Stress Distribution Analysis on Hyperspectral Corn Leaf Images for Improved Phenotyping Quality
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Handheld Hyperspectral Device and Plant Sampling
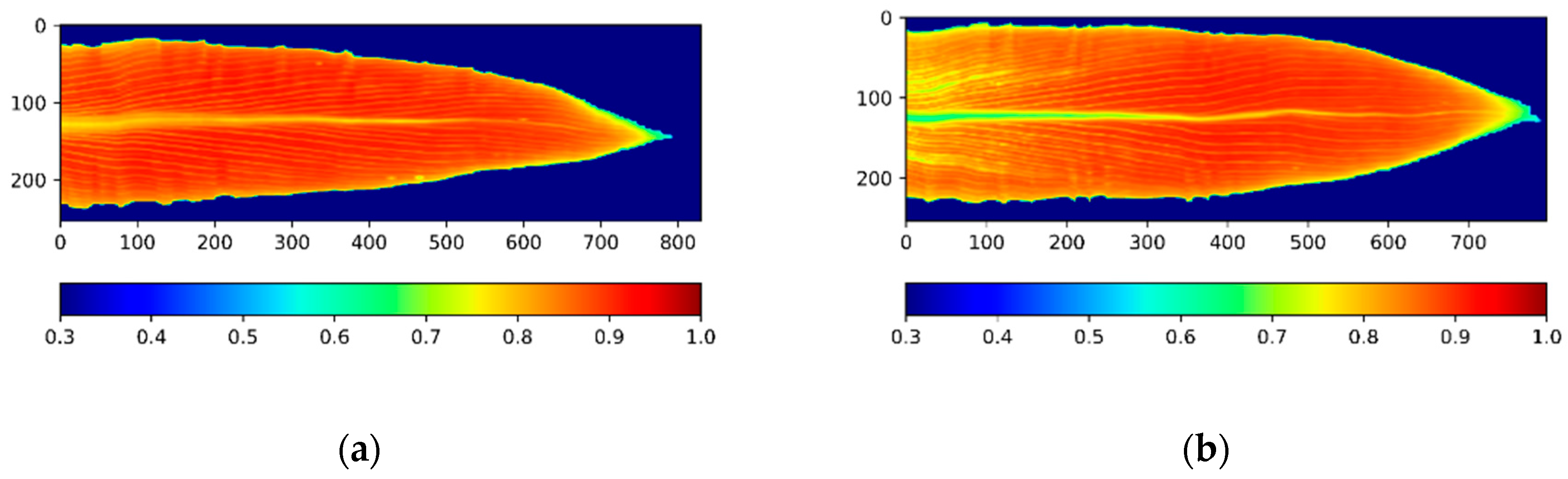
2.3. Image Processing and Segmentation
2.4. NDVI Calculation
2.5. Distribution Feature Estimation and Quantification
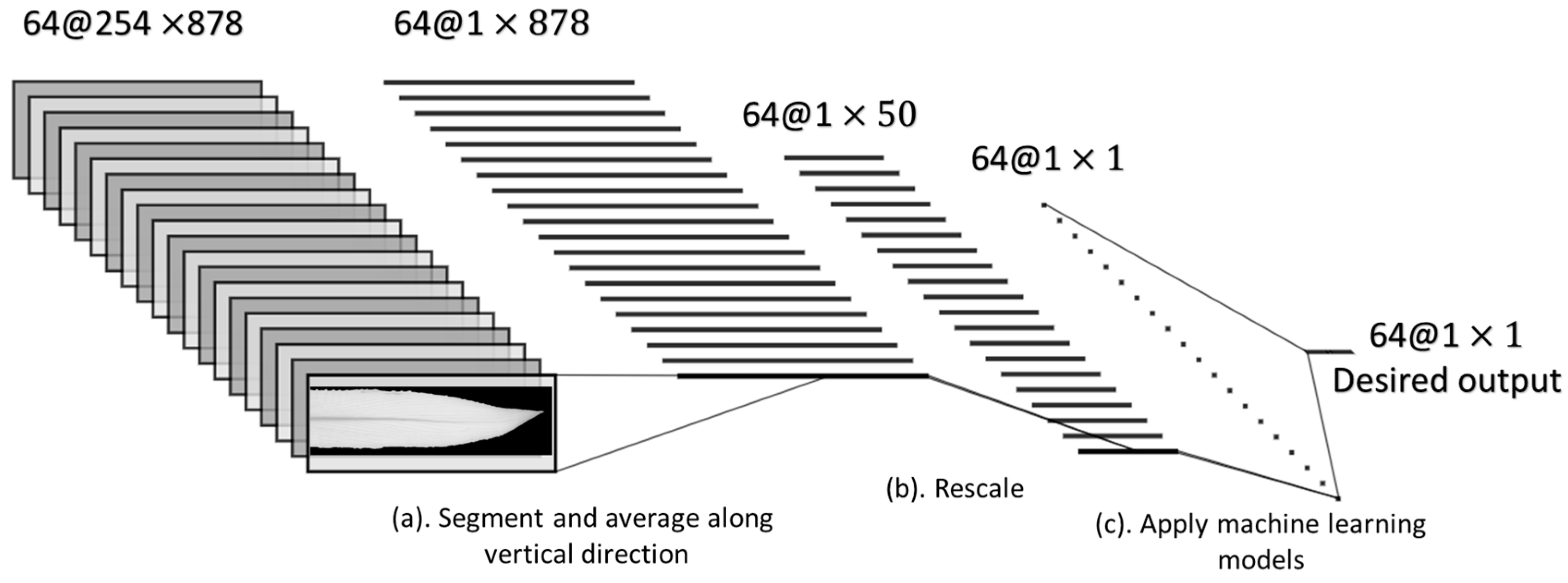
2.5.1. Preprocessing of NDVI Images
2.5.2. Application of Machine Learning Algorithms
2.5.3. Hyperparameter Optimization
2.5.4. Metrices for Model Evaluation
2.6. Software and Computation
3. Results
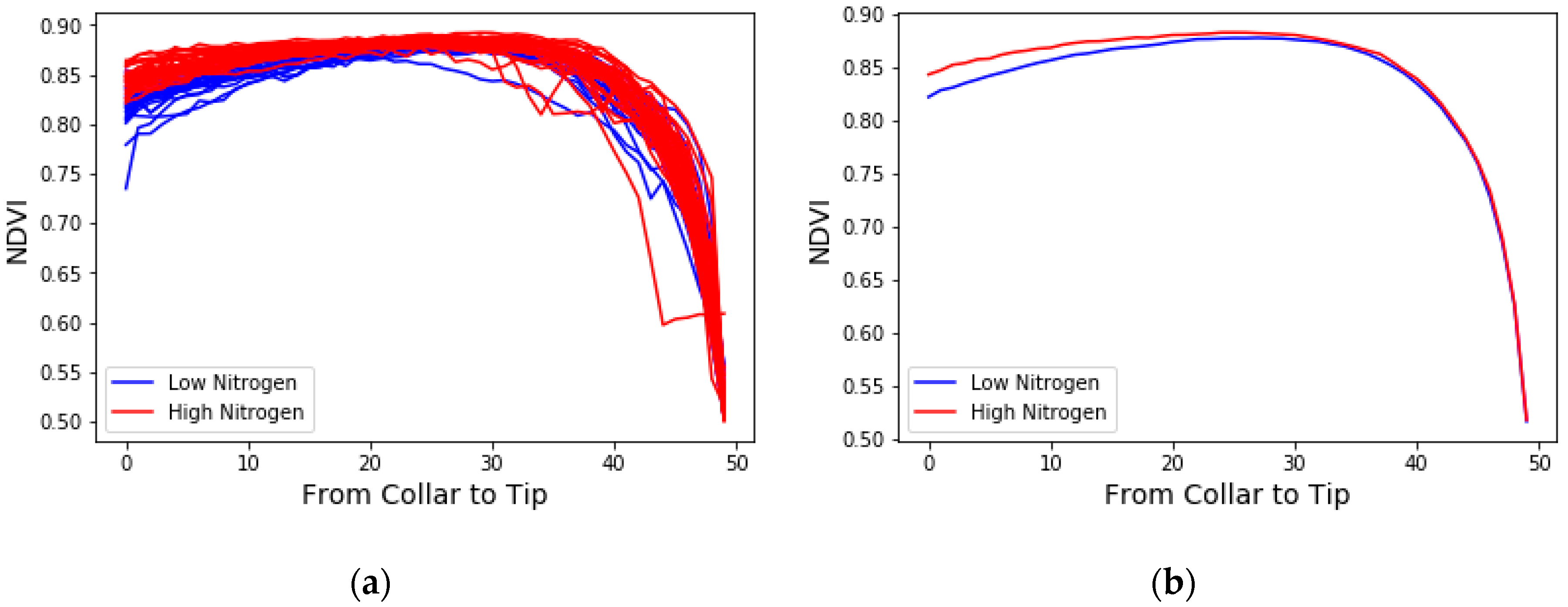
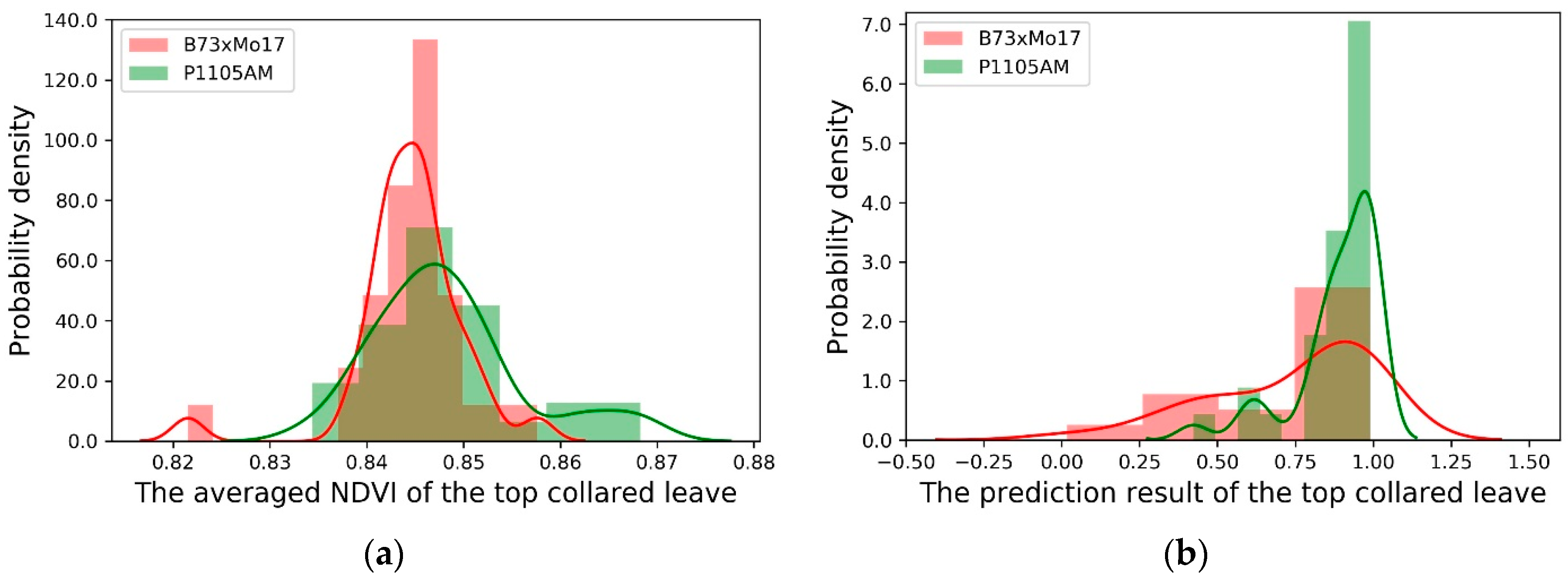
3.1. The Averaged NDVI Comparison
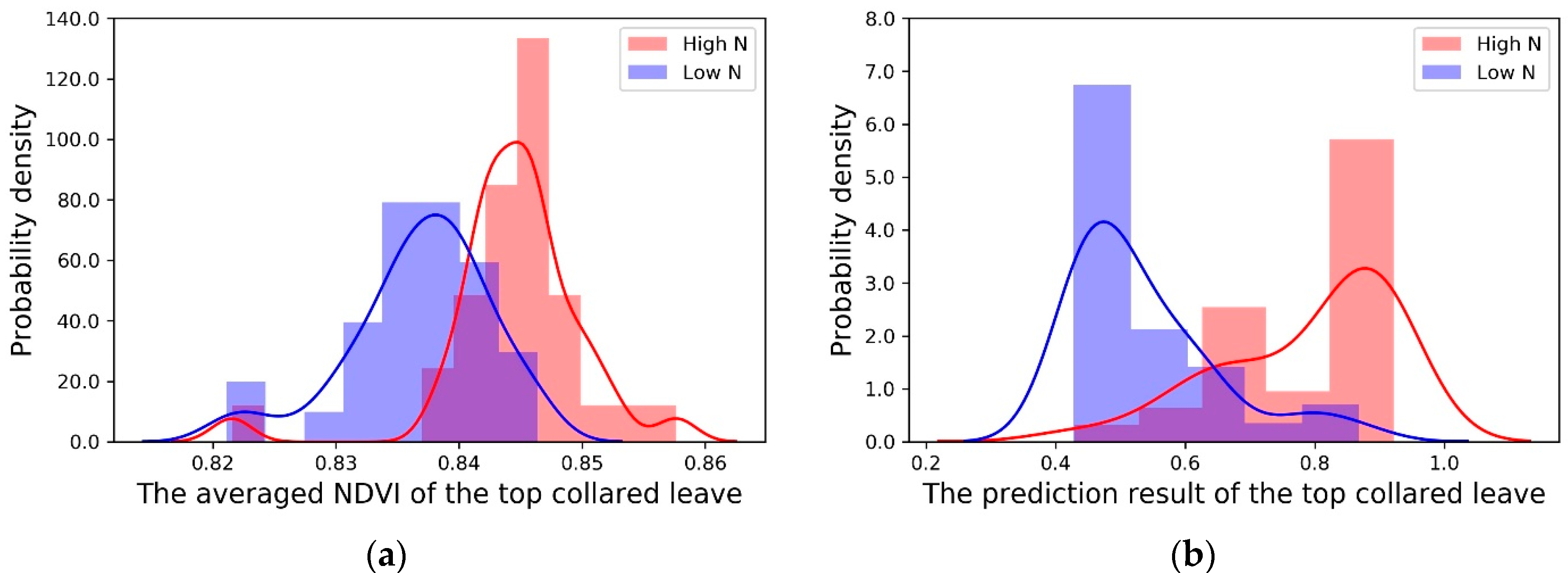
3.2. Results of Machine Learning Algorithms
3.3. Model Evaluation for Different Corn Genotypes
