Point-of-Care Strategies for Detection of Waterborne Pathogens
Abstract
1. Introduction
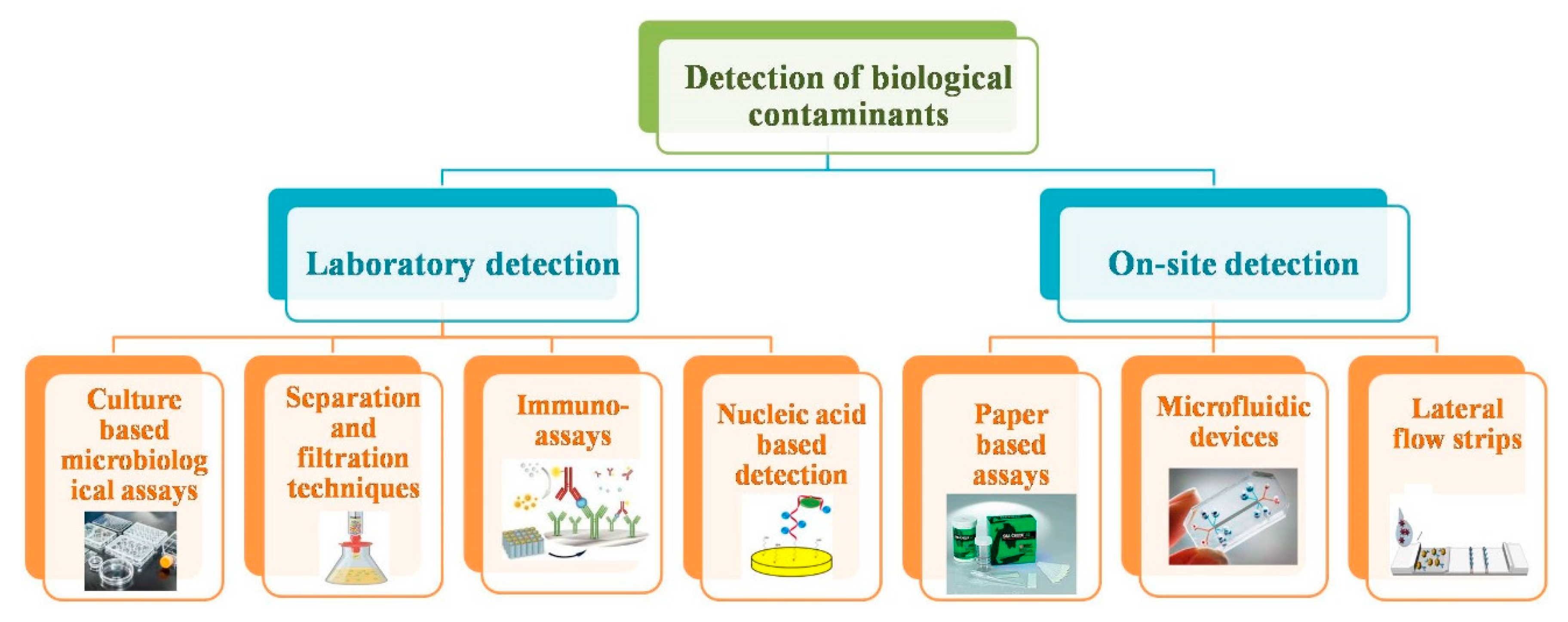
2. Conventional Methods to Detect Biological Contaminants
2.1. Microbiological Assays Technique
2.2. Separation and Filtration Techniques
2.3. Immunoassays Approach
2.4. Nucleic Acid-Based Detection
3. Point-Of-Care (POC) Devices for Biological Contaminants
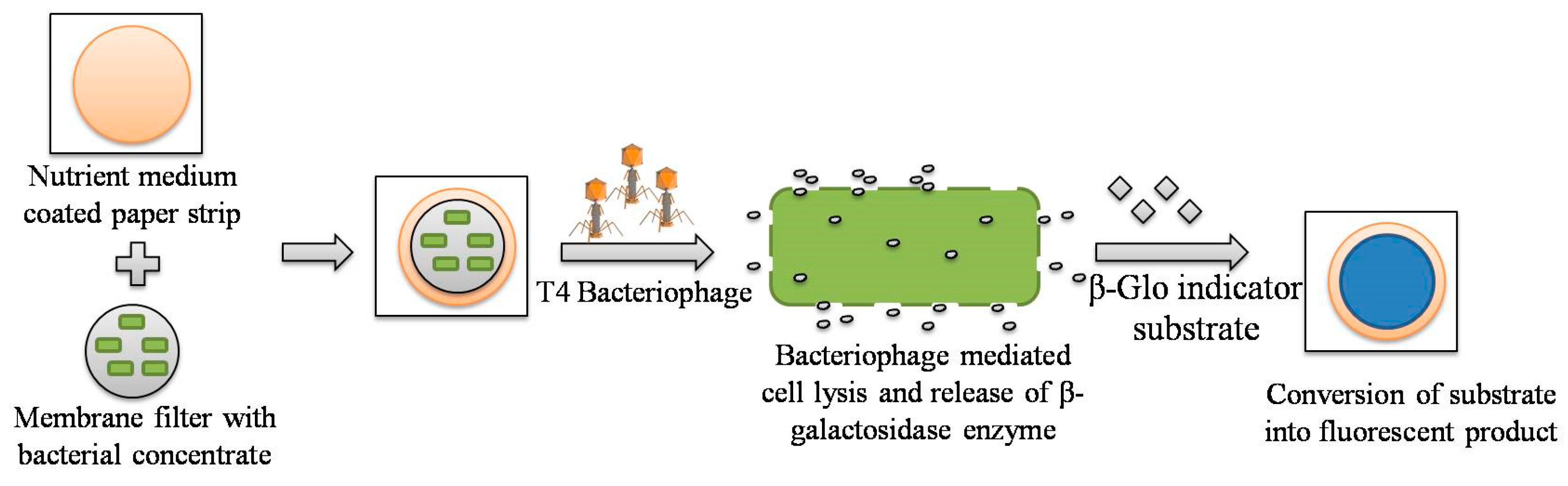
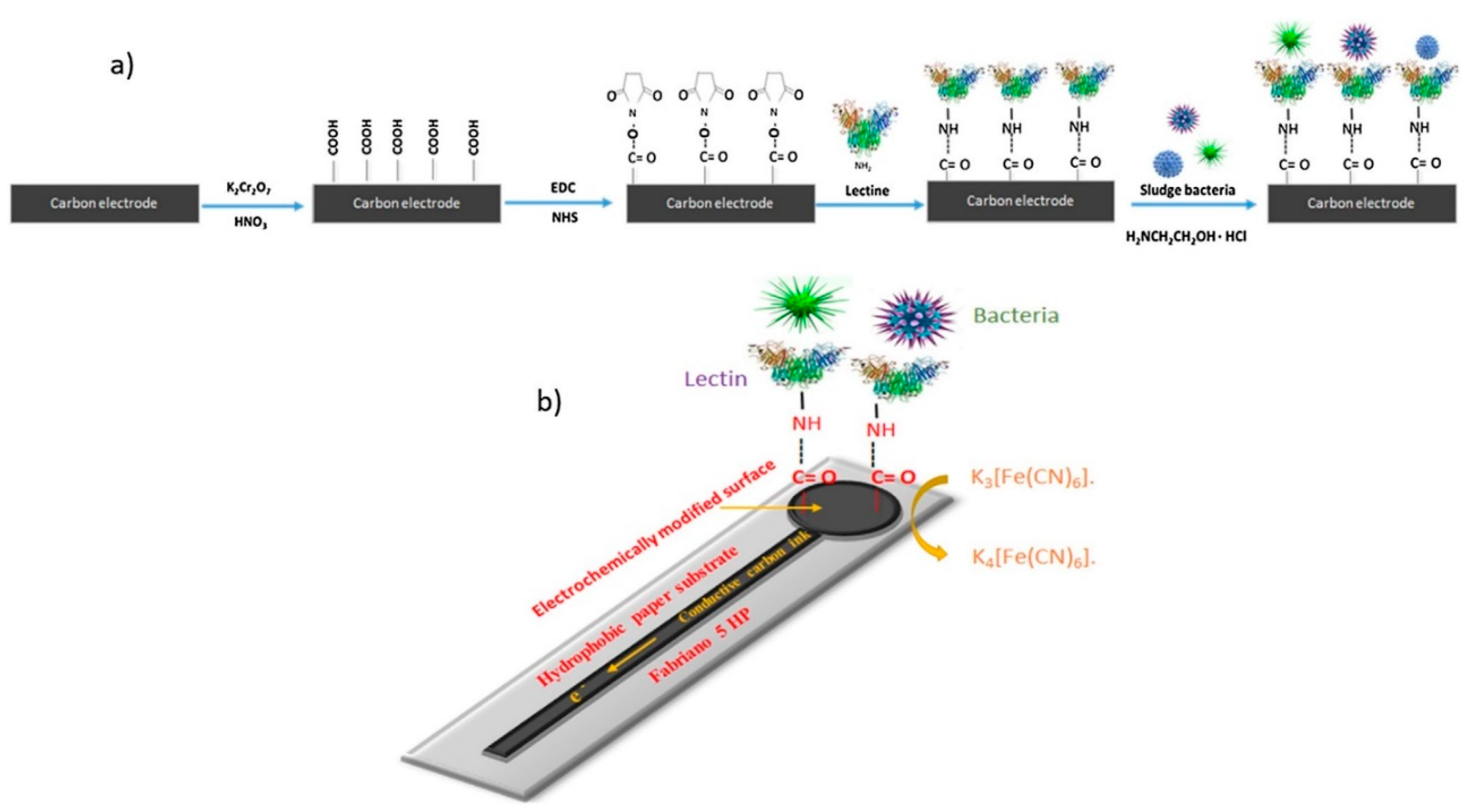
3.1. Paper-Based Assay Methodology
3.2. Microfluidic Detection Platforms for Water Quality Assessment
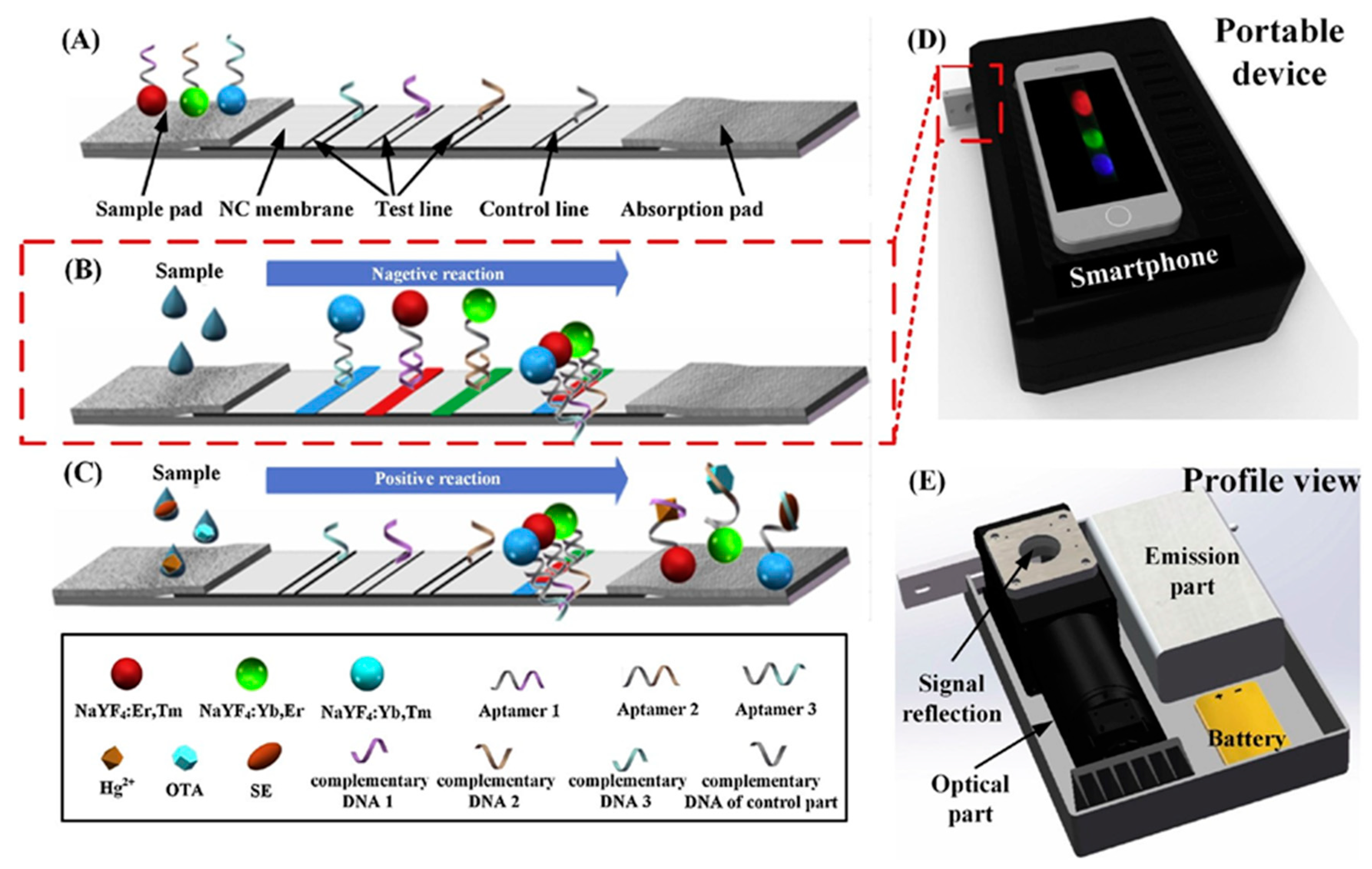
3.3. Lateral Flow Devices for Contaminant Detection
4. Conclusion and Future Prospects
Funding
Conflicts of Interest
References
- WHO Report. 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 17 August 2019).
- Kumar, S.; Bhanjana, G.; Dilbaghi, N.; Kumar, R.; Umar, A. Fabrication and characterization of highly sensitive and selective arsenic sensor based on ultra-thin graphene oxide nanosheets. Sens. Actuators B Chem. 2016, 227, 29–34. [Google Scholar] [CrossRef]
- Halden, R.U. Epistemology of contaminants of emerging concern and literature meta-analysis. J. Hazard. Mater. 2015, 282, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bhanjana, G.; Dilbaghi, N.; Umar, A. Zinc oxide nanocones as potential scaffold for the fabrication of ultra-high sensitive hydrazine chemical sensor. Ceram. Int. 2015, 41, 3101–3108. [Google Scholar] [CrossRef]
- Gruenberg, J.; van der Goot, F.G. Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 2006, 7, 495–504. [Google Scholar] [CrossRef]
- Cabral, J.P. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef]
- Ali, M.; Nelson, A.R.; Lopez, A.L.; Sack, D.A. Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 2015, 9, e0003832. [Google Scholar] [CrossRef]
