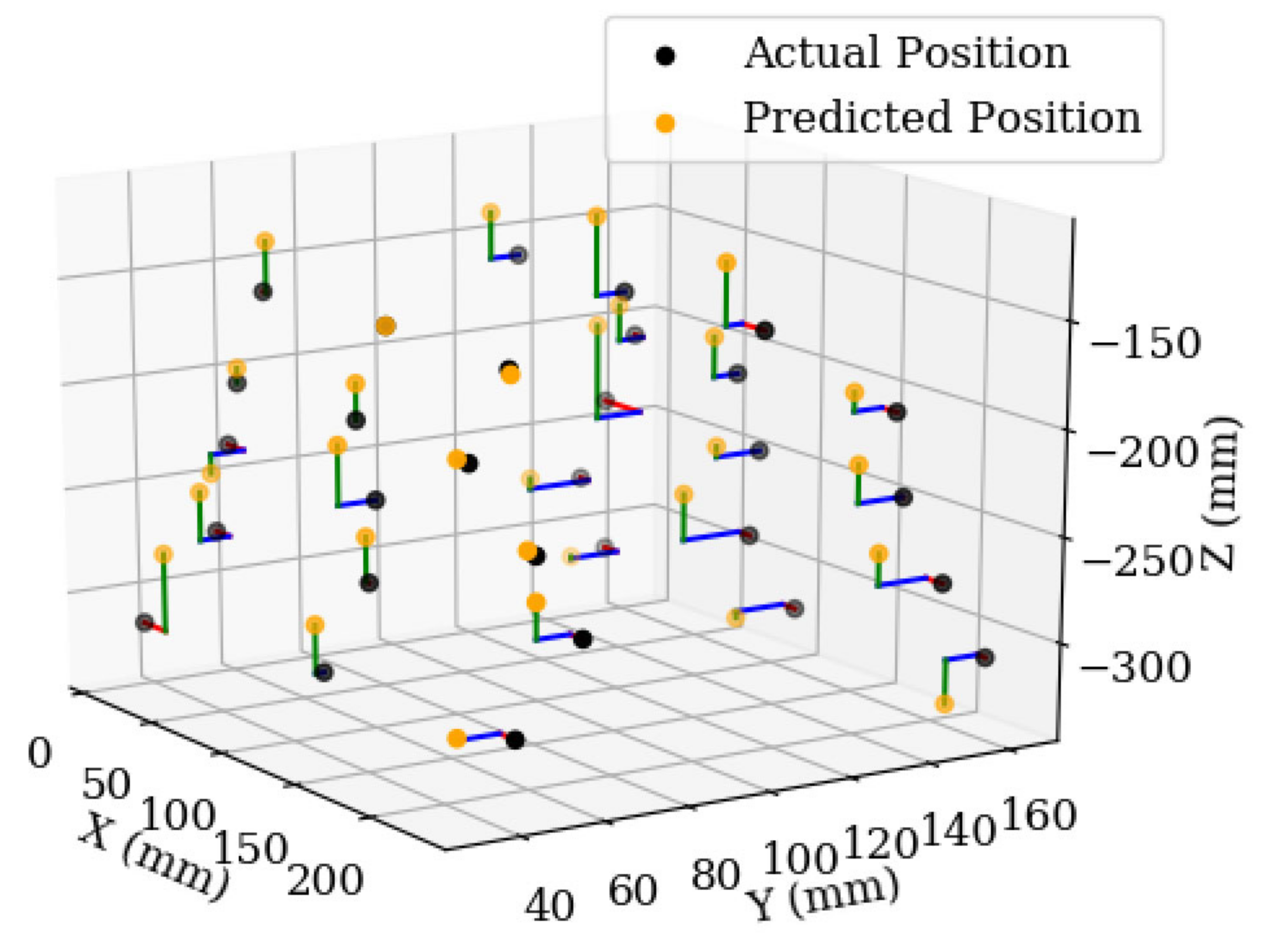
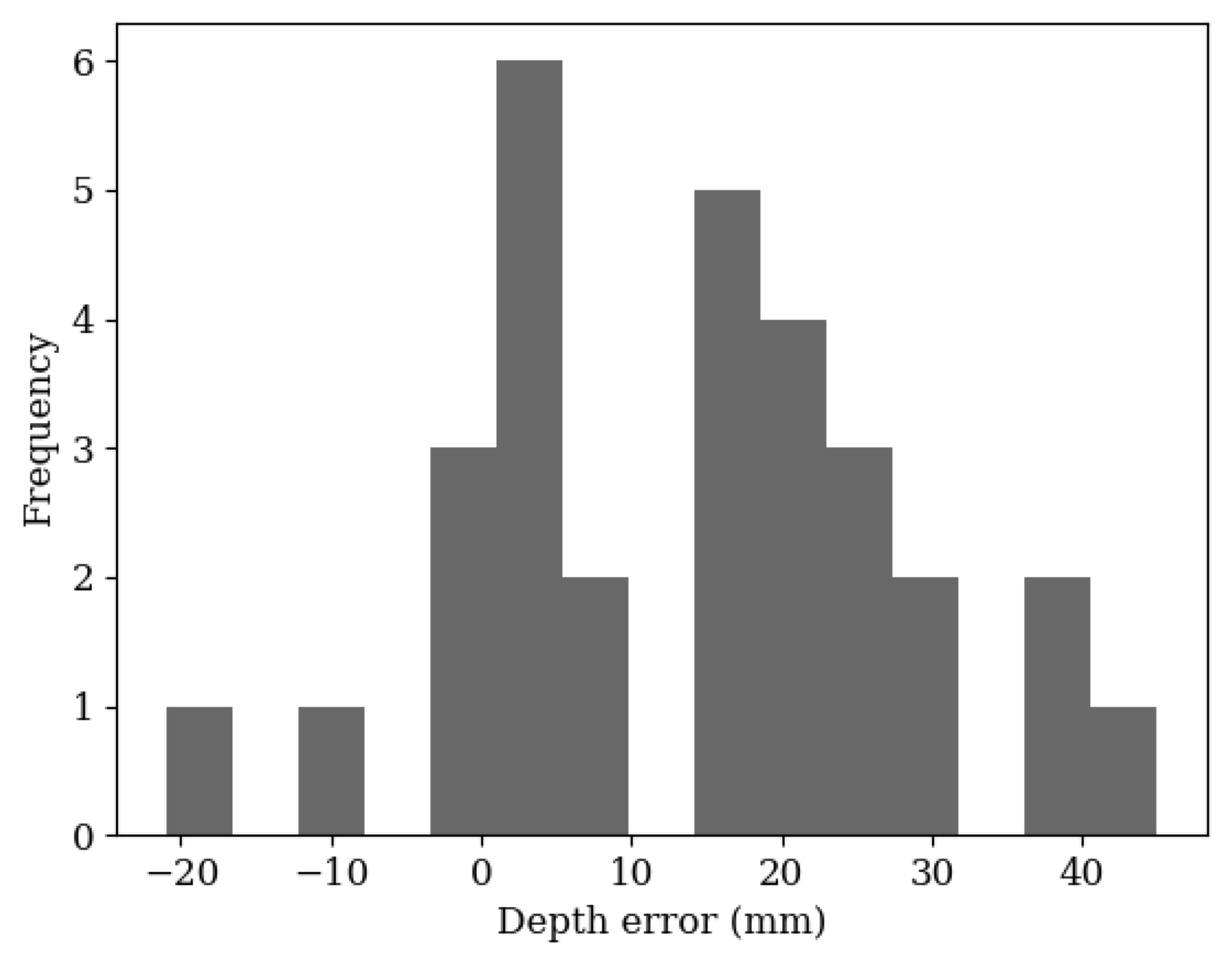
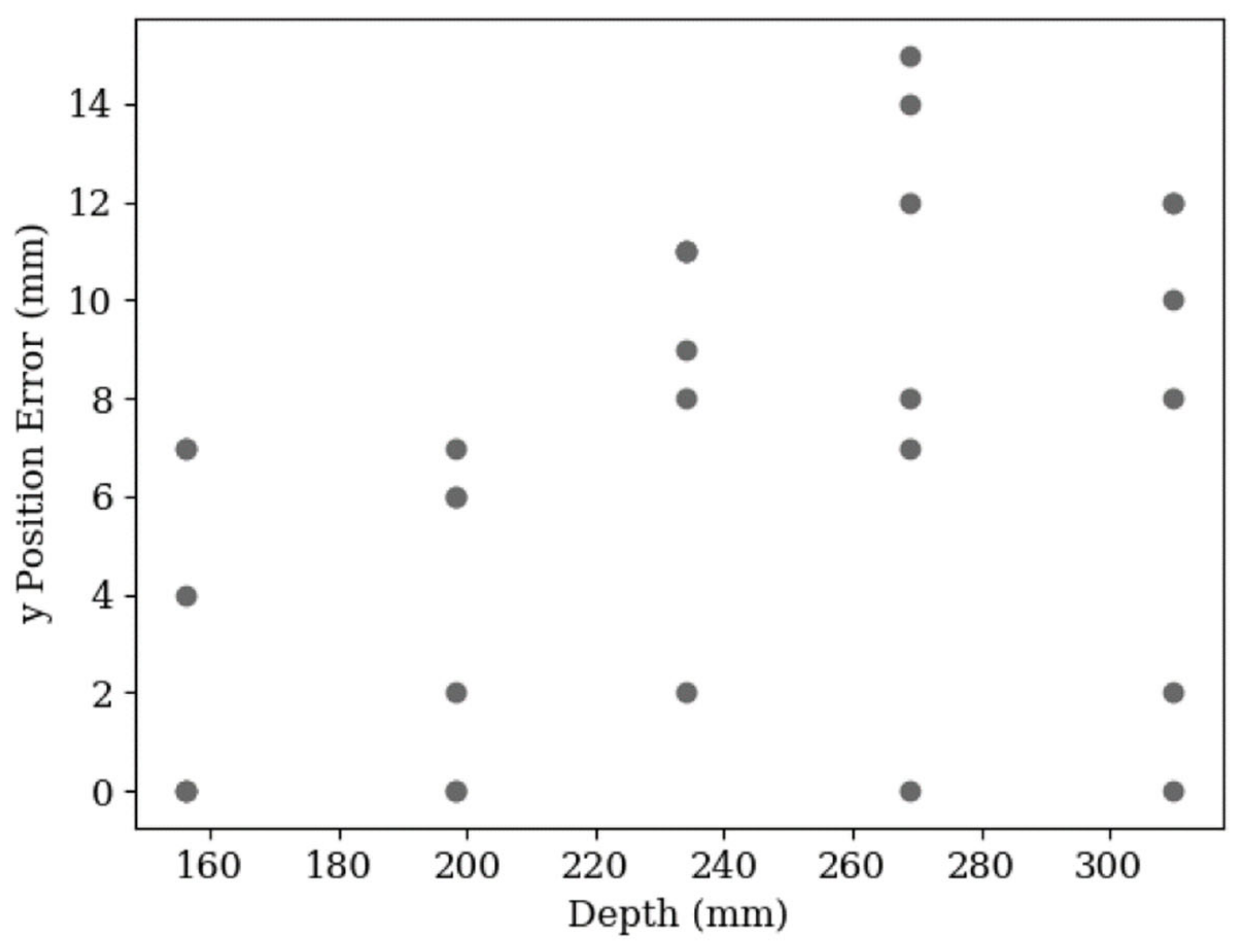
3. Results and Discussion
4. Conclusions
4.1. Neural Network
4.2. Proposed Position Estimation Technique
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erahaman, M.; Echen, D.; Egillani, Z.; Eklukas, C.; Chen, M. Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front. Plant Sci. 2015, 6, 619. [Google Scholar] [CrossRef] [Green Version]
- Karakaya, D.; Ulucan, O.; Turkan, M. Electronic Nose and Its Applications: A Survey. Int. J. Autom. Comput. 2019, 17, 179–209. [Google Scholar] [CrossRef] [Green Version]
- Webster, J.; Shakya, P.; Kennedy, E.; Caplan, M.; Rose, C.; Rosenstein, J.K. TruffleBot: Low-Cost Multi-Parametric Machine Olfaction. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2018. [Google Scholar]
- Burrell, T.; Fozard, S.; Holroyd, G.H.; French, A.P.; Pound, M.P.; Bigley, C.J.; Taylor, C.; Forde, B.G. The Microphenotron: A robotic miniaturized plant phenotyping platform with diverse applications in chemical biology. Plant Methods 2017, 13, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yalcin, H. Plant phenology recognition using deep learning: Deep-Pheno. In Proceedings of the 2017 6th International Conference on Agro-Geoinformatics, Fairfax, VA, USA, 7–10 August 2017; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2017. [Google Scholar]
- Namin, S.T.; Esmaeilzadeh, M.; Najafi, M.; Brown, T.B.; Borevitz, J.O. Deep phenotyping: Deep learning for temporal phenotype/genotype classification. Plant Methods 2018, 14, 66. [Google Scholar] [CrossRef] [Green Version]
