A LAPS-Based Differential Sensor for Parallelized Metabolism Monitoring of Various Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sensor Fabrication and Measurement Set-Up
2.2. Sample Preparation and Microorganism Cultivation
3. Results and Discussion
3.1. Monitoring the Cellular Metabolism of L. Brevis Bacteria
3.2. Determination of Calibration Curves for L. brevis, C. Glutamicum, and E. Coli
3.3. Simultaneous Measurements with L. Brevis, C. Glutamicum, and E. Coli Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Devanthi, P.V.P.; Gkatzionis, K. Soy sauce fermentation: Microorganisms, aroma formation, and process modification. Int. Food Res. J. 2019, 120, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Haile, M.; Kang, W.H. The role of microbes in coffee fermentation and their impact on coffee quality. J. Food Qual. 2019, 2019, 1–6. [Google Scholar] [CrossRef]
- Poszytek, K.; Pyzik, A. The effect of the source microorganisms on adaptation of hydrolytic consortia dedicated to anaerobic digestion of maize silage. Anaerobe 2017, 46, 46–55. [Google Scholar] [CrossRef] [PubMed]
- O’Mara, P.; Farrell, A.; Bones, J.; Twomey, K. Staying alive! Sensors used for monitoring cell health in bioreactors. Talanta 2018, 176, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, T.; Biechele, P.; Sobocinski, M.; Suhr, H.; Bakes, K.; Solle, D.; Jantunen, H.; Scheper, T.; Zimmermann, S. Continuous noninvasive monitoring of cell growth in disposable bioreactors. Sens. Actuators B Chem. 2017, 251, 1009–1017. [Google Scholar] [CrossRef][Green Version]