- Park, J.; Kim, J.S.; Kim, S.; Shin, E.; Oh, K.H.; Kim, Y.; Kim, C.H.; Hwang, M.A.; Jin, C.M.; Na, K.; et al. A waterborne outbreak of multiple diarrhoeagenic Escherichia coli infections associated with drinking water at a school camp. Int. J. Infect. Dis. 2018, 66, 45–50. [Google Scholar] [CrossRef]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 shiga toxin-producing Escherichia coli-a poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef]
- Karmali, M.A. Factors in the emergence of serious human infections associated with highly pathogenic strains of shiga toxin-producing Escherichia coli. Int. J. Med. Microbiol. 2018, 308, 1067–1072. [Google Scholar] [CrossRef]
- Van Nevel, S.; Koetzsch, S.; Proctor, C.R.; Besmer, M.D.; Prest, E.I.; Vrouwenvelder, J.S.; Knezev, A.; Boon, N.; Hammes, F. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring. Water Res. 2017, 113, 191–206. [Google Scholar] [CrossRef]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Nesatyy, V.J.; Suter, M.J.F. Proteomics for the analysis of environmental stress responses in organisms. Environ. Sci. Technol. 2007, 41, 6891–6900. [Google Scholar] [CrossRef] [PubMed]
- Párraga-Niño, N.; Quero, S.; Ventós-Alfonso, A.; Uria, N.; Castillo-Fernandez, O.; Ezenarro, J.J.; Muñoz, F.X.; Garcia-Nuñez, M.; Sabrià, M. New system for the detection of Legionella pneumophila in water samples. Talanta 2018, 189, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, O.; Casas, S.; Galvañ, C.; Lucena, F.; Bosch, A.; Galofré, B.; Mesa, J.; Jofre, J.; Bernat, X. Direct ultrafiltration performance and membrane integrity monitoring by microbiological analysis. Water Res. 2015, 83, 121–131. [Google Scholar] [CrossRef]