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dekker, A.; Phinn, S.; Roelfsema, C.; Anstee, J.; Brando, V. Mapping seagrass species, cover and biomass in shallow waters: An assessment of satellite multi-spectral and airborne hyper-spectral imaging systems in Moreton Bay (Australia). Remote Sens. Environ. 2008, 112, 3413–3425. [Google Scholar] [CrossRef]
- Prasad, A.K.; Chai, L.; Singh, R.P.; Kafatos, M. Crop yield estimation model for Iowa using remote sensing and surface parameters. Int. J. Appl. Earth Obs. Geoinf. 2006, 8, 26–33. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. Data mining and NIR spectroscopy in viticulture: Applications for plant phenotyping under field conditions. Sensors 2016, 16, 236. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Rehman, T.U.; Mahmud, M.S.; Chang, Y.K.; Jin, J.; Shin, J. Current and future applications of statistical machine learning algorithms for agricultural machine vision systems. Comput. Electron. Agric. 2019, 156, 585–605. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Hyperspectral band depth analysis for a better estimation of grass biomass (Cenchrus ciliaris) measured under controlled laboratory conditions. Int. J. Appl. Earth Obs. Geoinf. 2004, 5, 87–96. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Zhang, L.; Deng, M.; Qin, Z.; Zhao, S.; Lin, H. Estimation of crop LAI using hyperspectral vegetation indices and a hybrid inversion method. Remote Sens. Environ. 2015, 165, 123–134. [Google Scholar] [CrossRef]
- Zhang, L.; Maki, H.; Ma, D.; Sánchez-Gallego, J.A.; Mickelbart, M.V.; Wang, L.; Rehman, T.U.; Jin, J. Optimized angles of the swing hyperspectral imaging system for single corn plant. Comput. Electron. Agric. 2019, 156, 349–359. [Google Scholar] [CrossRef]