- Qui, J.; Arnold, M.A.; Murhammer, D.W. On-line near infrared bioreactor monitoring of cell density and concentrations of glucose and lactate during insect cell cultivation. J. Biotechnol. 2014, 173, 106–111. [Google Scholar]
- Marquard, D.; Schneider-Barthold, C.; Düsterloh, S.; Scheper, T.; Lindner, P. Online monitoring of cell concentration in high cell density Escherichia coli cultivations using in situ microscopy. J. Biotechnol. 2017, 259, 83–85. [Google Scholar] [CrossRef]
- Simmons, A.D.; Williams, C., III; Degoix, A.; Sikavitsas, V.L. Sensing metabolites for the monitoring of tissue engineered construct cellularity in perfusion bioreactors. Biosens. Bioelectron. 2017, 90, 443–449. [Google Scholar] [CrossRef]
- Buchenauer, A.; Hofmann, M.C.; Funke, M.; Büchs, J.; Mokwa, W.; Schnakenberg, U. Micro-biosensors for fed-batch fermentations with integrated online monitoring and microfluidic devices. Biosens. Bioelectron. 2009, 24, 1411–1416. [Google Scholar] [CrossRef]
- Hafeman, D.G.; Parce, J.W.; McConnell, H.M. Light-addressable potentiometric sensor for biochemical systems. Science 1988, 240, 1182–1185. [Google Scholar] [CrossRef] [PubMed]
- Owicki, J.C.; Bousse, L.J.; Hafeman, D.G.; Kirk, G.L.; Olson, J.D.; Wada, H.G.; Parce, J.W. The light-addressable potentiometric sensor: Principles and biological applications. Annu. Rev. Biophys. Biomol. Struc. 1994, 23, 87–113. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Schöning, M.J. Light-addressable potentiometric sensors (LAPS): Recent trends and applications. In Electrochemical Sensor Analysis; Alegret, S., Merkoci, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 87–128. [Google Scholar]
- Schöning, M.J. Playing around with field-effect sensors on the basis of EIS structures, LAPS and ISFETs. Sensors 2005, 5, 126–138. [Google Scholar] [CrossRef]
