The Efficiency of Color Space Channels to Quantify Color and Color Intensity Change in Liquids, pH Strips, and Lateral Flow Assays with Smartphones
Abstract
1. Introduction
1.1. General Introduction
1.2. Hyphenating Lateral Flow and Other Colorimetric Assays with Smartphones
1.3. Workflow Reported in This Study
2. Materials and Methods
2.1. Materials
2.2. Nanoparticle Synthesis
2.3. pH System (Color Change)
2.4. Nanoparticle Suspensions and Filter Paper Preparation
2.5. Liquid Assays Preparation
2.6. Sample Preparation and Picture Capturing
2.7. Commercial Assays
2.8. Scoring System
2.9. Software and Data Treatment
3. Results and Discussion
3.1. Comparing Channel Performance on a Huawei P8
3.2. Comparing Channel Performance between Phones
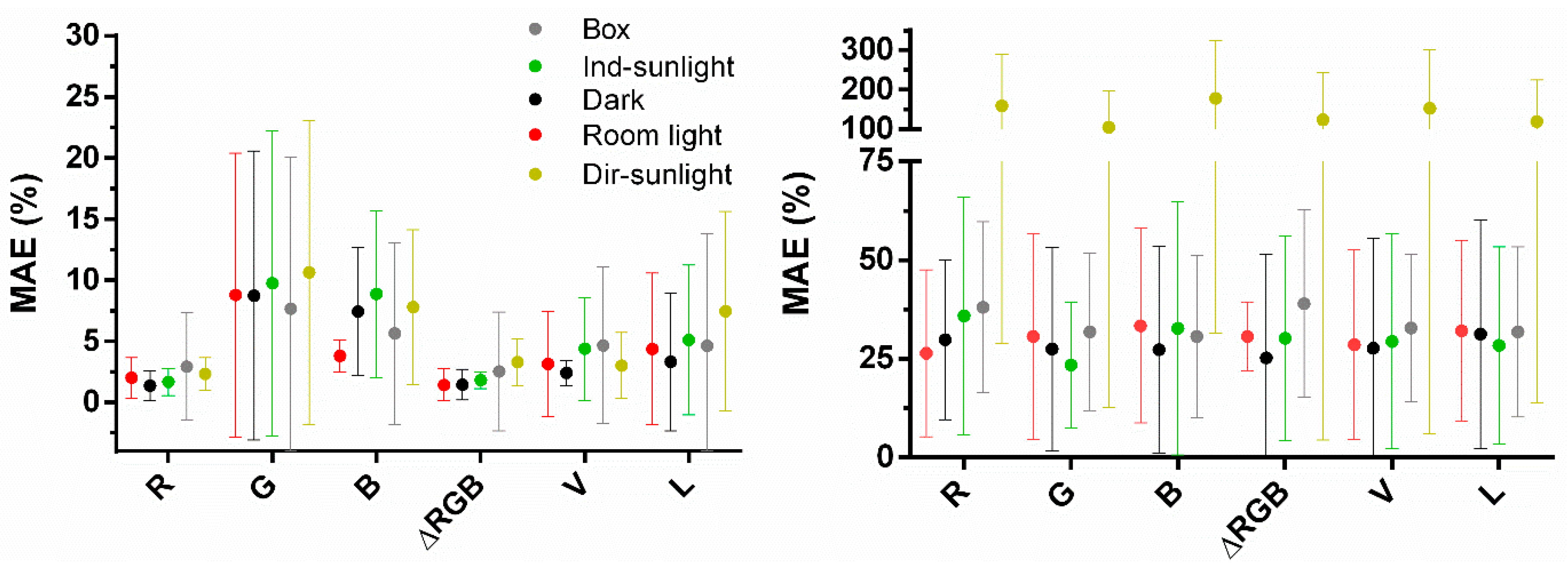
3.3. Comparing Box/No-Box Effects on Predictions in Various Background Settings
3.4. Channel Performance for Colored Liquids
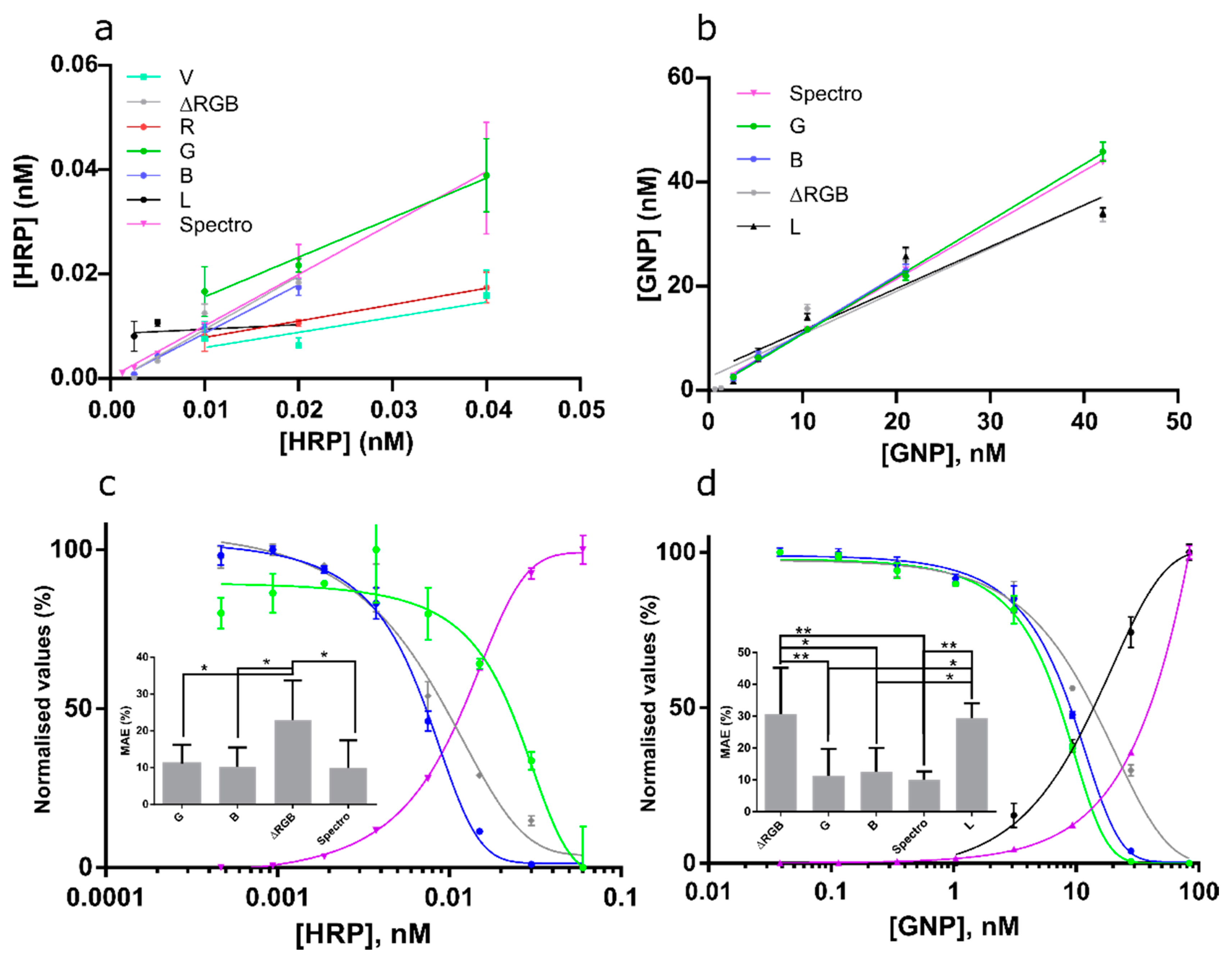
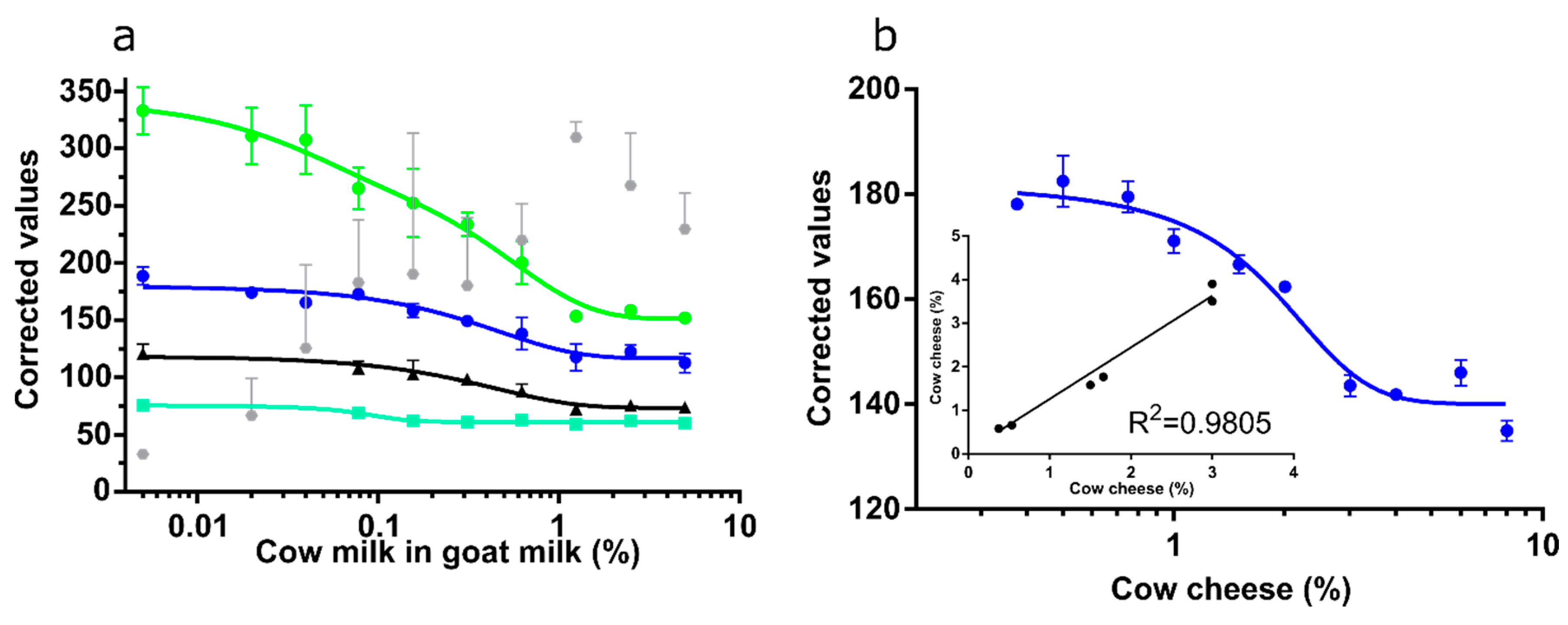
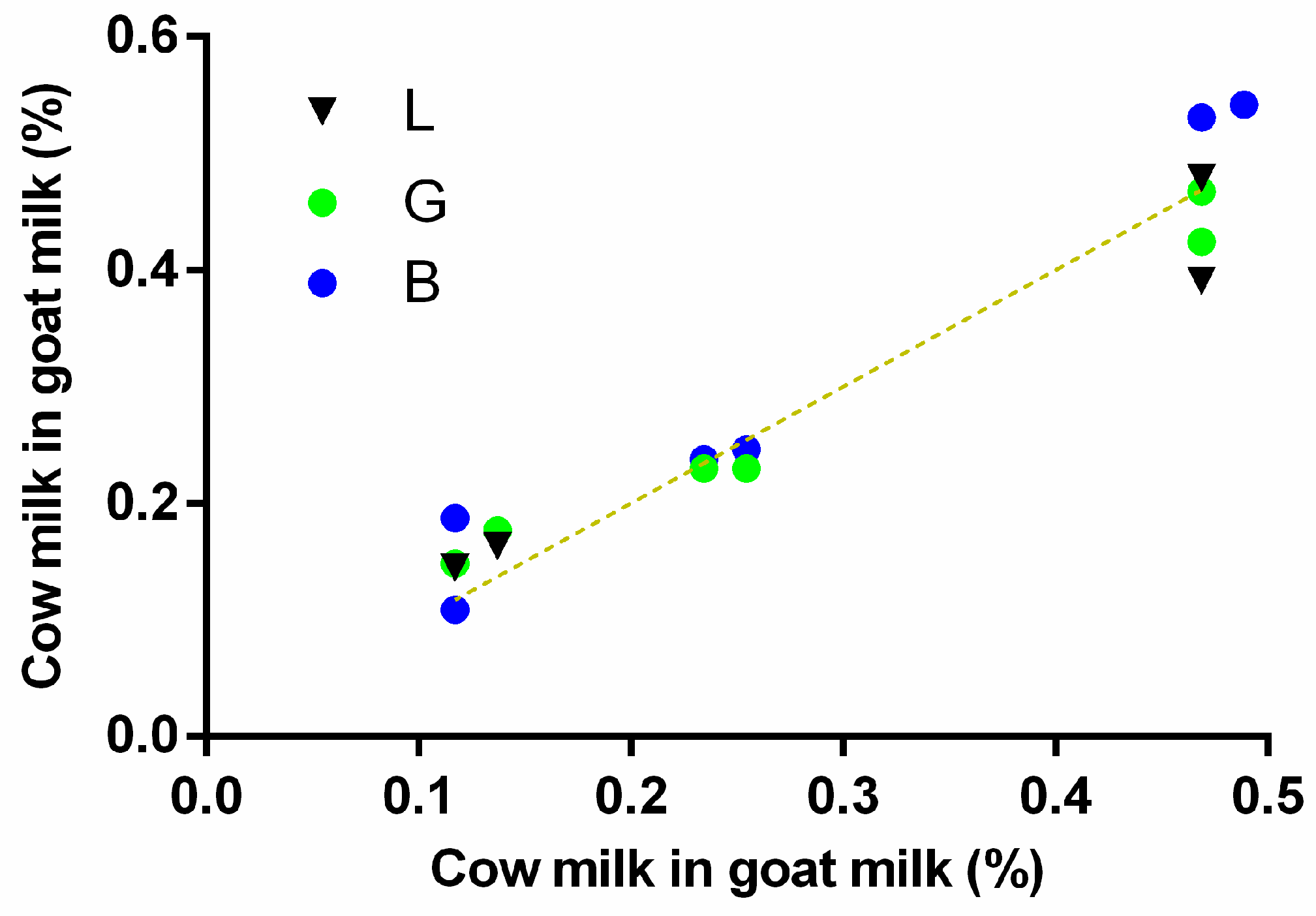
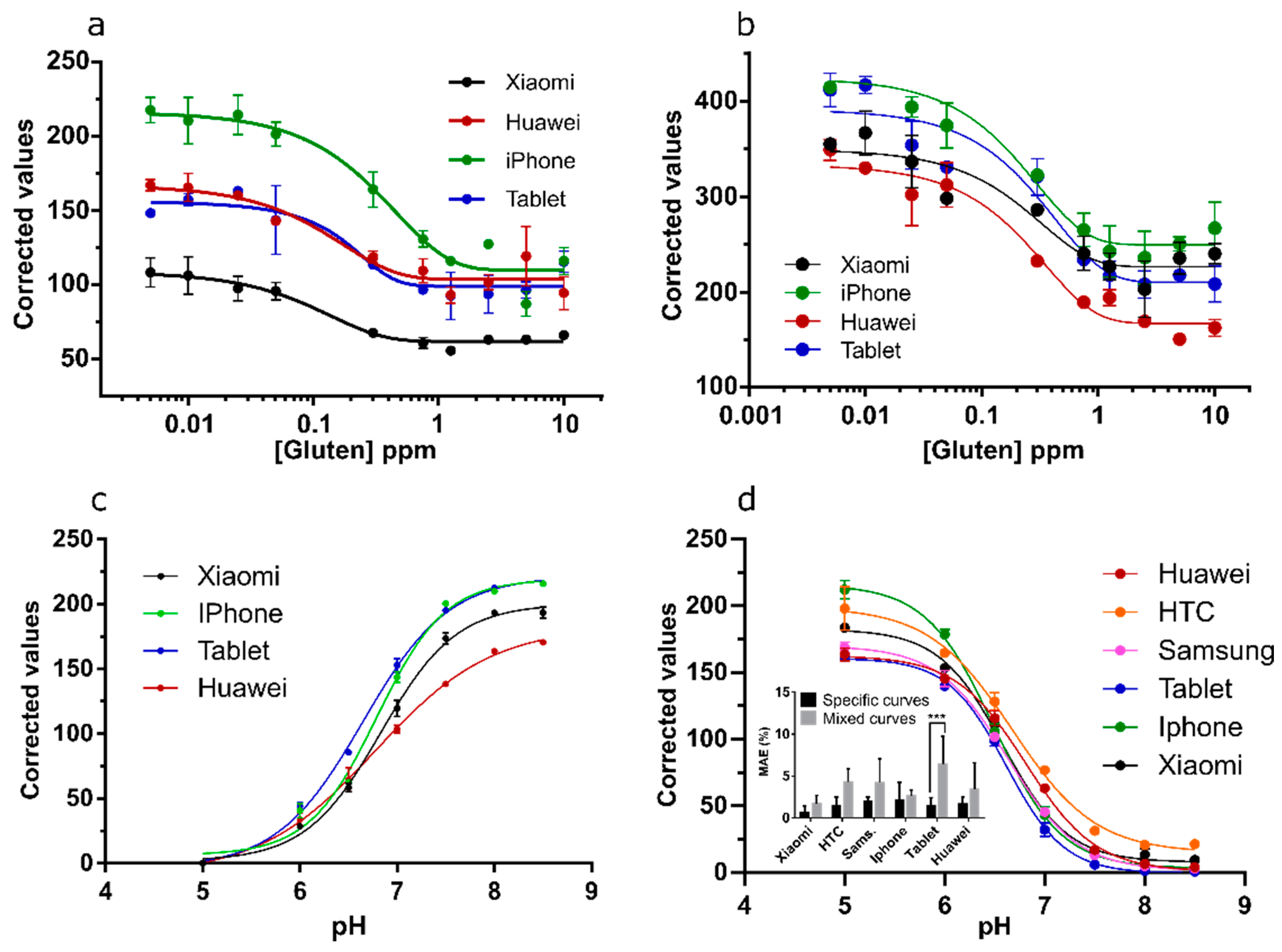
3.5. Channel Performance Comparison Using Commercial LFA for Milk Allergen Detection
3.6. Channel Performance Comparison Using Commercial LFA for Gluten and pH Strips for Soil pH Prediction
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malik, A.K.; Blasco, C.; Picó, Y. Liquid chromatography-mass spectrometry in food safety. J. Chromatogr. A 2010, 1217, 4018–4040. [Google Scholar] [CrossRef] [PubMed]
- Matumba, L.; Van Poucke, C.; Njumbe Ediage, E.; De Saeger, S. Keeping mycotoxins away from the food: Does the existence of regulations have any impact in Africa? Crit. Rev. Food Sci. Nutr. 2017, 57, 1584–1592. [Google Scholar] [CrossRef] [PubMed]
- The Rapid Alert System for Food and Feed (2017 Annual Report). Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/rasff_annual_report_2017.pdf (accessed on 20 November 2019).