- Kim, U.; Ghanbari, S.; Ravikumar, A.; Seubert, J.; Figueira, S. Rapid, affordable, and point-of-care water monitoring via a microfluidic DNA sensor and a mobile interface for global health. IEEE J. Transl. Eng. Health Med. 2013, 1, 3700207. [Google Scholar]
- Clark, K.D.; Purslow, J.A.; Pierson, S.A.; Nacham, O.; Anderson, J.L. Rapid preconcentration of viable bacteria using magnetic ionic liquids for PCR amplification and culture-based diagnostics. Anal. Bioanal. Chem. 2017, 409, 4983–4991. [Google Scholar] [CrossRef]
- Aw, T.G.; Rose, J.B. Detection of pathogens in water: From phylochips to qPCR to pyrosequencing. Curr. Opin. Biotechnol. 2012, 23, 422–430. [Google Scholar] [CrossRef]
- Zulkifli, S.N.; Rahim, H.A.; Lau, W.J. Detection of contaminants in water supply: A review on state-of-the-art monitoring technologies and their applications. Sens. Actuators B Chem. 2018, 255, 2657–2689. [Google Scholar] [CrossRef]
- Jimenez, M.; Miller, B.; Bridle, H.L. Efficient separation of small microparticles at high flowrates using spiral channels: Application to waterborne pathogens. Chem. Eng. Sci. 2017, 157, 247–254. [Google Scholar] [CrossRef]
- Hassan, A.H.A.; Bergua, J.F.; Morales-Narváez, E.; Mekoçi, A. Validity of a single antibody-based lateral flow immunoassay depending on graphene oxide for highly sensitive determination of E. coli O157: H7 in minced beef and river water. Food Chem. 2019, 297, 124965. [Google Scholar] [CrossRef]
- Wang, Y.; Salazar, J.K. Culture-independent rapid detection methods for bacterial pathogens and toxins in food matrices. Compr. Rev. Food Sci. Food Saf. 2016, 5, 183–205. [Google Scholar] [CrossRef]
