Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images
Abstract
:1. Introduction
- we introduce a high quality RGB image dataset named DND-SB for nutrient deficiencies classfication in sugar beets;
- we present a systematic evaluation of recent CNN-based architectures on the proposed dataset to encourage further research; and,
- we highlight the distinct impact of crop development stages on the classification results.
2. Related Work
3. Deep Nutrient Deficiency for Sugar Beet (DND-SB) Dataset
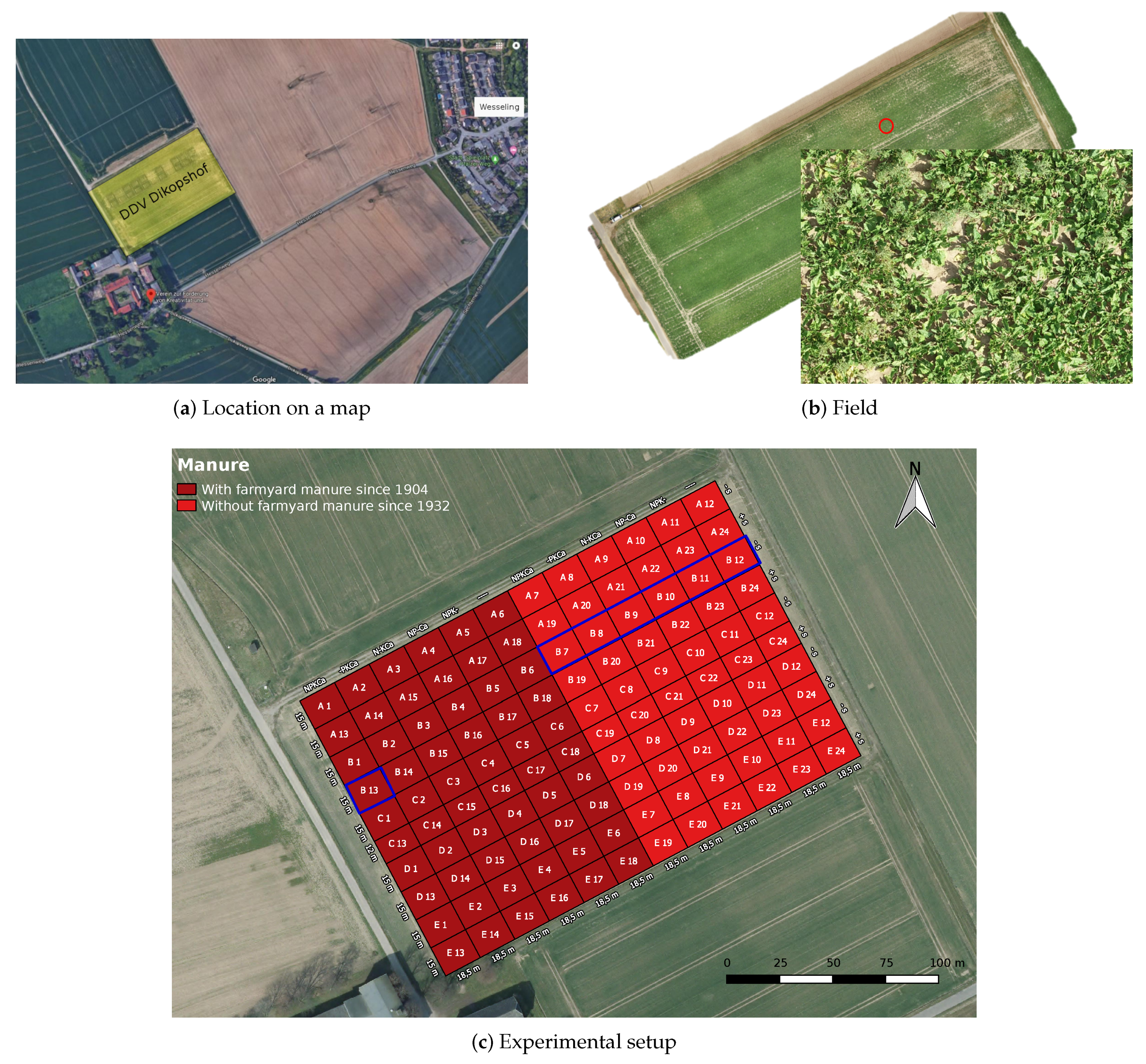
3.1. Experimental Site
3.2. Image Acquisition Protocol
4. Soil and Plant Nutrient Analyses
4.1. Soil Nutrient Analyses
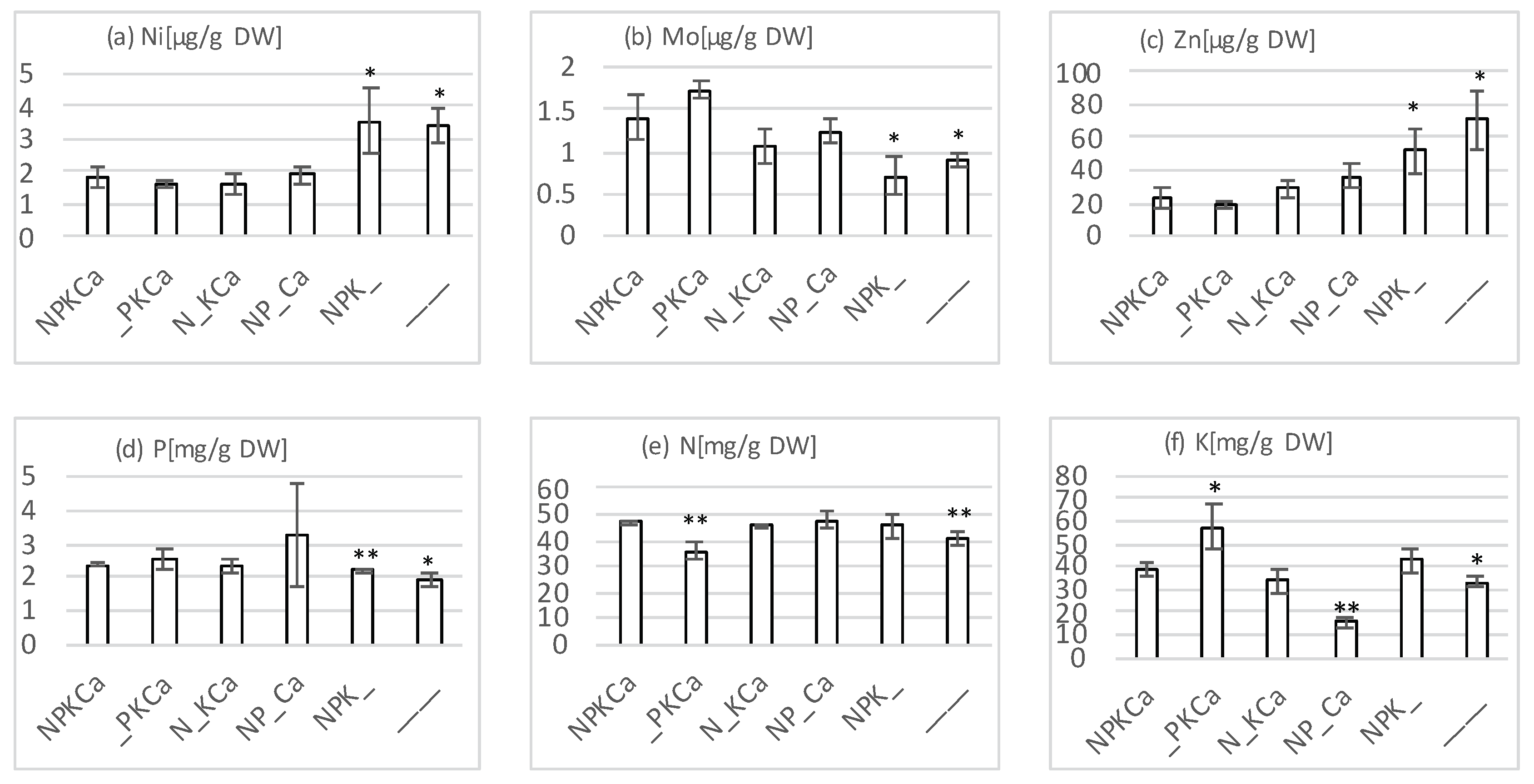
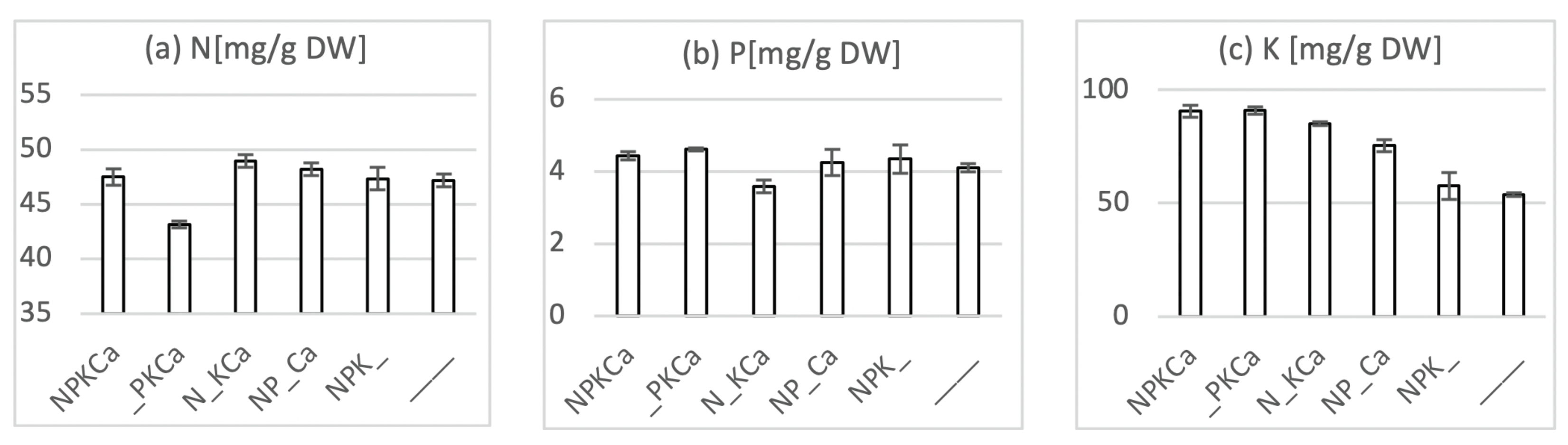
4.2. Plant Nutrient Analyses
5. Baselines
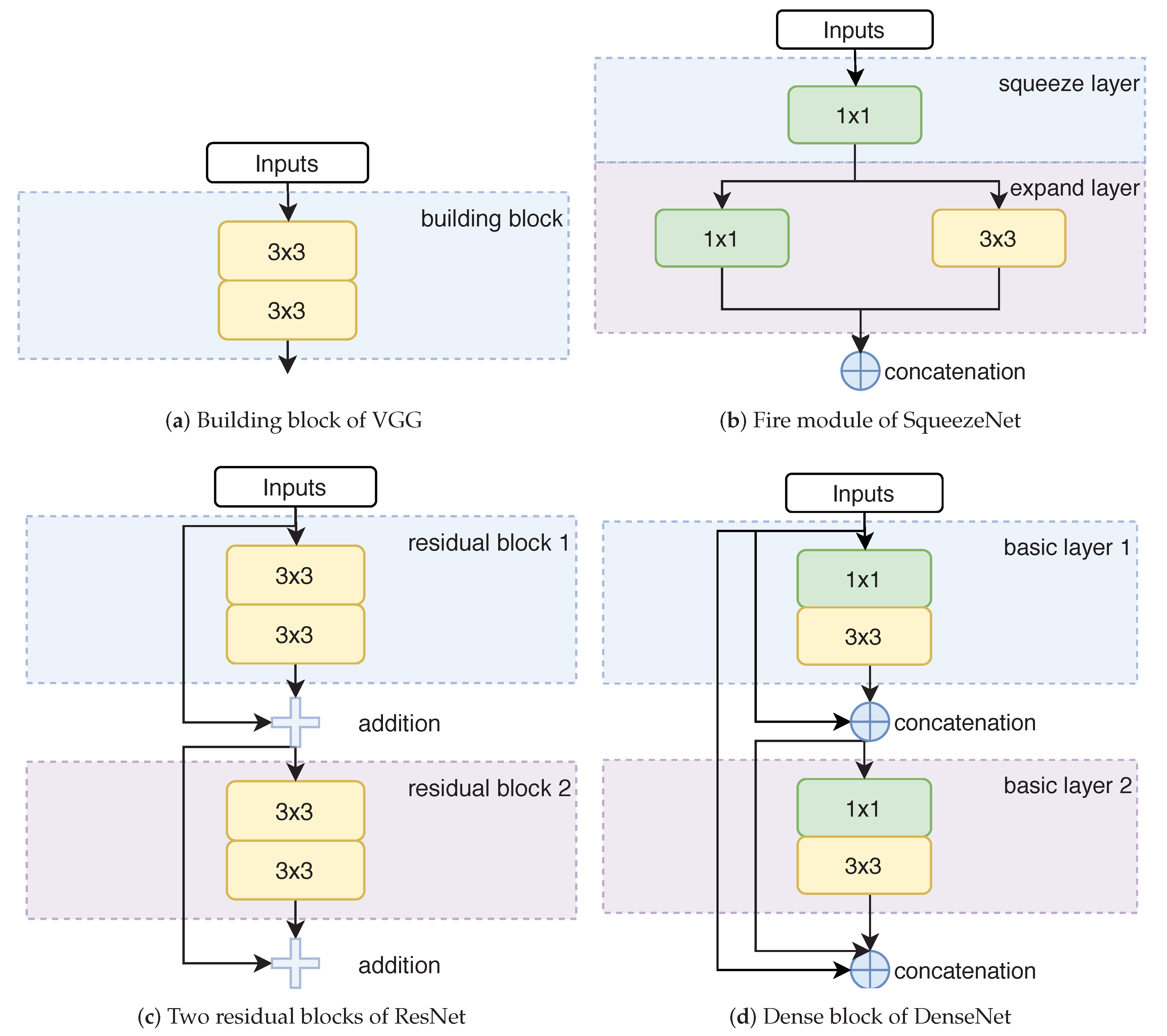
5.1. Convolutional Network Architectures
5.2. Details of Training
6. Experimental Results
6.1. Evaluation of CNNs for Nutrient Deficiency Detection
6.2. Impact of Data Augmentation
6.3. Evaluation across Crop Development Stages
6.4. Qualitative Results
7. Discussion
7.1. Dataset
7.2. Methodology
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marschner, H. Marschner’s Mineral Nutrition of Higher Plants; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Barker, A.V.; Pilbeam, D.J. Handbook of Plant Nutrition; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Vatansever, R.; Ozyigit, I.I.; Filiz, E. Essential and beneficial trace elements in plants, and their transport in roots: A review. Appl. Biochem. Biotechnol. 2017, 181, 464–482. [Google Scholar] [CrossRef]