- Ma, D.; Carpenter, N.; Maki, H.; Rehman, T.U.; Tuinstra, M.R.; Jin, J. Greenhouse environment modeling and simulation for microclimate control. Comput. Electron. Agric. 2019, 162, 134–142. [Google Scholar] [CrossRef]
- Ma, D.; Carpenter, N.; Amatya, S.; Maki, H.; Wang, L.; Zhang, L.; Neeno, S.; Tuinstra, M.R.; Jin, J. Removal of greenhouse microclimate heterogeneity with conveyor system for indoor phenotyping. Comput. Electron. Agric. 2019, 166, 104979. [Google Scholar] [CrossRef]
- Hu, J.; Li, C.; Wen, Y.; Gao, X.; Shi, F.; Han, L. Spatial distribution of SPAD value and determination of the suitable leaf for N diagnosis in cucumber. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Chongqing, China, 25–26 November 2017; p. 022001. [Google Scholar] [CrossRef]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-ul-karim, S.T.; Tian, Y.; Zhu, Y. Optimal Leaf Positions for SPAD Meter Measurement in Rice. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Ciganda, V.; Gitelson, A.; Schepers, J. Vertical profile and temporal variation of chlorophyll in maize canopy: Quantitative “crop vigor” indicator by means of reflectance-based techniques. Agron. J. 2008, 100, 1409–1417. [Google Scholar] [CrossRef]
- Hirzel, J. Pablo Undurraga Nutritional Management of Cereals Cropped Under Irrigation Conditions. In Proceedings of the Crop Production, London, UK, 3 July 2013; p. 13. [Google Scholar]
- Sack, L.; Streeter, C.M.; Holbrook, N.M. Hydraulic analysis of water flow through leaves of sugar maple and red oak. Plant Physiol. 2004, 134, 1824–1833. [Google Scholar] [CrossRef]
- Growth Potential Guide to Nutrient. Available online: https://www.pioneer.com/CMRoot/International/Australia_Intl/Publications/Corn_Workshop_Book.pdf (accessed on 9 June 2020).