- Yoshinobu, T.; Miyamoto, K.-I.; Werner, C.F.; Poghossian, A.; Wagner, T.; Schöning, M.J. Light-addressable potentiometric sensors for quantitative spatial imaging of chemical species. Annu. Rev. Anal. Chem. 2017, 10, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Yang, C.M.; Chen, T.C.; Lai, C.S. Analog micromirror-LAPS for chemical imaging and zoom-in application. Vacuum 2015, 118, 161–166. [Google Scholar] [CrossRef]
- Miyamoto, K.-I.; Wagner, T.; Yoshinobu, T.; Kanoh, S.; Schöning, M.J. Phase-mode LAPS and its application to chemical imaging. Sens. Actuator B Chem. 2011, 154, 28–32. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Jiang, S.; Kunze, J.; Schmuki, P.; Krause, S. High resolution LAPS and SPIM. Electrochem. Commun. 2010, 12, 758–760. [Google Scholar] [CrossRef]
- Schöning, M.J.; Poghossian, A. Bio FEDs (field-effect devices), state-of-the-art and new directions. Electroanalysis 2006, 18, 1893–1900. [Google Scholar] [CrossRef]
- Hizawa, T.; Sawada, K.; Takao, H.; Ishida, M. Fabrication of a two-dimensional pH image sensor using a charge transfer technique. Sens. Actuators B Chem. 2006, 117, 509–515. [Google Scholar] [CrossRef]
- Dantism, S.; Takenaga, S.; Wagner, P.; Wagner, T.; Schöning, M.J. Determination of the extracellular acidification of Escherichia coli K12 with a multi-chamber-based LAPS system. Phys. Status Solidi A 2016, 213, 1479–1485. [Google Scholar] [CrossRef]
- Sasaki, Y.; Kanai, Y.; Uchida, H.; Katsube, T. Highly sensitive taste sensor with a new differential LAPS method. Sens. Actuators B Chem. 1995, 24–25, 819–822. [Google Scholar] [CrossRef]