- Adams, F. Soil Acidity and Liming; Number 631.821 S683s; Soil Science Society of America: Madison, WI, USA, 1990. [Google Scholar]
- Kennedy, I.R. Acid Soil and Acid Rain; Number Ed. 2; Research Studies Press Ltd.: Taunton, UK, 1992. [Google Scholar]
- Rengel, Z. Handbook of Soil Acidity; CRC Press: Boca Raton, FL, USA, 2003; Volume 94. [Google Scholar]
- Samborski, S.M.; Tremblay, N.; Fallon, E. Strategies to make use of plant sensors-based diagnostic information for nitrogen recommendations. Agron. J. 2009, 101, 800–816. [Google Scholar] [CrossRef]
- Padilla, F.M.; Gallardo, M.; Peña-Fleitas, M.T.; De Souza, R.; Thompson, R.B. Proximal optical sensors for nitrogen management of vegetable crops: A review. Sensors 2018, 18, 2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.; Al-Ani, A.; Eamus, D.; Tan, D.K. Leaf nitrogen determination using non-destructive techniques—A review. J. Plant Nutr. 2017, 40, 928–953. [Google Scholar] [CrossRef]
- Amaral, L.R.; Molin, J.P.; Portz, G.; Finazzi, F.B.; Cortinove, L. Comparison of crop canopy reflectance sensors used to identify sugarcane biomass and nitrogen status. Precis. Agric. 2015, 16, 15–28. [Google Scholar] [CrossRef]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front. Plant Sci. 2017, 8, 1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 6, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Chiu, M.T.; Xu, X.; Wei, Y.; Huang, Z.; Schwing, A.G.; Brunner, R.; Khachatrian, H.; Karapetyan, H.; Dozier, I.; Rose, G.; et al. Agriculture-vision: A large aerial image database for agricultural pattern analysis. arXiv 2020, arXiv:2001.01306. [Google