- Hall, D.; McCool, C.; Dayoub, F.; Sunderhauf, N.; Upcroft, B. Evaluation of Features for Leaf Classification in Challenging Conditions. In Proceedings of the 2015 IEEE Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 5–9 January 2015; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2015. [Google Scholar]
- Sa, I.; Ge, Z.; Dayoub, F.; Upcroft, B.; Perez, T.; McCool, C. DeepFruits: A Fruit Detection System Using Deep Neural Networks. Sensors 2016, 16, 1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazen, F.M.A.; Nashat, A.A. Ripeness Classification of Bananas Using an Artificial Neural Network. Arab. J. Sci. Eng. 2019, 44, 6901–6910. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, R.J.; Varcalcel, F.E. Understanding Microsoft Kinect for application in 3-D vision processes. Rev. Clepsidra 2014, 10, 45–53. [Google Scholar] [CrossRef]
- El-Laithy, R.A.; Huang, J.; Yeh, M. Study on the use of Microsoft Kinect for robotics applications. In Proceedings of the Proceedings of the 2012 IEEE/ION Position, Location and Navigation Symposium, Myrtle Beach, SC, USA, 23–26 April 2012; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2012. [Google Scholar]
- A Nair, S.K.; Joladarashi, S.; Ganesh, N. Evaluation of ultrasonic sensor in robot mapping. In Proceedings of the 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 23–25 April 2019; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2019. [Google Scholar]