- Wang, J.; Campos, I.; Wu, F.; Zhu, J.; Su, H.; Stellberg, G.B.; Palma, M.; Watkinson, M.; Krause, S. The effect of gold nanoparticles on the impedance of microcapsules visualized by scanning photo-induced impedance microscopy. Electrochim. Acta 2016, 208, 39–46. [Google Scholar] [CrossRef]
- Yoshinobu, T.; Krause, S.; Miyamoto, K.-I.; Werner, C.F.; Poghossian, A.; Wagner, T.; Schöning, M.J. (Bio-) chemical Sensing and Imaging by LAPS and SPIM. In Label-Free Biosensing; Schöning, M.J., Poghossian, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 103–132. [Google Scholar]
- Werner, C.F.; Krumbe, C.; Schumacher, K.; Groebel, S.; Spelthahn, H.; Stellberg, M.; Wagner, T.; Yoshinobu, T.; Selmer, T.; Keusgen, M.; et al. Determination of the extracellular acidification of Escherichia coli by a light-addressable potentiometric sensor. Phys. Status Solidi A 2011, 208, 1340–1344. [Google Scholar] [CrossRef]
- Werner, C.F.; Groebel, S.; Krumbe, C.; Wagner, T.; Selmer, T.; Yoshinubo, T.; Baumann, M.E.M.; Keusgen, M.; Schöning, M.J. Nutrient concentration-sensitive microorganism-based biosensor. Phys. Status Solidi A 2012, 209, 900–904. [Google Scholar] [CrossRef]
- Dantism, S.; Röhlen, D.; Wagner, T.; Wagner, P.; Schöning, M.J. Optimization of cell-based multi-chamber LAPS measurements utilizing FPGA-controlled laser-diode modules. Phys. Status Solidi A 2018, 215, 1–8. [Google Scholar] [CrossRef]
- Dantism, S.; Takenaga, S.; Wagner, T.; Wagner, P.; Schöning, M.J. Differential imaging of the metabolism of bacteria and eukaryotic cells based on light-addressable potentiometric sensors. Elecrochim. Acta 2017, 246, 234–241. [Google Scholar] [CrossRef]