Scholar]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallau, L.; Neumann, M.; Klatt, B.; Kleinhenz, B.; Klein, T.; Kuhn, C.; Röhrig, M.; Bauckhage, C.; Kersting, K.; Mahlein, A.K.; et al. Automated identification of sugar beet diseases using smartphones. Plant Pathol. 2018, 67, 399–410. [Google Scholar] [CrossRef]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-time blob-wise sugar beets vs weeds classification for monitoring fields using convolutional neural networks. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Suh, H.K.; Ijsselmuiden, J.; Hofstee, J.W.; van Henten, E.J. Transfer learning for the classification of sugar beet and volunteer potato under field conditions. Biosyst. Eng. 2018, 174, 50–65. [Google Scholar] [CrossRef]
- Rueda-Ayala, V.; Ahrends, H.E.; Siebert, S.; Gaiser, T.; Hüging, H.; Ewert, F. Impact of nutrient supply on the expression of genetic improvements of cereals and row crops—A case study using data from a long-term fertilization experiment in Germany. Eur. J. Agron. 2018, 96, 34–46. [Google Scholar] [CrossRef]
- Olsen, A.; Konovalov, D.A.; Philippa, B.; Ridd, P.; Wood, J.C.; Johns, J.; Banks, W.; Girgenti, B.; Kenny, O.; Whinney, J. DeepWeeds: A multiclass weed species image dataset for deep learning. Sci. Rep. 2019, 9, 2058. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Ramcharan, A.; McCloskey, P.; Baranowski, K.; Mbilinyi, N.; Mrisho, L.; Ndalahwa, M.; Legg, J.; Hughes, D.P. A mobile-based deep learning model for cassava disease diagnosis. Front. Plant Sci. 2019, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Han, K.A.M.; Watchareeruetai, U. Classification of nutrient deficiency in black gram using deep convolutional neural networks. In Proceedings of the International Joint Conference on Computer Science and Software Engineering, Chonburi, Thailand, 10–12 July 2019; pp. 277–282. [Google Scholar]