- Sivadasan, S.; Efstathiou, J.; Calinescu, A.; Huatuco, L.H. Advances on measuring the operational complexity of supplier-customer systems. Eur. J. Oper. Res. 2006, 171, 208–226. [Google Scholar] [CrossRef]
- Knowles, T.; Moody, R.; McEachern, M. European food scares and their impact on EU food policy. Br. Food J. 2007, 109, 43–67. [Google Scholar] [CrossRef]
- Ludwig, S.K.; Tokarski, C.; Lang, S.N.; van Ginkel, L.A.; Zhu, H. Calling Biomarkers in Milk Using a Protein Microarray on Your Smartphone. PLoS One 2015, 10, e0134360. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.K.J.; Zhu, H.; Phillips, S.; Shiledar, A.; Feng, S.; Tseng, D.; van Ginkel, L.A.; Nielen, M.W.F.; Ozcan, A. Cellphone-based detection platform for rbST biomarker analysis in milk extracts using a microsphere fluorescence immunoassay. Anal. Bioanal. Chem. 2014, 406, 6857–6866. [Google Scholar] [CrossRef]
- Zeinhom, M.M.A.; Wang, Y.; Song, Y.; Zhu, M.-J.; Lin, Y.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157:H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef]
- Fang, J.; Qiu, X.; Wan, Z.; Zou, Q.; Su, K.; Hu, N.; Wang, P. A sensing smartphone and its portable accessory for on-site rapid biochemical detection of marine toxins. Anal. Methods 2016, 8, 6895–6902. [Google Scholar] [CrossRef]
- Lee, S.; Kim, G.; Moon, J. Performance improvement of the one-dot lateral flow immunoassay for aflatoxin b1 by using a smartphone-based reading system. Sensors 2013, 13, 5109–5116. [Google Scholar] [CrossRef]
- Ross, G.M.S.; Bremer, M.G.E.G.; Wichers, J.H.; Van Amerongen, A.; Nielen, M.W.F. Rapid antibody selection using surface plasmon resonance for high-speed and sensitive hazelnut lateral flow prototypes. Biosensors 2018, 8, 130. [Google Scholar] [CrossRef]
- Coskun, A.F.; Wong, J.; Khodadadi, D.; Nagi, R.; Tey, A.; Ozcan, A. A personalized food allergen testing platform on a cellphone. Lab Chip 2013, 13, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Tsagkaris, A.S.; Nelis, J.L.D.; Ross, G.M.S.; Jafari, S.; Guercetti, J.; Kopper, K.; Zhao, Y.; Rafferty, K.; Salvador, J.P.; Migliorelli, D.; et al. Critical assessment of recent trends related to screening and confirmatory analytical methods for selected food contaminants and allergens. TrAC Trends Anal. Chem. 2019, 121, 115688. [Google Scholar] [CrossRef]