- Dantism, S.; Röhlen, D.; Selmer, T.; Wagner, T.; Wagner, P.; Schöning, M.J. Quantitative differential monitoring of the metabolic activity of Corynebacterium glutamicum cultures utilizing a light-addressable potentiometric sensor system. Biosens. Bioelectron. 2019, 139, 11332. [Google Scholar] [CrossRef]
- Zhang, D.W.; Wu, F.; Wang, J.; Watkinson, M.; Krause, S. Image detection of yeast Saccharomyces cerevisiae by light-addressable potentiometric sensors (LAPS). Elechtrochem. Commun. 2016, 72, 41–45. [Google Scholar] [CrossRef]
- Du, L.; Wang, J.; Chen, W.; Zhao, L.; Wu, C.; Wang, P. Dual functional extracellular recording using a light-addressable potentiometric sensor for bitter signal transduction. Anal. Chim. Acta 2018, 1022, 106–112. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, N.; Zhang, F.; Wang, H.; Ye, W.; Wang, P. Neurosecretory cell-based biosensor: Monitoring secretion of adrenal chromaffin cells by local extracellular acidification using light-addressable potentiometric sensor. Biosens. Bioelectron. 2012, 35, 421–424. [Google Scholar] [CrossRef]
- Udaka, S. The discovery of Corynebacterium glutamicum and birth of amino acid fermentation industry in Japan. In Corynebacteria: Genomics and Molecular Biology; Burkovski, A., Ed.; Caister Academic Press: Norfolk, UK, 2008. [Google Scholar]
- Rönkä, E.; Malinen, E.; Saarela, M.; Rinta-Koski, M.; Aarnikunnas, J.; Palva, A. Probiotic and milk technological properties of Lactobacillus brevis. Int. J. Food Microbiol. 2003, 83, 63–74. [Google Scholar] [CrossRef]
- Schöning, M.J.; Brinkmann, D.; Rolka, D.; Demuth, C.; Poghossian, A. CIP (cleaning-in-place) suitable “non-glass” pH sensor based on Ta2O5-gate EIS structure. Sens. Actuators B Chem. 2005, 111, 423–429. [Google Scholar] [CrossRef]