- Mandal, P.K.; Biswas, A.K.; Choi, K.; Pal, U.K. Methods for rapid detection of foodborne pathogens: An overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. Rapid detection of Escherichia coli O157: H7 and Salmonella Typhimurium in foods using an electrochemical immunosensor based on screen-printed interdigitated microelectrode and immunomagnetic separation. Talanta 2016, 148, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Skjerve, E.; Rørvik, L.M.; Olsvik, O. Detection of Listeria monocytogenes in foods by immunomagnetic separation. Appl. Environ. Microbiol. 1990, 56, 3478–3481. [Google Scholar] [PubMed]
- Hyeon, J.Y.; Deng, X. Rapid detection of Salmonella in raw chicken breast using real-time PCR combined with immunomagnetic separation and whole genome amplification. Food Microbiol. 2017, 63, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Kim, M.; Schlesinger, D.; Kranz, C.; Ha, S.; Ha, J.; Slauch, J.; Baek, S.; Moe, C. Immunomagnetic separation combined with RT-qPCR for determining the efficacy of disinfectants against human noroviruses. J. Infect. Public Health 2015, 8, 145–154. [Google Scholar] [CrossRef]
- Mao, Y.; Huang, X.; Xiong, S.; Xu, H.; Aguilar, Z.P.; Xiong, Y. Large-volume immunomagnetic separation combined with multiplex PCR assay for simultaneous detection of Listeria monocytogenes and Listeria ivanovii in lettuce. Food Control 2016, 59, 601–608. [Google Scholar] [CrossRef]
- Dwivedi, H.P.; Jaykus, L.A. Detection of pathogens in foods: The current state-of-the-art and future directions. Crit. Rev. Microbiol. 2011, 37, 40–63. [Google Scholar] [CrossRef]
- Musso, D.; La Scola, B. Laboratory diagnosis of leptospirosis: A challenge. J. Microbiol. Immunol. Infect. 2013, 46, 245–252. [Google Scholar] [CrossRef]
- Connelly, J.T.; Baeumner, A.J. Biosensors for the detection of waterborne pathogens. Anal. Bioanal. Chem. 2012, 402, 117–127. [Google Scholar] [CrossRef]