- Nelis, J.L.D.; Tsagkaris, A.S.; Zhao, Y.; Lou-Franco, J.; Nolan, P.; Zhou, H.; Cao, C.; Rafferty, K.; Hajslova, J.; Elliott, C.T.; et al. The end user sensor tree: An end-user friendly sensor database. Biosens. Bioelectron. 2019, 130, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Nelis, J.; Elliott, C.; Campbell, K. “The Smartphone’s Guide to the Galaxy”: In Situ Analysis in Space. Biosensors 2018, 8, 96. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar]
- Roda, A.; Michelini, E.; Zangheri, M.; Di Fusco, M.; Calabria, D.; Simoni, P. Smartphone-based biosensors: A critical review and perspectives. TrAC Trends Anal. Chem. 2016, 79, 317–325. [Google Scholar] [CrossRef]
- Rateni, G.; Dario, P.; Cavallo, F. Smartphone-Based Food Diagnostic Technologies: A Review. Sensors 2017, 17, 1453. [Google Scholar] [CrossRef]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards Lateral Flow Quantitative Assays: Detection Approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef]
- Suaifan, G.A.R.Y.; Zourob, M. Portable paper-based colorimetric nanoprobe for the detection of Stachybotrys chartarum using peptide labeled magnetic nanoparticles. Microchim. Acta 2019, 186, 230. [Google Scholar] [CrossRef]
- Zhang, X.; Zhi, X.; Zhang, C.; Wang, K.; Cui, D.; Li, D.; Wang, C. A CCD-based reader combined quantum dots-labeled lateral flow strips for ultrasensitive quantitative detection of anti-HBs antibody. J. Biomed. Nanotechnol. 2012, 8, 372–379. [Google Scholar] [CrossRef]
- Murdock, R.C.; Shen, L.; Griffin, D.K.; Kelley-Loughnane, N.; Papautsky, I.; Hagen, J.A. Optimization of a paper-based ELISA for a human performance biomarker. Anal. Chem. 2013, 85, 11634–11642. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.; You, J.B.; Zhang, B.; Nguyen, B.; Tarlan, F.; Jarvi, K.; Sinton, D. Accessory-free quantitative smartphone imaging of colorimetric paper-based assays. Lab Chip 2019, 19, 1991–1999. [Google Scholar] [CrossRef] [PubMed]
- Masawat, P.; Harfield, A.; Namwong, A. An iPhone-based digital image colorimeter for detecting tetracycline in milk. Food Chem. 2015, 184, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Turkevich, J. Colloidal gold. Part I. Gold Bull. 1985, 18, 125–131. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Mcvey, C.; Logan, N.; Thanh, N.T.K.; Elliott, C.; Cao, C. Unusual switchable peroxidase-mimicking nanozyme for the deter- mination of proteolytic biomarker. Nano Res. 2019, 12, 1–8. [Google Scholar] [CrossRef]