- Lakovic, N.; Brkic, M.; Batinic, B.; Bajic, J.; Rajs, V.; Kulundzic, N. Application of low-cost VL53L0X ToF sensor for robot environment detection. In Proceedings of the 2019 18th International Symposium INFOTEH-JAHORINA (INFOTEH), East Sarajevo, Bosnia and Herzegovina, 20–22 March 2019; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2019. [Google Scholar]
- Falkenhagen, L. Depth Estimation from Stereoscopic Image Pairs Assuming Piecewise Continuos Surfaces. In Workshops in Computing; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1995; pp. 115–127. [Google Scholar]
- Yaguchi, H.; Nagahama, K.; Hasegawa, T.; Inaba, M. Development of an autonomous tomato harvesting robot with rotational plucking gripper. In Proceedings of the 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Daejeon, Korea, 9–14 October 2016; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2016. [Google Scholar]
- Kaczmarek, A.L. Improving depth maps of plants by using a set of five cameras. J. Electron. Imaging 2015, 24, 23018. [Google Scholar] [CrossRef] [Green Version]
- Laga, H.; Miklavcic, S.J. Curve-based stereo matching for 3D modeling of plants. In Proceedings of the 20th International Congress on Modelling and Simulation, Adelaide, SA, Australia, 1–6 December 2013. [Google Scholar]
- Quan, L.; Tan, P.; Zeng, G.; Yuan, L.; Wang, J.; Kang, S.B. Image-based plant modelling. In ACM SIGGRAPH 2006 Research Posters on SIGGRAPH ’06; Association for Computing Machinery: New York, NY, USA, 2006; Volume 25, p. 599. [Google Scholar] [CrossRef]