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant Phenomics, From Sensors to Knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Víg, R.; Huzsvai, L.; Dobos, A.; Nagy, J. Systematic measurement methods for the determination of the SPAD values of maize (Zea mays L.) canopy and potato (Solanum tuberosum L.). Commun. Soil Sci. Plant Anal. 2012, 43, 1684–1693. [Google Scholar] [CrossRef]
- Debaeke, P.; Rouet, P.; Justes, E. Relationship between the normalized SPAD index and the nitrogen nutrition index: Application to durum wheat. J. Plant Nutr. 2006, 29, 75–92. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, H.H.; Wang, X. Machine learning for Big Data analytics in plants. Trends Plant Sci. 2014, 19, 798–808. [Google Scholar] [CrossRef]
- Hand, D.J. Principles of Data Mining; Springer Publishing: London, UK, 2007. [Google Scholar]
- Morellos, A.; Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.; Tziotzios, G.; Wiebensohn, J.; Bill, R.; Mouazen, A.M. Machine learning based prediction of soil total nitrogen, organic carbon and moisture content by using VIS-NIR spectroscopy. Biosyst. Eng. 2016, 152, 104–116. [Google Scholar] [CrossRef]
- Maresma, Á.; Ariza, M.; Martínez, E.; Lloveras, J.; Martínez-Casasnovas, J.A. Analysis of vegetation indices to determine nitrogen application and yield prediction in maize (zea mays l.) from a standard uav service. Remote Sens. 2016, 8, 973. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.L.; Mouazen, A.M. Wheat yield prediction using machine learning and advanced sensing techniques. Comput. Electron. Agric. 2016, 121, 57–65. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine learning in agriculture: A review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Polder, G.; Blok, P.M.; De Villiers, H.A.C.; van der Wolf, J.M.; Kamp, J. Potato virus Y detection in seed potatoes using deep learning on hyperspectral images. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Rosa, A.T.; Ruiz Diaz, D.A. Fertilizer Placement and Tillage Interaction in Corn and Soybean Production. Kansas Agric. Exp. Stn. Res. Rep. 2015, 1. [Google Scholar] [CrossRef]
- Wang, L.; Jin, J.; Song, Z.; Wang, J.; Zhang, L.; Rehman, T.U.; Ma, D.; Carpenter, N.R.; Tuinstra, M.R. LeafSpec: An accurate and portable hyperspectral corn leaf imager. Comput. Electron. Agric. 2020, 169, 105209. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Molero, G.; Stellacci, A.; Bort, J.; Nogués, S.; Araus, J. NDVI as a potential tool for predicting biomass, plant nitrogen content and growth in wheat genotypes subjected to different water and nitrogen conditions. Cereal Res. Commun. 2011, 39, 147–159. [Google Scholar] [CrossRef]
- Wang, L.; Duan, Y.; Zhang, L.; Rehman, T.U.; Ma, D.; Jin, J. Precise Estimation of NDVI with a Simple NIR Sensitive RGB Camera and Machine Learning Methods for Corn Plants. Sensors 2020, 20, 3208. [Google Scholar] [CrossRef] [PubMed]
- Schafleitner, R.; Gutierrez, R.; Espino, R.; Gaudin, A.; Pérez, J.; Martínez, M.; Domínguez, A.; Tincopa, L.; Alvarado, C.; Numberto, G.; et al. Field screening for variation of drought tolerance in Solanum tuberosum L. by agronomical, physiological and genetic analysis. Potato Res. 2007, 50, 71–85. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; De Colstoun, E.B.; McMurtrey Iii, J.E. Estimating corn leaf chlorophyll concentration from leaf and canopy reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Wang, J.; Song, Z.; Rehman, T.U.; Bureetes, T.; Ma, D.; Chen, Z.; Neeno, S.; Jin, J. Leaf Scanner: A portable and low-cost multispectral corn leaf scanning device for precise phenotyping. Comput. Electron. Agric. 2019, 167, 105069. [Google Scholar] [CrossRef]
- Klikauer, T. Scikit-learn: Machine Learning in Python. TripleC 2016, 14, 260–264. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Press, S.J.; Wilson, S.; Press, S.J.; Wilson, S. Choosing Between Logistic Regression and Discriminant Analysis. J. Am. Stat. Assoc. 2007, 73, 699–705. [Google Scholar] [CrossRef]
- Höskuldsson, A. PLS regression methods. J. Chemom. 1988, 2, 211–228. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar] [CrossRef]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE)? -Arguments against avoiding RMSE in the literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; ISBN 1441412697. [Google Scholar]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Oliphant, T.E. A Guide to NumPy; Trelgol Publishing: USA, 2006. Available online: http://citebay.com/how-to-cite/numpy/ (accessed on 13 May 2019).