- Shen, L.; Hagen, J.A.; Papautsky, I. Point-of-care colorimetric detection with a smartphone. Lab Chip 2012, 12, 4240–4243. [Google Scholar] [CrossRef]
- Cantrell, K.; Erenas, M.M.; de Orbe-Payá, I.; Capitán-Vallvey, L.F. Use of the Hue Parameter of the Hue, Saturation, Value Color Space as a Quantitative Analytical Parameter for Bitonal Optical Sensors. Anal. Chem. 2010, 82, 531–542. [Google Scholar] [CrossRef]
- Kim, S.D.; Koo, Y.; Yun, Y. A smartphone-based automatic measurement method for colorimetric pH detection using a color adaptation algorithm. Sensors 2017, 17, 1604. [Google Scholar] [CrossRef]
- Zhao, Y.; Choi, S.Y.; Nelis, J.L.D.; Zhou, H.; Cao, C.; Campbell, K.; Elliott, C.; Rafferty, K. Smartphone Modulated Colorimetric Reader with Color Subtraction. In Proceedings of the IEEE Sensors 2019 Conference, Montreal, QC, Canada, 27–30 October 2019. [Google Scholar]
- Skandarajah, A.; Reber, C.D.; Switz, N.A.; Fletcher, D.A. Quantitative imaging with a mobile phone microscope. PLoS ONE 2014, 9, e96906. [Google Scholar] [CrossRef]
- Saisin, L.; Amarit, R.; Somboonkaew, A.; Gajanandana, O.; Himananto, O.; Sutapun, B. Significant Sensitivity Improvement for Camera-Based Lateral Flow Immunoassay Readers. Sensors 2018, 18, 4026. [Google Scholar] [CrossRef] [PubMed]

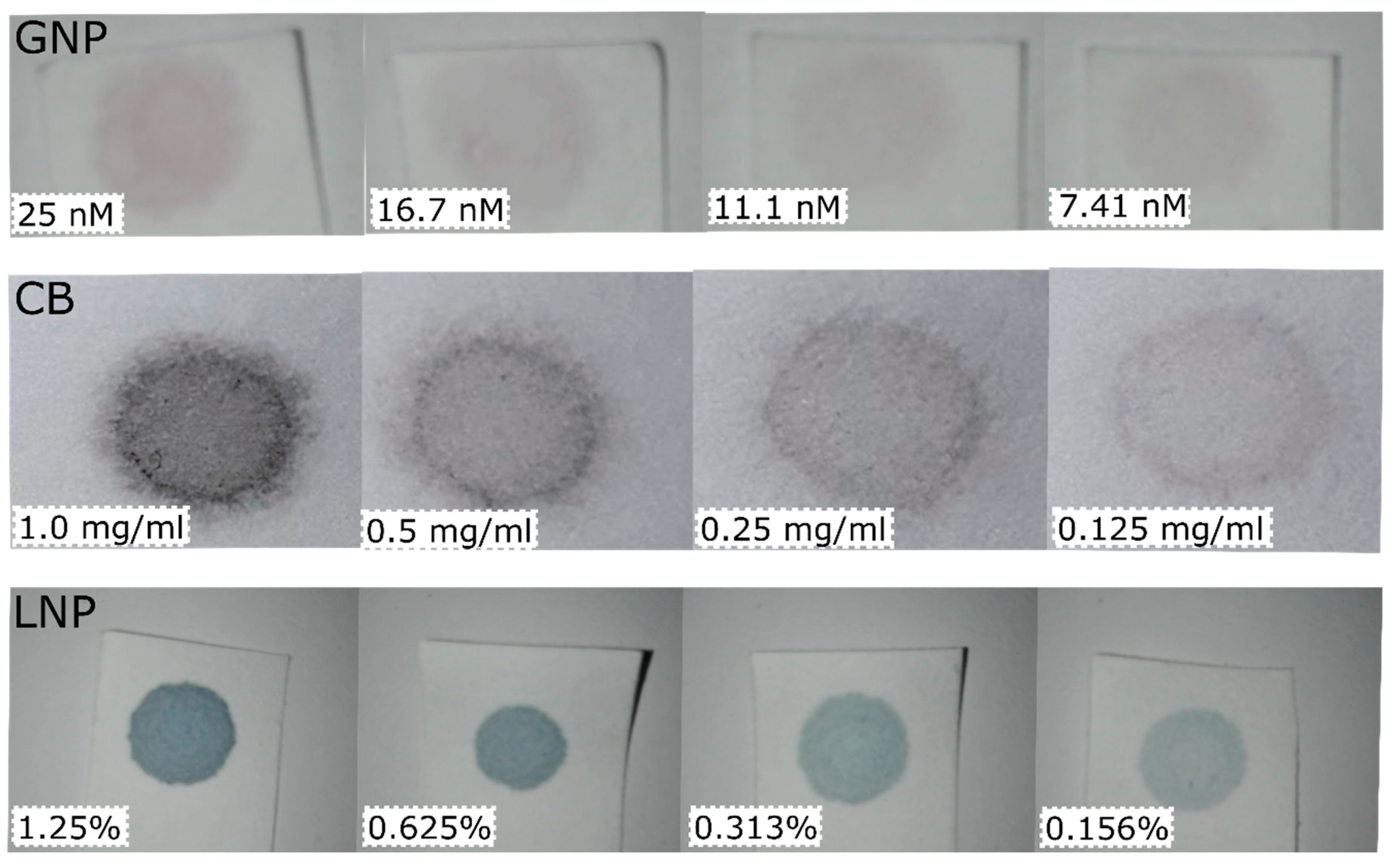
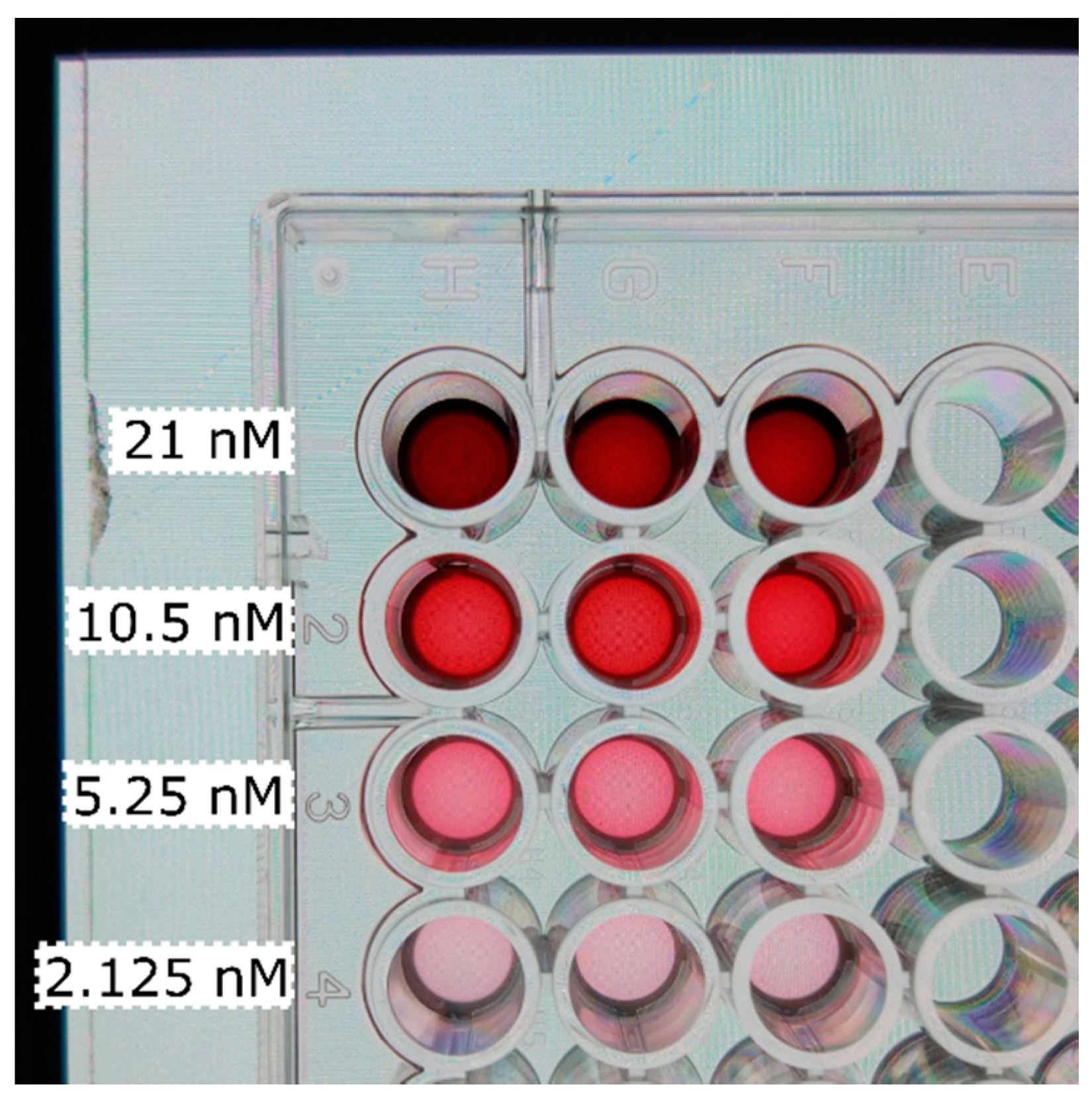


| GNP | CB | LNP | pH | |||||
|---|---|---|---|---|---|---|---|---|
| Channel | R2 | Function | R2 | Function | R2 | Function | R2 | Function |
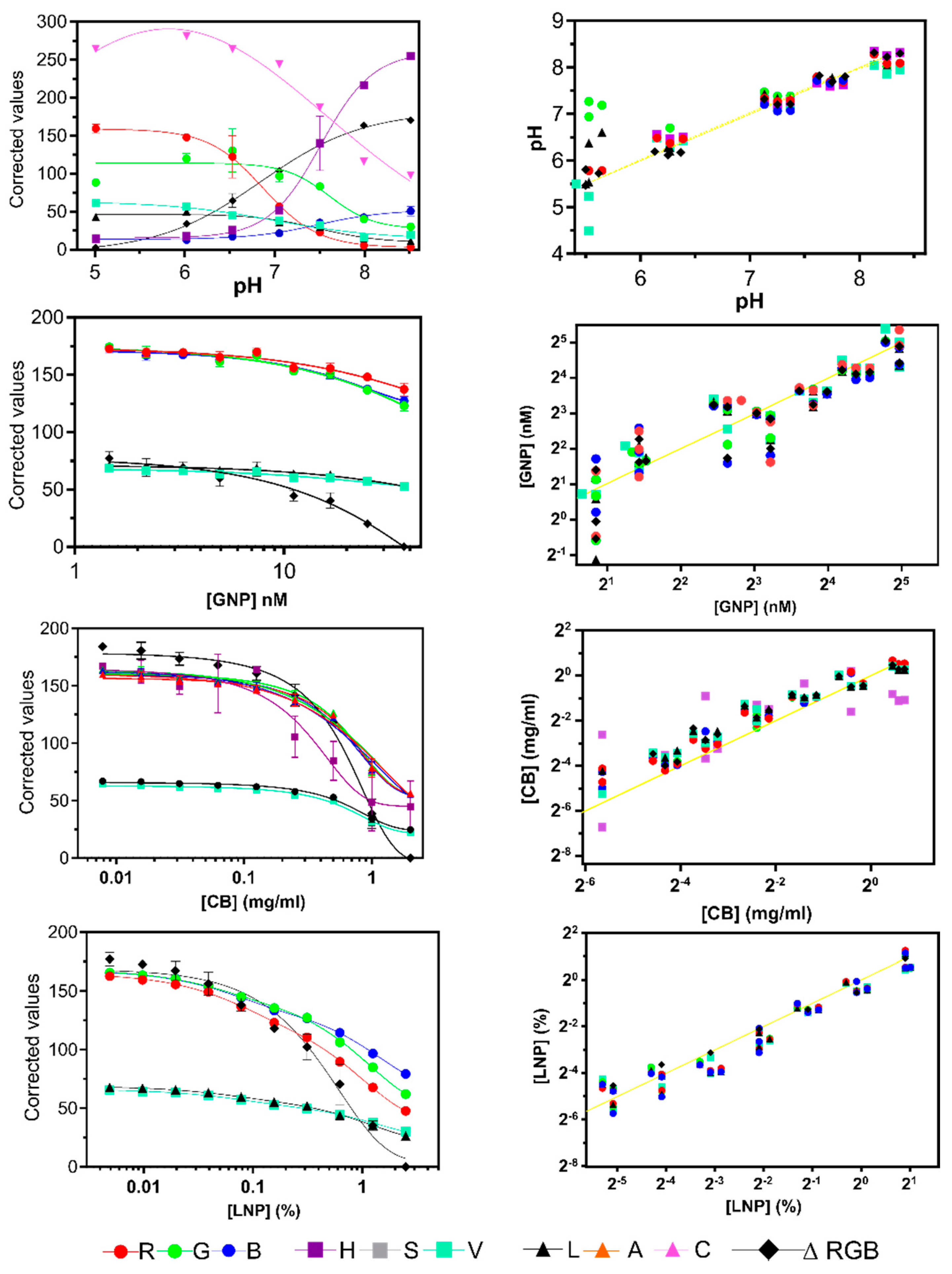
| R | 0.8871 | Sigmoidal | 0.9856 | Sigmoidal | 0.9847 | Two phase decay | 0.9795 | Sigmoidal |
| G | 0.9365 | Sigmoidal | 0.9859 | Sigmoidal | 0.9830 | Two phase decay | 0.8350 | Sigmoidal |
| B | 0.9383 | Sigmoidal | 0.9877 | 0.9818 | Two phase decay | 0.9531 | Sigmoidal | |
| ΔRGB | 0.9374 | Sigmoidal | 0.9870 | Sigmoidal | 0.9867 | Two phase decay | 0.9958 | Sigmoidal |
| H | - | - | 0.8862 | Sigmoidal | 0.6042 | Two phase decay | 0.9844 | Sigmoidal |