- Ahlin, K.; Joffe, B.; Hu, A.-P.; McMurray, G.; Sadegh, N. Autonomous Leaf Picking Using Deep Learning and Visual-Servoing. IFAC-PapersOnLine 2016, 49, 177–183. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous systems. arXiv 2016, arXiv:1603.044672015. [Google Scholar]
- How to Train an Object Detection Classifier for Multiple Objects Using TensorFlow (GPU) on Windows 10. Available online: https://github.com/EdjeElectronics/TensorFlow-Object-Detection-API-Tutorial-Train-Multiple-Objects-Windows-10 (accessed on 3 October 2020).
- TensorFlow 1 Detection Model Zoo. Available online: https://github.com/tensorflow/models/blob/master/research/object_detection/g3doc/tf1_detection_zoo.md (accessed on 3 October 2020).








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beadle, J.; Taylor, C.J.; Ashworth, K.; Cheneler, D. Plant Leaf Position Estimation with Computer Vision. Sensors 2020, 20, 5933. https://doi.org/10.3390/s20205933
Beadle J, Taylor CJ, Ashworth K, Cheneler D. Plant Leaf Position Estimation with Computer Vision. Sensors. 2020; 20(20):5933. https://doi.org/10.3390/s20205933
Chicago/Turabian StyleBeadle, James, C. James Taylor, Kirsti Ashworth, and David Cheneler. 2020. "Plant Leaf Position Estimation with Computer Vision" Sensors 20, no. 20: 5933. https://doi.org/10.3390/s20205933
APA StyleBeadle, J., Taylor, C. J., Ashworth, K., & Cheneler, D. (2020). Plant Leaf Position Estimation with Computer Vision. Sensors, 20(20), 5933. https://doi.org/10.3390/s20205933