- Tran, T.T.; Choi, J.W.; Le, T.T.H.; Kim, J.W. A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, A.; Hills, F.J. Sugar Beet Nutrient Deficiency Symptoms: A Color Atlas And Chemical Guide; UC Press: Berkeley, CA, USA, 1969. [Google Scholar]
- Holthusen, D.; Jänicke, M.; Peth, S.; Horn, R. Physical properties of a Luvisol for different long-term fertilization treatments: I. Mesoscale capacity and intensity parameters. J. Plant Nutr. Soil Sci. 2012, 175, 4–13. [Google Scholar] [CrossRef]
- Heinrichs, H.; Brumsack, H.J.; Loftfield, N.; König, N. Verbessertes Druckaufschlußsystem für biologische und anorganische Materialien. Zeitschrift Für Pflanzenernährung Und Bodenkd. 1986, 149, 350–353. [Google Scholar] [CrossRef]
- Singh, D.K.; Sale, P.W. Phosphorus supply and the growth of frequently defoliated white clover (Trifolium repens L.) in dry soil. Plant Soil 1998, 205, 155–162. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Huh, M.; Agrawal, P.; Efros, A.A. What makes ImageNet good for transfer learning? arXiv 2016, arXiv:1608.08614. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- He, K.; Girshick, R.; Dollár, P. Rethinking imagenet pre-training. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 4918–4927. [Google Scholar]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Grossberger, L. UMAP: Uniform manifold approximation and projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Maaten, L.V.D.