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics-technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhu, H.; Feng, J.; Shen, C.; Zhang, H.; Zhou, J.T. Deep Clustering With Sample-Assignment Invariance Prior. IEEE Trans. Neural Networks Learn. Syst. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Feng, J.; Xiao, S.; Yau, W.Y.; Zhou, J.T.; Yang, S. Structured autoencoders for subspace clustering. IEEE Trans. Image Process. 2018, 27, 5076–5086. [Google Scholar] [CrossRef]


| Parameters | LeafSpec |
|---|---|
| Camera model | BFLY-U3-05S2M-CS |
| Spectrograph | Customized |
| Frame rate (FPS) | 20 |
| Exposure time (ms) | 50 |
| Spectral resolution | 676 |
| Spatial resolution (pixels) | 878 |
| Spectral range (nm) | 450–900 |
| Scan speed (mm/s) | 5.08 |
| Regression Algorithm | Description | Reference |
|---|---|---|
| AdaBoost | Adaptive Boosting (AdaBoost) is a generalized boost method which is an ensemble technique that attempts to create a strong classifier from several weak classifiers. | [40] |
| Logistic Regression | Logistic Regression is a predictive analysis method usually used when the dependent variable is dichotomous (binary). | [41] |
| PLSR | Partial Least Squares Regression (PLSR) is a method that performs least squares regression on new components after reducing original predictors to a smaller set of uncorrelated components. | [42] |
| Random Forest | Random Forest is an ensemble technique performing both regression and classification tasks with the use of multiple decision trees with Bootstrap Aggregation. | [43] |
| Regression Algorithms | Nitrogen Treatment | Samples # | Mean of the Prediction Results | Standard Deviation | -Log (p-Value) |
|---|---|---|---|---|---|
| Averaged NDVI | High N | 32 | 0.845 | 0.006 | 5.845 |
| Low N | 32 | 0.837 | 0.006 | ||
| AdaBoost | High N | 32 | 0.744 | 0.267 | 7.995 |
| Low N | 32 | 0.281 | 0.284 | ||
| Logistic Regression | High N | 32 | 0.506 | 0.008 | 7.066 |
| Low N | 32 | 0.493 | 0.009 | ||
| PLSR | High N | 32 | 0.738 | 0.225 | 9.375 |
| Low N | 32 | 0.242 | 0.298 | ||
| Random Forest | High N | 32 | 0.727 | 0.263 | 9.519 |
| Low N | 32 | 0.246 | 0.242 |
| Methods | Corn Genotypes | Nitrogen Treatment | Sample # | Mean | Standard Deviation | p-Value |
|---|---|---|---|---|---|---|
| Averaged NDVI | B73xMo17 | High N | 32 | 0.845 | 0.006 | 0.035 |
| P1105AM | High N | 32 | 0.848 | 0.008 | ||
| Prediction result from Random Forest | B73xMo17 | High N | 32 | 0.727 | 0.263 | 0.004 |
| P1105AM | High N | 32 | 0.886 | 0.137 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, D.; Wang, L.; Zhang, L.; Song, Z.; U. Rehman, T.; Jin, J. Stress Distribution Analysis on Hyperspectral Corn Leaf Images for Improved Phenotyping Quality. Sensors 2020, 20, 3659. https://doi.org/10.3390/s20133659
Ma D, Wang L, Zhang L, Song Z, U. Rehman T, Jin J. Stress Distribution Analysis on Hyperspectral Corn Leaf Images for Improved Phenotyping Quality. Sensors. 2020; 20(13):3659. https://doi.org/10.3390/s20133659
Chicago/Turabian StyleMa, Dongdong, Liangju Wang, Libo Zhang, Zhihang Song, Tanzeel U. Rehman, and Jian Jin. 2020. "Stress Distribution Analysis on Hyperspectral Corn Leaf Images for Improved Phenotyping Quality" Sensors 20, no. 13: 3659. https://doi.org/10.3390/s20133659
APA StyleMa, D., Wang, L., Zhang, L., Song, Z., U. Rehman, T., & Jin, J. (2020). Stress Distribution Analysis on Hyperspectral Corn Leaf Images for Improved Phenotyping Quality. Sensors, 20(13), 3659. https://doi.org/10.3390/s20133659