- Poghossian, A.; Schöning, M.J. Silicon-based chemical and biological field-effect sensors. In Encyclopedia of Sensors; Grimes, C.A., Dickey, E.C., Pishko, M.V., Eds.; American Scientific Publisher: Stevenson Ranch, CA, USA, 2006; pp. 463–534. [Google Scholar]
- Schöning, M.J.; Arzdorf, M.; Mulchandani, P.; Chen, W.; Mulchandani, A. A capacitive field-effect sensor for the direct determination of organophosphorus pesticides. Sens. Actuators B Chem. 2003, 91, 92–97. [Google Scholar] [CrossRef]
- Poghossian, A.; Thust, M.; Schroth, P.; Steffen, A.; Lüth, H.; Schöning, M.J. Penicillin detection by means of silicon-based field-effect structures. Sens. Mater. 2001, 13, 207–223. [Google Scholar]
- Werner, C.F.; Wagner, T.; Yoshinubo, T.; Keusgen, M.; Schöning, M.J. Frequency behaviour of light-addressable potentiometric sensors. Phys. Status Solidi A 2011, 210, 884–891. [Google Scholar] [CrossRef]
- Poghossian, A.; Werner, C.F.; Buniatyan, V.V.; Wagner, T.; Miyamoto, K.-I.; Yoshinubo, T.; Schöning, M.J. Towards addressability of light-addressable potentiometric sensors: Shunting effect of non-illuminated region and cross-talk. Sens. Actuators B Chem. 2017, 244, 1071–1079. [Google Scholar] [CrossRef]
- Yoshinobu, T.; Ecken, H.; Poghossian, A.; Simonis, A.; Iwasaki, H.; Lüth, H.; Schöning, M.J. Constant-current-mode LAPS (CLAPS) for the detection of penicillin. Electroanalysis 2001, 13, 733–736. [Google Scholar] [CrossRef]