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Panareda Busto, P.; Iqbal, A.; Gall, J. Open set domain adaptation for image and action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 413–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itti, L.; Koch, C.; Niebur, E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1254–1259. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.M.; Mitra, N.J.; Huang, X.; Torr, P.H.; Hu, S.M. Global contrast based salient region detection. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 37, 569–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Lai, Q.; Fu, H.; Shen, J.; Ling, H.; Yang, R. Salient object detection in the deep learning era: An in-depth survey. arXiv 2019, arXiv:1904.09146. [Google Scholar]
- Geirhos, R.; Rubisch, P.; Michaelis, C.; Bethge, M.; Wichmann, F.A.; Brendel, W. ImageNet-trained CNNs are biased towards texture; increasing shape bias improves accuracy and robustness. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]


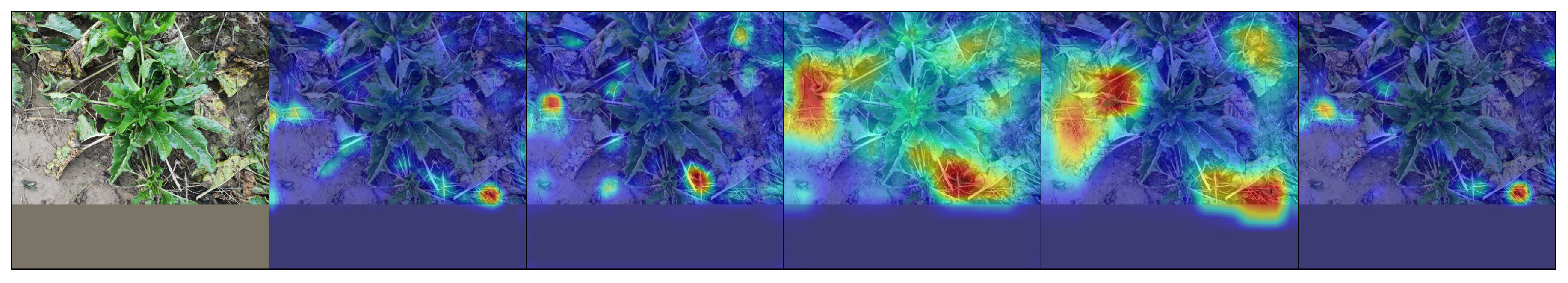
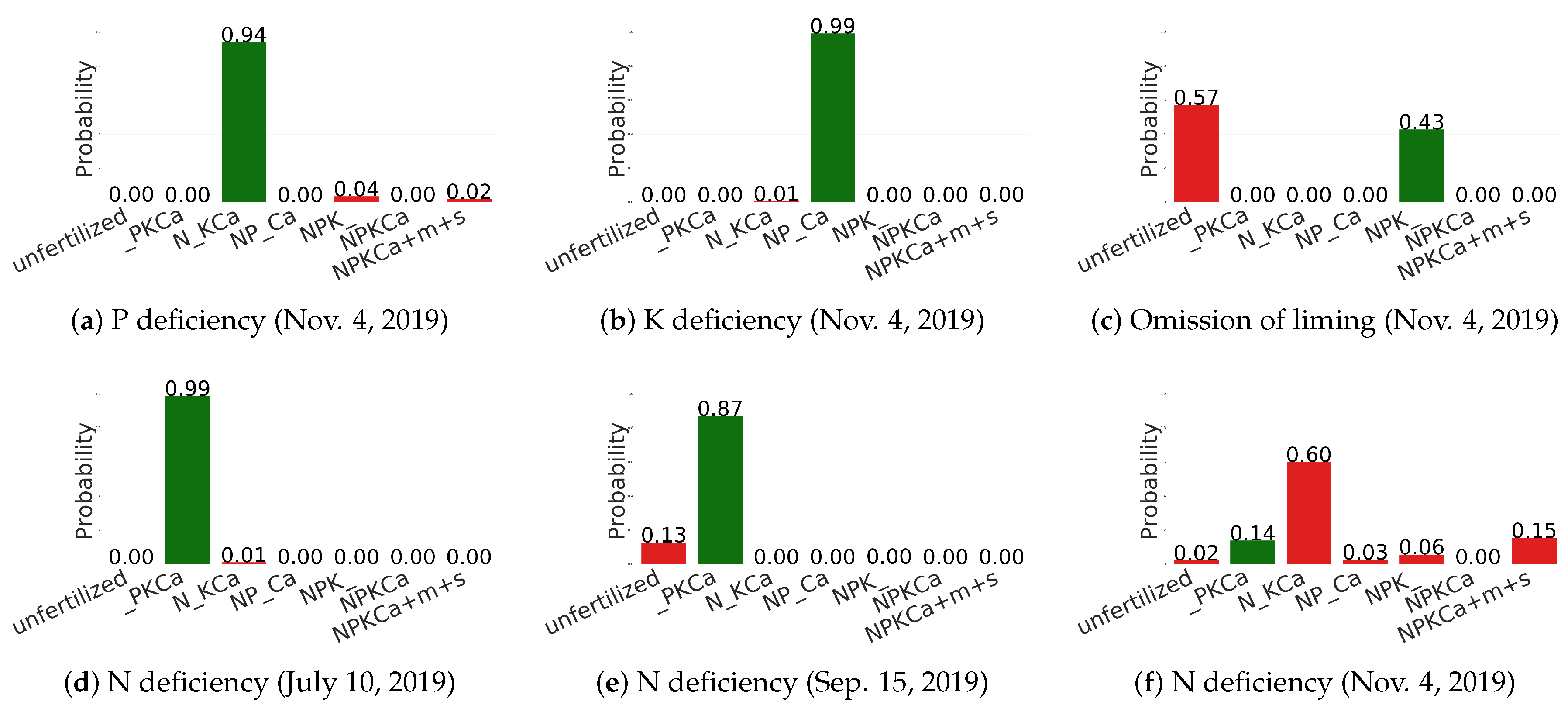
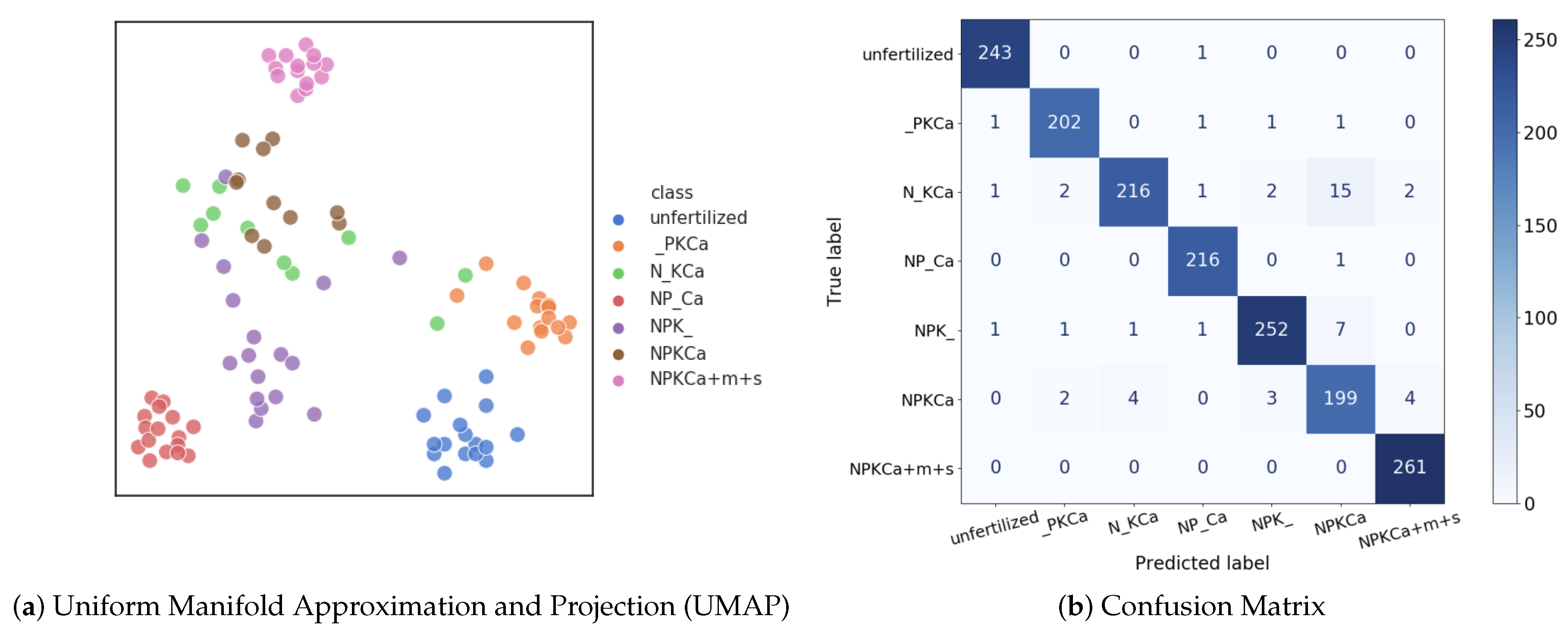
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class/Date | 07/10 | 07/19 | 07/30 | 08/04 | 08/15 | 08/26 | 09/03 | 09/15 | 10/03 | 11/04 | Total |
| unfertilized | 85 | 56 | 50 | 111 | 97 | 119 | 59 | 70 | 145 | 76 | 868 |
| _PKCa | 86 | 56 | / | 105 | 64 | 111 | 72 | 69 | 55 | 90 | 708 |
| N_KCa | 63 | 61 | 66 | 116 | 89 | 124 | 49 | 71 | 79 | 90 | 808 |
| NP_Ca | 68 | 64 | 63 | 118 | 79 | 108 | 55 | 62 | 83 | 94 | 794 |