| S | 0.6895 | Sigmoidal | - | - | 0.7707 | Sigmoidal | 0.1878 | Sigmoidal |
| V | 0.8794 | Sigmoidal | 0.9836 | Sigmoidal | 0.9803 | Two phase decay | 0.9600 | Sigmoidal |
| L | 0.9330 | Sigmoidal | 0.9861 | Sigmoidal | 0.9811 | Two phase decay | 0.9610 | Sigmoidal |
| A | - | - | - | - | - | - | 0.7979 | Sigmoidal |
| C | - | - | - | - | - | - | 0.9941 | Sigmoidal |
| GNP | CB | Latex | pH | Total Scores | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | Ch | R2 | Slope | Ch | R2 | Slope | Ch | R2 | Slope | Ch | R2 | Slope | Ch | Score |
| 1 | Δ | 0.9173 | 1.008 | B | 0.979 | 0.801 | V | 0.951 | 1,012 | Δ | 0.987 | 0.974 | B | 37 |
| 2 | L | 0.9008 | 0.996 | R | 0.977 | 0.814 | L | 0.949 | 1,049 | R | 0.996 | 0.860 | R | 36 |
| 3 | G | 0.9164 | 0.972 | G | 0.973 | 0.795 | G | 0.947 | 1.028 | H | 0.977 | 0.874 | Δ | 35 |
| 4 | B | 0.8856 | 0.996 | L | 0.974 | 0.658 | B | 0.944 | 1.051 | V | 0.927 | 1.043 | V | 34 |
| 5 | R | 0.895 | 1.168 | Δ | 0.971 | 0.677 | R | 0.927 | 1.074 | B | 0.973 | 1.239 | L | 34 |
| 6 | V | 0.8678 | 1.058 | V | 0.972 | 0.676 | Δ | 0.762 | 1.178 | L | 0.898 | 0.603 | G | 32 |
| 7 | H | - | - | H | 0.462 | 0.354 | H | - | - | G | 0.712 | 0.307 | H | 12 |
| GNP | CB | LNP | pH | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Channel | Score | Channel | Score | Channel | Score | Channel | Score | Channel | Score |
| ΔRGB | 12 | B | 13 | V | 14 | ΔRGB | 13 | B | 37 |
| L | 12 | R | 13 | L | 11 | R | 11 | R | 36 |
| G | 10 | G | 9 | G | 11 | H | 10 | ΔRGB | 35 |
| B | 9 | L | 7 | B | 8 | V | 9 | V | 34 |
| R | 6 | ΔRGB | 6 | R | 6 | B | 7 | L | 34 |
| V | 5 | V | 6 | ΔRGB | 4 | L | 4 | G | 32 |
| H | 0 | H | 2 | H | 0 | G | 2 | H | 12 |
| GNP | CB | LNP | pH Original | |||||
|---|---|---|---|---|---|---|---|---|
| Source | Var | p-Value | Var | p-Value | Var | p-Value | Var | p-Value |
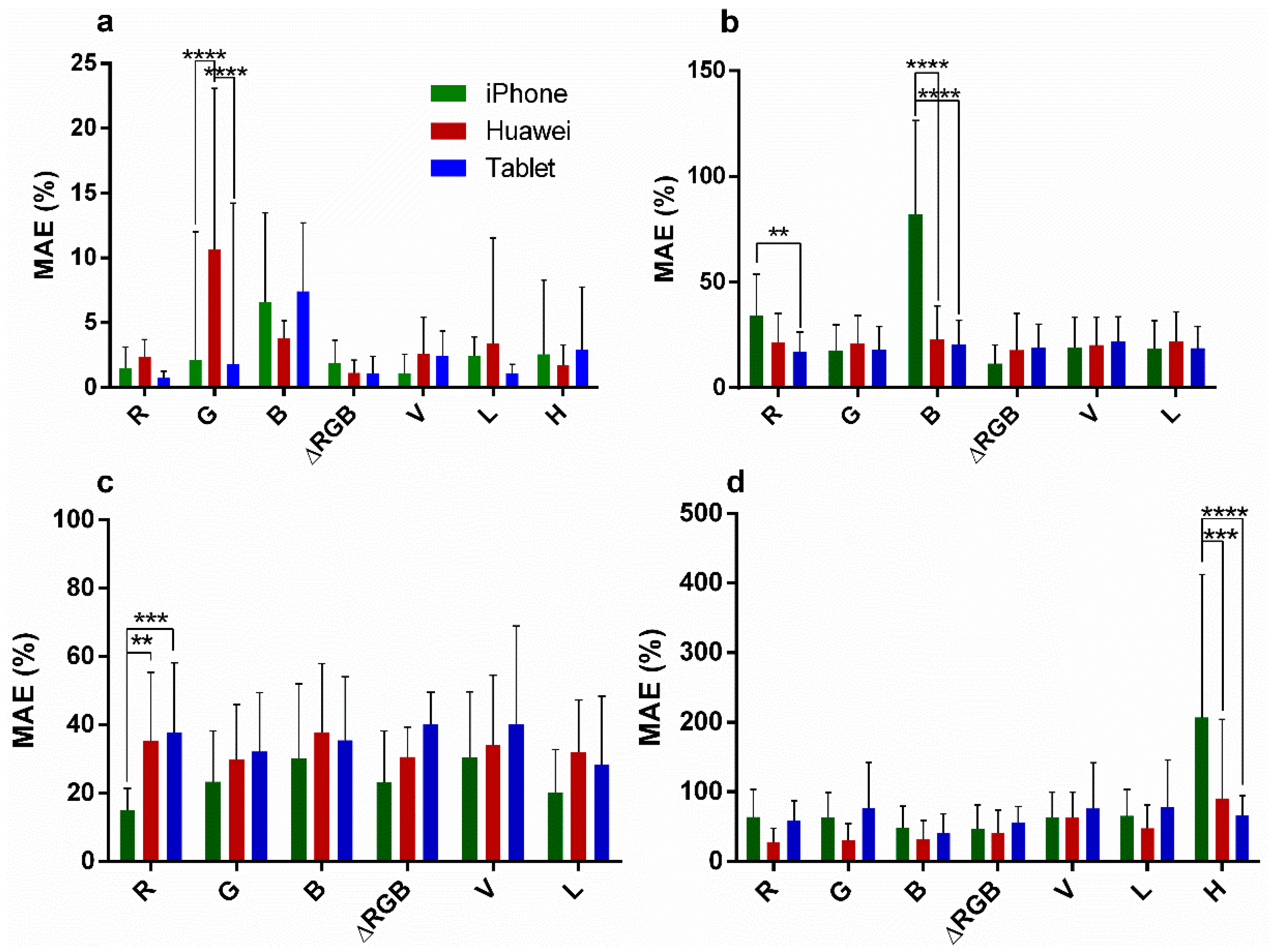
| Interaction | 2.8 | 0.45 | 8.7 | 0.0001 | 25.7 | <0.0001 | 8.1 | 0.0058 |
| Channel | 2.6 | 0.10 | 7.1 | <0.0001 | 15.0 | <0.0001 | 7.8 | 0.0002 |
| Phone | 7.9 | <0.0001 | 5.7 | <0.0001 | 5.2 | <0.0001 | 0.8 | 0.24 |
| pH | GNP | |||
|---|---|---|---|---|
| Source | Var | p-Value | Var | p-Value |
| Interaction | 1.652 | 0.9893 | 2.177 | 0.6248 |
| Channels | 13.24 | <0.0001 | 0.6425 | 0.4002 |