- Poghossian, A.; Mai, D.-T.; Mourzina, Yu.; Schöning, M.J. Impedance effect of an ion-sensitive membrane: Characterization of an EMIS sensor by impedance spectroscopy, capacitance-voltage and constant-capacitance method. Sens. Actuators B Chem. 2004, 103, 423–428. [Google Scholar] [CrossRef]
- Schöning, M.J.; Wagner, T.; Wang, C.; Otto, R.; Yoshinubo, T. Development of a handheld 16 channel pen-type LAPS for electrochemical sensing. Sens. Actuators B Chem. 2005, 108, 808–814. [Google Scholar] [CrossRef]
- Wagner, T.; Rao, C.; Kloock, J.P.; Yoshinobu, T.; Otto, R.; Keusgen, M.; Schöning, M.J. “LAPS Card”—A novel chip card-based light-addressable potentiometric sensor (LAPS). Sens. Actuators B Chem. 2006, 118, 33–40. [Google Scholar] [CrossRef]
- Schöning, M.J.; Näther, N.; Auger, V.; Poghossian, A.; Koudelka-Hep, M. Miniaturised flow-through cell with integrated capacitive EIS sensor fabricated at wafer level using Si and SU-8 technologies. Sens. Actuators B Chem. 2005, 108, 986–992. [Google Scholar] [CrossRef]
- Huck, C.; Schiffels, J.; Herrera, C.N.; Schelden, M.; Selmer, T.; Poghossian, A.; Baumann, M.E.M.; Wagner, P.; Schöning, M.J. Metabolic responses of Escherichia coli upon glucose pulses captured by a capacitive field-effect sensor. Phys. Status Solidi A 2013, 210, 926–931. [Google Scholar] [CrossRef]

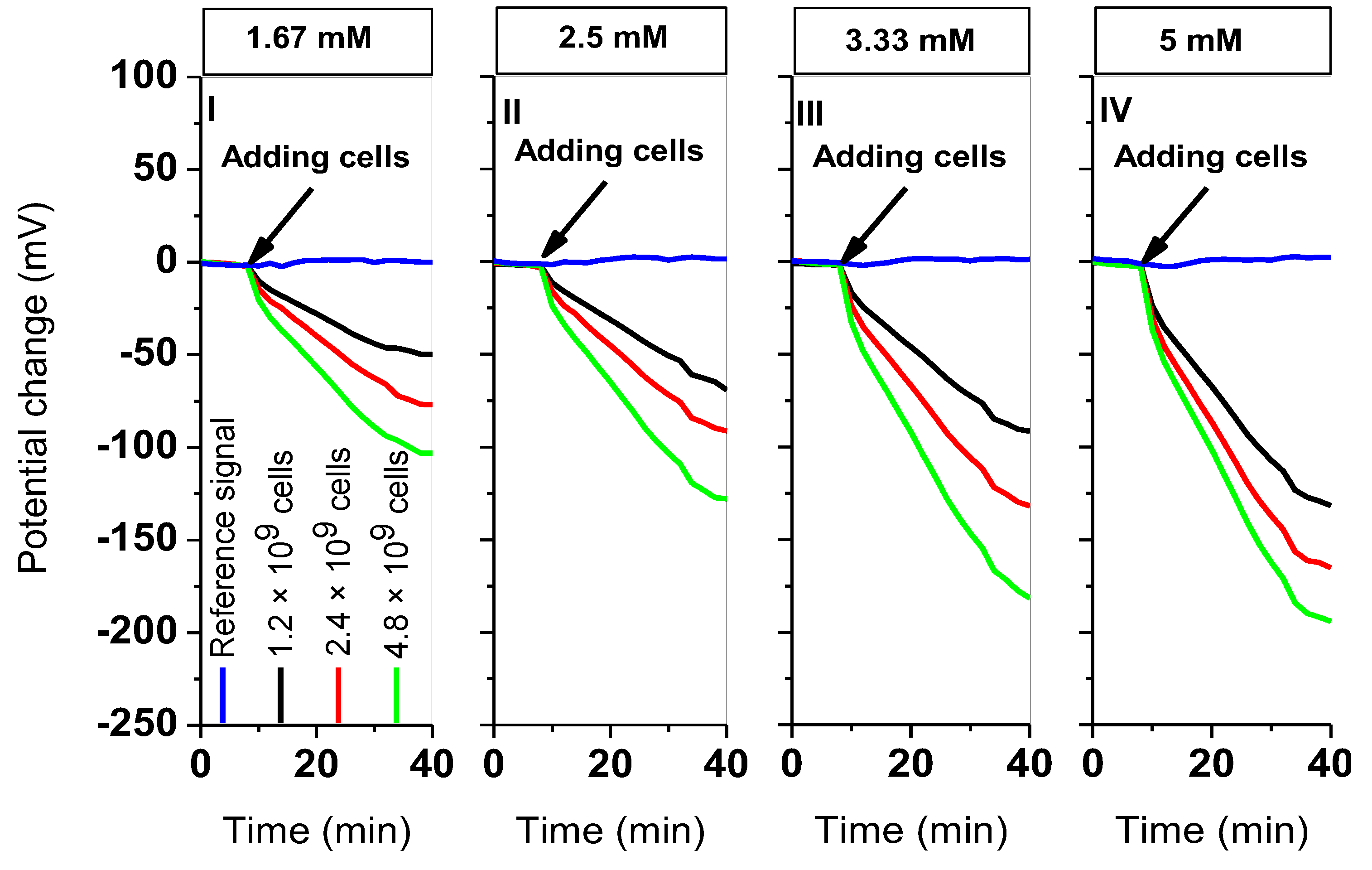

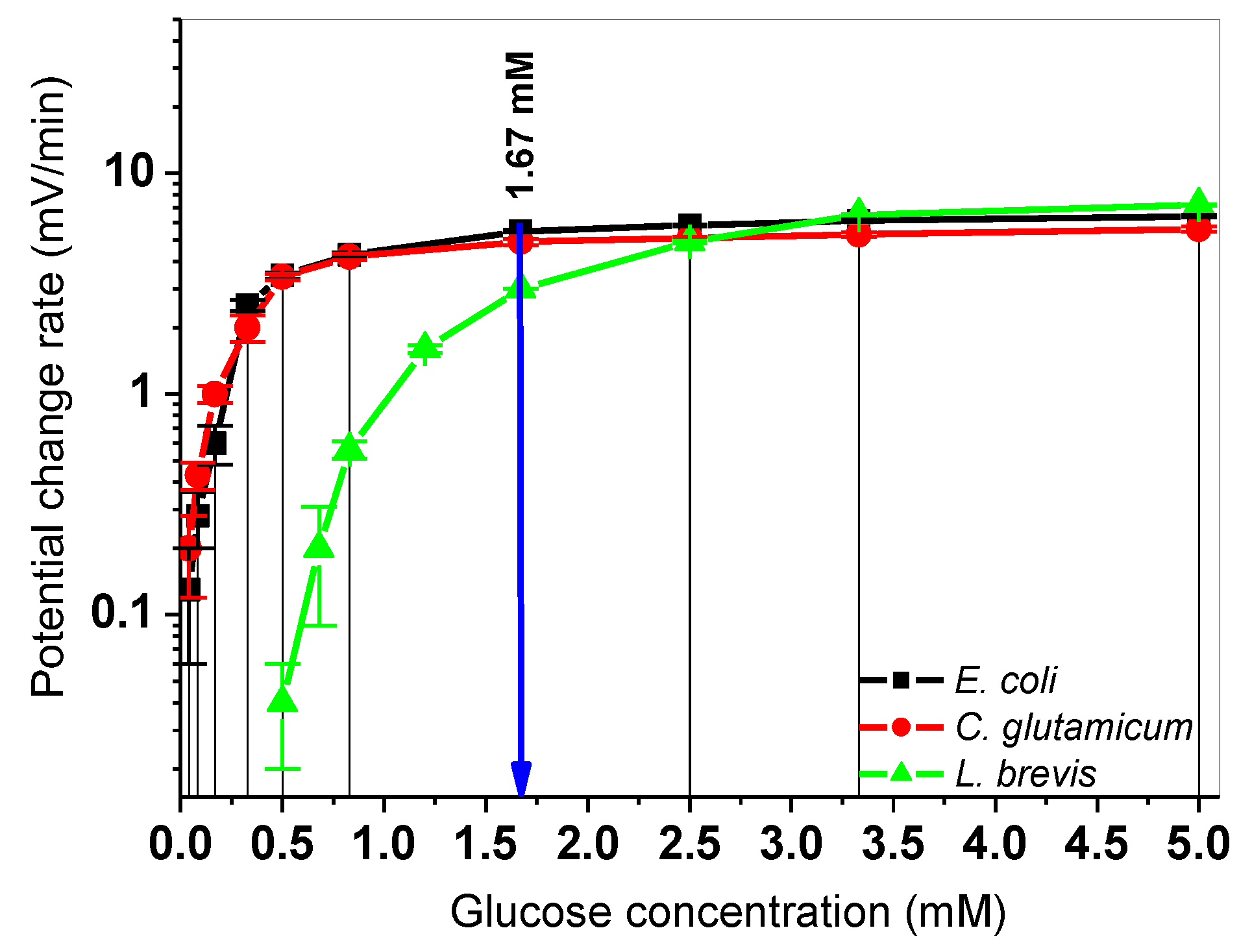
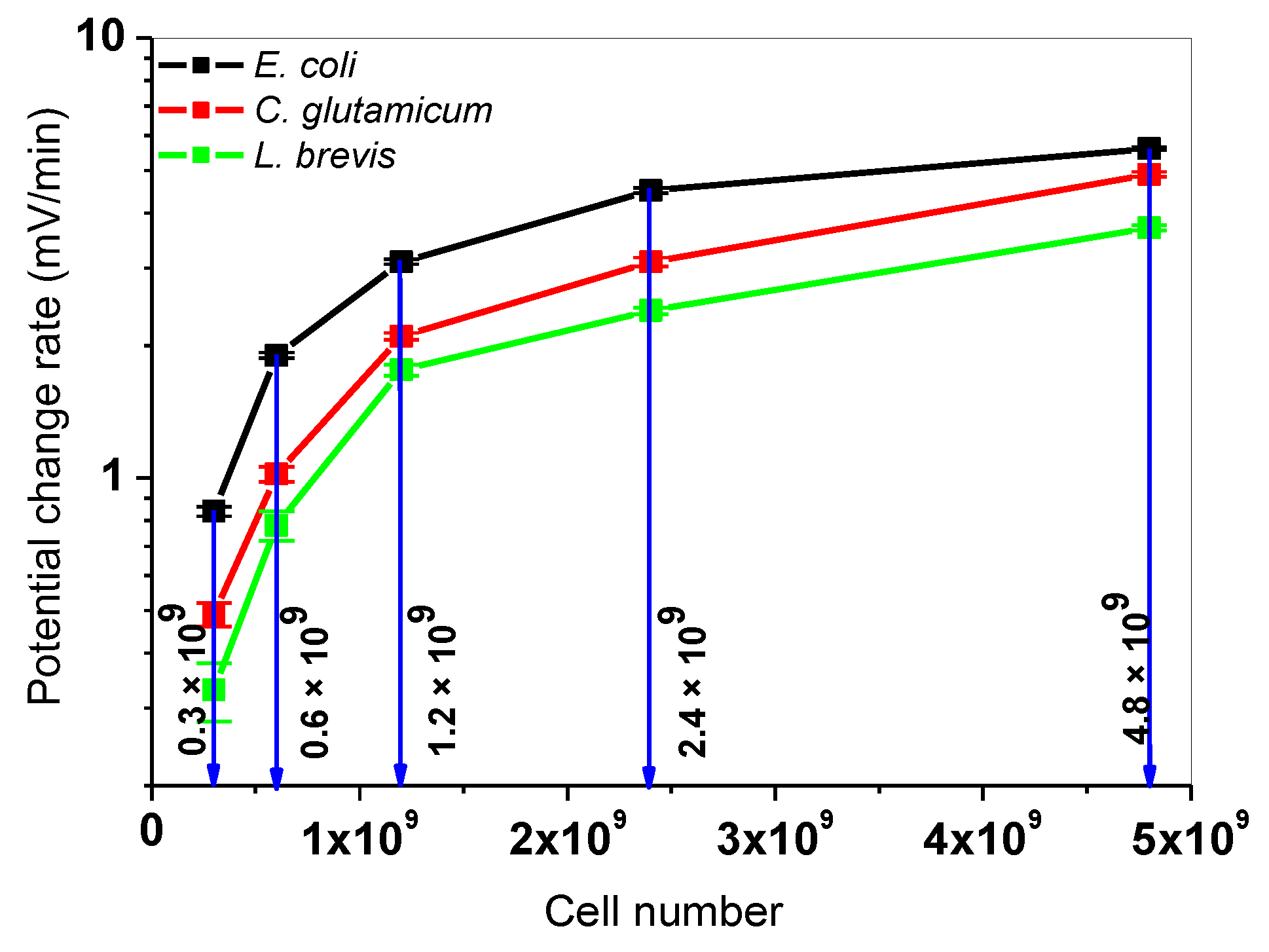
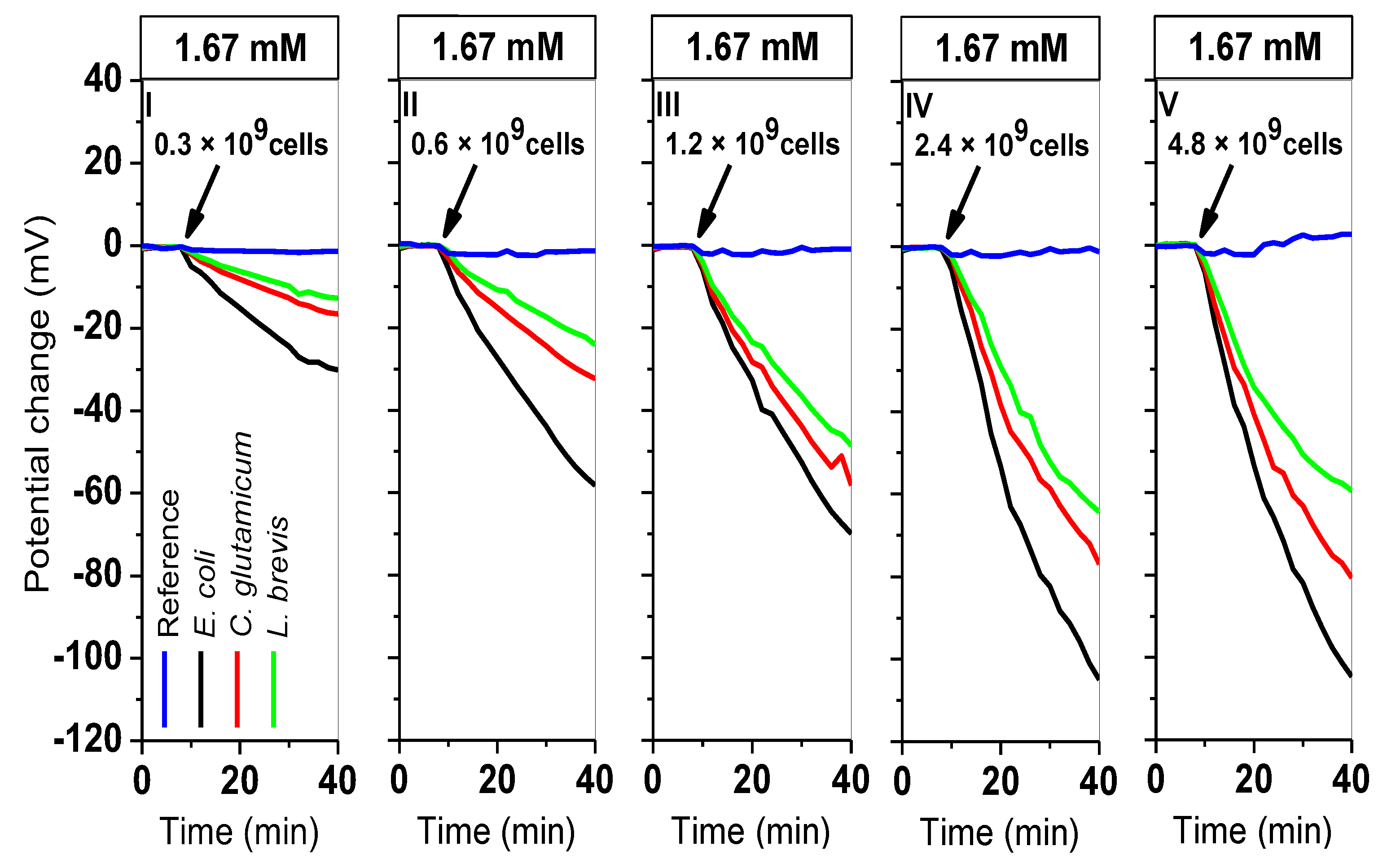
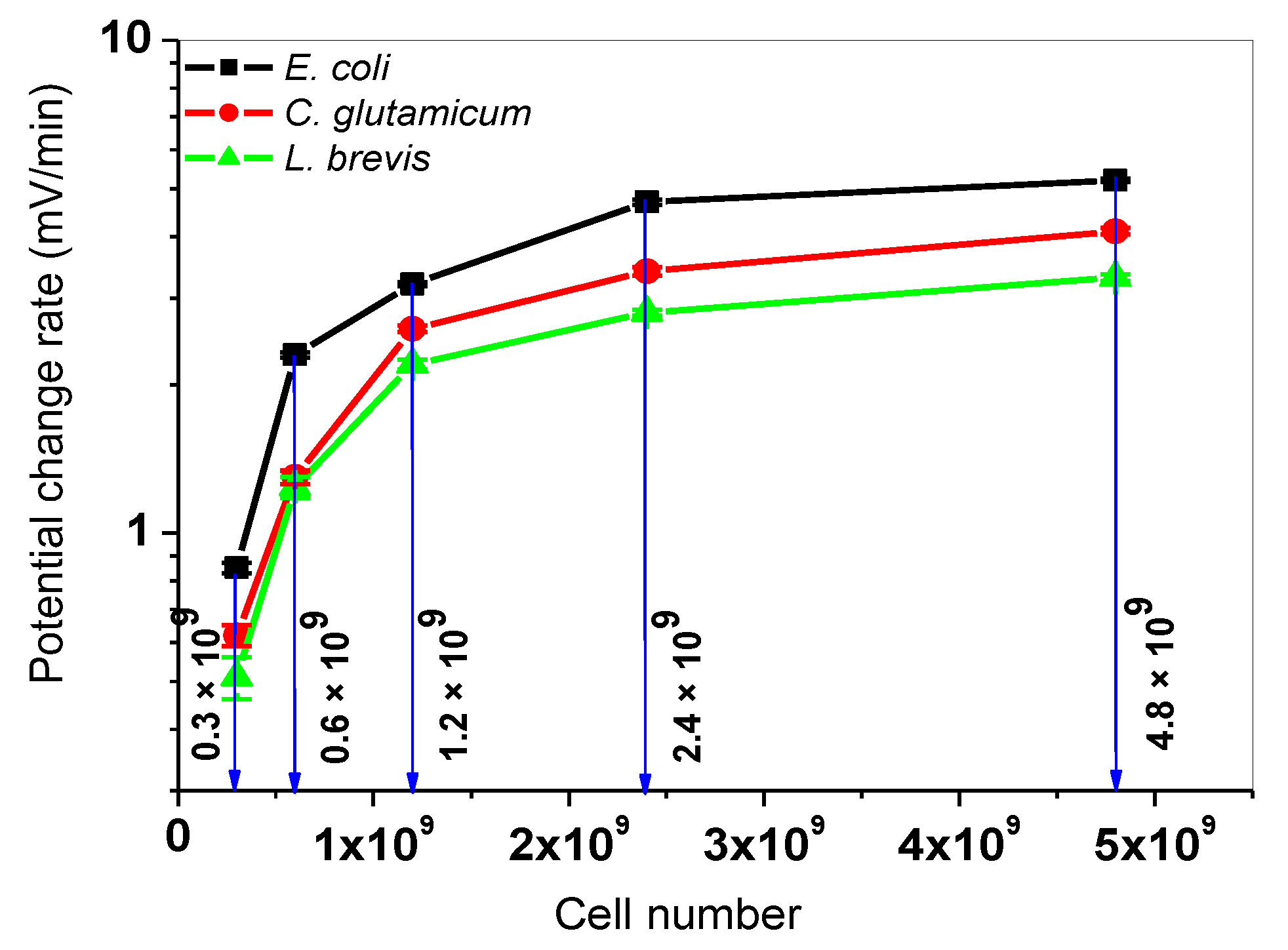
| Cells In 200 µL | Type Of Bacteria | |Sequential PCR| (mV/min) | |Simultaneous PCR| (mV/min) | |Δ PCR| (mV/min) |
|---|---|---|---|---|
| E. Coli | 0.84 ± 0.13 | 0.85 ± 0.02 | 0.01 | |
| 0.3 × 109 | C. Glutamicum | 0.49 ± 0.05 | 0.62 ± 0.03 | 0.13 |