- Jain, S.; Chattopadhyay, S.; Jackeray, R.; Abid, C.Z.; Kohli, G.S.; Singh, H. Highly sensitive detection of Salmonella typhi using surface aminated polycarbonate membrane enhanced-ELISA. Biosens. Bioelectron. 2012, 31, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.L.; Chuang, C.C.; Liang, C.C.; Chiao, D.J.; Wu, H.L.; Wu, Y.P.; Lin, F.P.; Shyu, R.H. Rapid and sensitive detection of Yersinia pestis by lateral-flow assay in simulated clinical samples. BMC Infect. Dis. 2018, 18, 402. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Blanch, A.R. Feasibility of methods based on nucleic acid amplification techniques to fulfil the requirements for microbiological analysis of water quality. J. Appl. Microbiol. 2010, 109, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Saccà, M.L.; Ferrero, V.E.V.; Loos, R.; Di Lenola, M.; Tavazzi, S.; Grenni, P.; Ademollo, N.; Patrolecco, L.; Huggett, J.; Caracciolo, A.B.; et al. Chemical mixtures and fluorescence in situ hybridization analysis of natural microbial community in the Tiber river. Sci. Total Environ. 2019, 673, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, H.; Wu, S.; Liu, Y.; Wei, W.; Zhang, Y.; Wei, M.; Liu, S. Manifold methods for telomerase activity detection based on various unique probes. TrAC Trends Anal. Chem. 2018, 105, 404–412. [Google Scholar] [CrossRef]
- Silva, D.M.; Domingues, L. On the track for an efficient detection of Escherichia coli in water: A review on PCR-based methods. Ecotoxicol. Environ. Saf. 2015, 113, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef]
- Brandt, J.; Albertsen, M. Investigation of detection limits and the influence of DNA extraction and primer choice on the observed microbial communities in drinking water samples using 16S rRNA gene amplicon sequencing. Front. Microbiol. 2018, 9, 2140. [Google Scholar] [CrossRef]