| NPK_ | 79 | 61 | 57 | 119 | 100 | 105 | 52 | 67 | 153 | 100 | 893 |
| NPKCa | 58 | 53 | / | 118 | 71 | 117 | 79 | 90 | / | 90 | 676 |
| NPKCa+m+s | 75 | 54 | / | 128 | 86 | 131 | 77 | 86 | 172 | 92 | 901 |
| total | 514 | 405 | 236 | 815 | 586 | 815 | 443 | 515 | 687 | 632 | 5648 |
| Content | Sampling Date | Unfertilized | _PKCa | N_KCa | NP_Ca | NPK_ | NPKCa |
|---|---|---|---|---|---|---|---|
| Mineral N | 05/16 | 13.5 | 10.1 | 76.5 | 74.0 | 109.5 | 22.8 |
| 06/13 | 20.7 | 5.2 | 18.8 | 26.6 | 36.2 | 9.4 | |
| 07/10 | 6.8 | 3.1 | 2.7 | 3.4 | 18.2 | 6.8 | |
| 09/10 | 3.2 | 1.8 | 4.0 | 8.1 | 3.6 | 4.1 | |
| 05/16 | 18 | 168 | 30 | 90 | 57 | 142 | |
| 06/13 | 35 | 182 | 36 | 88 | 67 | 158 | |
| 07/10 | 47 | 166 | 34 | 61 | 82 | 84 | |
| 09/10 | 18 | 147 | 28 | 91 | 65 | 147 | |
| 05/16 | 108 | 228 | 226 | 121 | 162 | 219 | |
| 06/13 | 126 | 243 | 198 | 100 | 144 | 211 | |
| 07/10 | 122 | 239 | 216 | 117 | 137 | 275 | |
| 09/10 | 76 | 320 | 285 | 84 | 117 | 462 | |
| pH value | 05/16 | 5.8 | 6.9 | 6.4 | 6.6 | 5.5 | 6.5 |
| 06/13 | 5.6 | 7.0 | 6.7 | 6.8 | 5.8 | 6.8 | |
| 07/10 | 5.5 | 6.9 | 6.4 | 6.8 | 5.8 | 6.3 | |
| 09/10 | 5.7 | 6.8 | 6.7 | 6.7 | 5.5 | 6.6 |
| Model | Pre-Trained | From Scratch | Test Time (s/image) | Parameters (M) | ||
|---|---|---|---|---|---|---|
| Accuracy (%) | Training Time (min) | Accuracy (%) | Training Time (min) | |||
| random | 14.3 | - | 14.3 | - | - | - |
| AlexNet | 87.4 | 27 | 62.4 | 41 | 0.009 | 57 |
| VGG-16 | 89.7 | 105 | 66.8 | 466 | 0.046 | 134 |
| ResNet-101 | 95.0 | 103 | 80.3 | 293 | 0.050 | 43 |
| DenseNet-161 | 98.4 | 129 | 90.7 | 240 | 0.043 | 27 |