| L. Brevis | 0.33 ± 0.04 | 0.51 ± 0.05 | 0.18 | |
| E. Coli | 1.90 ± 0.12 | 2.30 ± 0.03 | 0.40 | |
| 0.6 × 109 | C. Glutamicum | 1.02 ± 0.04 | 1.30 ± 0.04 | 0.28 |
| L. Brevis | 0.78 ± 0.06 | 1.24 ± 0.06 | 0.46 | |
| E. Coli | 3.10 ± 0.11 | 3.20 ± 0.03 | 0.10 | |
| 1.2 × 109 | C. Glutamicum | 2.10 ± 0.04 | 2.60 ± 0.04 | 0.50 |
| L. Brevis | 1.76 ± 0.05 | 2.20 ± 0.05 | 0.44 | |
| E. Coli | 4.50 ± 0.10 | 4.70 ± 0.06 | 0.20 | |
| 2.4 × 109 | C. Glutamicum | 3.10 ± 0.22 | 3.40 ± 0.07 | 0.30 |
| L. Brevis | 2.40 ± 0.06 | 2.80 ± 0.04 | 0.40 | |
| E. Coli | 5.60 ± 0.15 | 5.20 ± 0.04 | 0.40 | |
| 4.8 × 109 | C. Glutamicum | 4.90 ± 0.04 | 4.10 ± 0.06 | 0.80 |
| L. Brevis | 3.70 ± 0.02 | 3.30 ± 0.05 | 0.40 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dantism, S.; Röhlen, D.; Wagner, T.; Wagner, P.; Schöning, M.J. A LAPS-Based Differential Sensor for Parallelized Metabolism Monitoring of Various Bacteria. Sensors 2019, 19, 4692. https://doi.org/10.3390/s19214692
Dantism S, Röhlen D, Wagner T, Wagner P, Schöning MJ. A LAPS-Based Differential Sensor for Parallelized Metabolism Monitoring of Various Bacteria. Sensors. 2019; 19(21):4692. https://doi.org/10.3390/s19214692
Chicago/Turabian StyleDantism, Shahriar, Désirée Röhlen, Torsten Wagner, Patrick Wagner, and Michael J. Schöning. 2019. "A LAPS-Based Differential Sensor for Parallelized Metabolism Monitoring of Various Bacteria" Sensors 19, no. 21: 4692. https://doi.org/10.3390/s19214692
APA StyleDantism, S., Röhlen, D., Wagner, T., Wagner, P., & Schöning, M. J. (2019). A LAPS-Based Differential Sensor for Parallelized Metabolism Monitoring of Various Bacteria. Sensors, 19(21), 4692. https://doi.org/10.3390/s19214692