- Meredith, N.A.; Quinn, C.; Cate, D.M.; Reilly, T.H.; Volckens, J.; Henry, C.S. Paper-based analytical devices for environmental analysis. Analyst 2016, 141, 1874–1887. [Google Scholar] [CrossRef]
- Carrell, C.; Kava, A.; Nguyen, M.; Menger, R.; Munshi, Z.; Call, Z.; Nussbaum, M.; Henry, C. Beyond the lateral flow assay: A review of paper-based microfluidics. Microelectron. Eng. 2019, 206, 45–54. [Google Scholar] [CrossRef]
- Ding, X.; Mauk, M.G.; Yin, K.; Kadimisetty, K.; Liu, C. Interfacing pathogen detection with smartphones for point-of-care applications. Anal. Chem. 2018, 91, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.I.G.; Jayawardane, B.M.; Kolev, S.D.; McKelvie, I.D. Developments of microfluidic paper-based analytical devices (μPADs) for water analysis: A review. Talanta 2018, 177, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Kaneta, T.; Alahmad, W.; Varanusupakul, P. Microfluidic paper-based analytical devices with instrument-free detection and miniaturized portable detectors. Appl. Spectrosc. Rev. 2019, 54, 117–141. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Zhao, F.; Ge, L.; Zhao, X.; Zhang, H.; Huang, X.; Zeng, J.; Lv, J. Establishment of a highly sensitive lateral-flow test method for the determination of Vibrio parahemolyticus. J. Food Saf. Qual. 2018, 9, 910–913. [Google Scholar]
- Melnik, S.; Neumann, A.C.; Karongo, R.; Dirndorfer, S.; Stübler, M.; Ibl, V.; Niessner, R.; Knopp, D.; Stoger, E. Cloning and plant-based production of antibody MC 10E7 for a lateral flow immunoassay to detect [4-arginine] microcystin in freshwater. Plant Biotechnol. J. 2018, 16, 27–38. [Google Scholar] [CrossRef]
- Akter, S.; Kustila, T.; Leivo, J.; Muralitharan, G.; Vehniäinen, M.; Lamminmäki, U. Noncompetitive chromogenic lateral-flow immunoassay for simultaneous detection of microcystins and nodularin. Biosensors 2019, 9, 79. [Google Scholar] [CrossRef]