| SqueezeNet | 89.3 | 14 | 65.4 | 20 | 0.011 | 0.7 |
| Model | Resize (R) | Crop (C) | Accuracy (%) | Resize (R) + Padding | Accuracy (%) |
|---|---|---|---|---|---|
| DenseNet-161 | 256 | 224 | 82.4 | 224 | 86.1 |
| DenseNet-161 | 480 | 224 | 64.5 | ||
| DenseNet-161 | 592 | 224 | 61.0 | ||
| DenseNet-161 | 416 | 384 | 90.1 | 384 | 91.5 |
| DenseNet-161 | 624 | 592 | 93.2 | 592 | 95.8 |
| DenseNet-161 | 832 | 800 | 89.1 | 800 | 76.4 |
| Model | Flipping | Rotation | Color Jitter | Noise | Accuracy (%) |
|---|---|---|---|---|---|
| DenseNet-161 | - | - | - | - | 95.8 |
| DenseNet-161 | √ | - | - | - | 96.5 |
| DenseNet-161 | - | √ | - | - | 98.3 |
| DenseNet-161 | - | - | √ | - | 94.9 |
| DenseNet-161 | - | - | - | √ | 94.5 |
| DenseNet-161 | √ | √ | - | - | 98.4 |
| DenseNet-161 | √ | √ | √ | - | 97.5 |
| DenseNet-161 | √ | √ | √ | √ | 97.2 |
| Training Set | Test Set | #training:#Test | Accuracy (%) | #Training:#Test | Accuracy (%) |
|---|---|---|---|---|---|
| (Random) | (Random) | ||||
| w/o 1 | 1 | 5134:514 (10:1) | 74.7 | 5134:514 (10:1) | 98.4 |
| w/o 2 | 2 | 5243:405 (13:1) | 84.4 | 5243:405 (13:1) | 98.5 |
| w/o 3 | 3 | 5412:236 (23:1) | 80.9 | 5412:236 (23:1) | 98.5 |
| w/o 4 | 4 | 4833:815 (6:1) | 78.4 | 4833:815 (6:1) | 97.9 |
| w/o 5 | 5 | 5062:586 (9:1) | 80.6 | 5062:586 (9:1) | 97.6 |
| w/o 6 | 6 | 4833:815 (6:1) | 76.8 | 4833:815 (6:1) | 97.9 |
| w/o 7 | 7 | 5205:443 (12:1) | 78.3 | 5205:443 (12:1) | 98.6 |
| w/o 8 | 8 | 5133:515 (10:1) | 78.8 | 5133:515 (10:1) | 98.4 |
| w/o 9 | 9 | 4961:687 (7:1) | 80.1 | 4961:687 (7:1) | 98.3 |
| w/o 10 | 10 | 5016:632 (8:1) | 48.5 | 5016:632 (8:1) | 98.1 |