| Light conditions | 0.9718 | 0.3046 | 39.7 | <0.0001 |
| Prediction Curves | Calibration Curves (nM) | ||||||
|---|---|---|---|---|---|---|---|
| Solution Type | Channel | R2 | Slope | R2 | LOD | Linear Range | IC50 |
| GNP | G | 0.9966 | 1.086 | 0.9969 | 1.5 | 3.1-12.8 | 7.4 |
| GNP | B | 0.9890 | 1.089 | 0.9973 | 1.8 | 3.7–16.2 | 9.0 |
| GNP | ∆RGB | 0.9094 | 0.824 | 0.9890 | 1.6 | 4.1–32.7 | 13.8 |
| GNP | L | 0.913 | 0.8009 | 0.8947 | - | - | - |
| GNP | spectro | 0.9989 | 1.039 | 0.9996 | 7.5 | 15.3–65.7 | 39.7 |
| HRP | B | 0.9742 | 0.932 | 0.9957 | 0.0025 | 0.0038–0.012 | 0.0073 |
| HRP | R | 0.6087 | 0.601 | 0.829 | NA | 0.010–0.038 | 0.027 |
| HRP | G | 0.8365 | 0.506 | 0.9377 | NA | 0.0083–0.036 | 0.023 |
| HRP | ∆RGB | 0.9099 | 0.734 | 0.9804 | 0.0023 | 0.0039–0.020 | 0.010 |
| HRP | L | 0.07789 | 0.08 | 0.9307 | - | - | - |
| V | 0.532 | 0.29 | 0.8992 | - | - | - | |
| HRP | spectro | 0.9752 | 1.153 | 0.9982 | 0.0034 | 0.0057–0.021 | 0.012 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelis, J.L.D.; Bura, L.; Zhao, Y.; Burkin, K.M.; Rafferty, K.; Elliott, C.T.; Campbell, K. The Efficiency of Color Space Channels to Quantify Color and Color Intensity Change in Liquids, pH Strips, and Lateral Flow Assays with Smartphones. Sensors 2019, 19, 5104. https://doi.org/10.3390/s19235104
Nelis JLD, Bura L, Zhao Y, Burkin KM, Rafferty K, Elliott CT, Campbell K. The Efficiency of Color Space Channels to Quantify Color and Color Intensity Change in Liquids, pH Strips, and Lateral Flow Assays with Smartphones. Sensors. 2019; 19(23):5104. https://doi.org/10.3390/s19235104
Chicago/Turabian StyleNelis, Joost Laurus Dinant, Laszlo Bura, Yunfeng Zhao, Konstantin M. Burkin, Karen Rafferty, Christopher T. Elliott, and Katrina Campbell. 2019. "The Efficiency of Color Space Channels to Quantify Color and Color Intensity Change in Liquids, pH Strips, and Lateral Flow Assays with Smartphones" Sensors 19, no. 23: 5104. https://doi.org/10.3390/s19235104
APA StyleNelis, J. L. D., Bura, L., Zhao, Y., Burkin, K. M., Rafferty, K., Elliott, C. T., & Campbell, K. (2019). The Efficiency of Color Space Channels to Quantify Color and Color Intensity Change in Liquids, pH Strips, and Lateral Flow Assays with Smartphones. Sensors, 19(23), 5104. https://doi.org/10.3390/s19235104