| Training Set | Test Set | #Training:#Test | Accuracy (%) | #Training:#Test | Accuracy (%) |
|---|---|---|---|---|---|
| (Random) | (Random) | ||||
| 1-9 | 10 | 5016:632 (8:1) | 48.5 | 5016:632 (8:1) | 98.1 |
| 1-8 | 9-10 | 4329:1319 (3:1) | 54.0 | 4329:1319 (3:1) | 95.9 |
| 1-7 | 8-10 | 3814:1834 (2:1) | 61.6 | 3814:1834 (2:1) | 96.8 |
| 1-6 | 7-10 | 3371:2277 (1.5:1) | 75.4 | 3371:2277 (1.5:1) | 95.0 |
| 1-5 | 6-10 | 2556:3092 (0.8:1) | 47.5 | 2556:3092 (0.8:1) | 93.0 |
| 1-4 | 5-10 | 1970:3678 (0.5:1) | 43.6 | 1970:3678 (0.5:1) | 91.5 |
| 1-3 | 4-10 | 1155:4493 (0.26:1) | 44.2 | 1155:4493 (0.26:1) | 87.2 |
| 1-2 | 3-10 | 919:4729 (0.2:1) | 41.5 | 919:4729 (0.2:1) | 84.6 |
| 1 | 2-10 | 514:5134 (0.1:1) | 31.8 | 514:5134 (0.1:1) | 79.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, J.; Krusenbaum, L.; Unger, P.; Hüging, H.; Seidel, S.J.; Schaaf, G.; Gall, J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors 2020, 20, 5893. https://doi.org/10.3390/s20205893
Yi J, Krusenbaum L, Unger P, Hüging H, Seidel SJ, Schaaf G, Gall J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors. 2020; 20(20):5893. https://doi.org/10.3390/s20205893
Chicago/Turabian StyleYi, Jinhui, Lukas Krusenbaum, Paula Unger, Hubert Hüging, Sabine J. Seidel, Gabriel Schaaf, and Juergen Gall. 2020. "Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images" Sensors 20, no. 20: 5893. https://doi.org/10.3390/s20205893
APA StyleYi, J., Krusenbaum, L., Unger, P., Hüging, H., Seidel, S. J., Schaaf, G., & Gall, J. (2020). Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors, 20(20), 5893. https://doi.org/10.3390/s20205893