- Taitt, C.R.; Anderson, G.P.; Ligler, F.S. Evanescent wave fluorescence biosensors: Advances of the last decade. Biosens. Bioelectron. 2016, 76, 103–112. [Google Scholar] [CrossRef]
- Kudo, H.; Yamada, K.; Watanabe, D.; Suzuki, K.; Citterio, D. Paper-based analytical device for zinc ion quantification in water samples with power-free analyte concentration. Micromachines 2017, 8, 127. [Google Scholar] [CrossRef]
- Mettakoonpitak, J.; Boehle, K.; Nantaphol, S.; Teengam, P.; Adkins, J.A.; Srisa-Art, M.; Henry, C.S. Electrochemistry on paper-based analytical devices: A review. Electroanalysis 2016, 28, 1420–1436. [Google Scholar] [CrossRef]
- Bhanjana, G.; Dilbaghi, N.; Kumar, R.; Kumar, S. Zinc oxide quantum dots as efficient electron mediator for ultrasensitive and selective electrochemical sensing of mercury. Electrochim. Acta 2015, 178, 361–367. [Google Scholar] [CrossRef]
- Bhanjana, G.; Rana, P.; Chaudhary, G.R.; Dilbaghi, N.; Kim, K.H.; Kumar, S. Manganese oxide nanochips as a novel electrocatalyst for direct redox sensing of hexavalent chromium. Sci. Rep. 2019, 9, 8050. [Google Scholar] [CrossRef] [PubMed]
- Burnham, S.; Hu, J.; Anany, H.; Brovko, L.; Deiss, F.; Derda, R.; Griffiths, M.W. Towards rapid on-site phage-mediated detection of generic Escherichia coli in water using luminescent and visual readout. Anal. Bioanal. Chem. 2014, 406, 5685–5693. [Google Scholar] [CrossRef] [PubMed]
- Morales-Narváez, E.; Naghdi, T.; Zor, E.; Merkoçi, A. Photoluminescent lateral-flow immunoassay revealed by graphene oxide: Highly sensitive paper-based pathogen detection. Anal. Chem. 2015, 87, 8573–8577. [Google Scholar] [CrossRef]
- Ma, S.; Tang, Y.; Liu, J.; Wu, J. Visible paper chip immunoassay for rapid determination of bacteria in water distribution system. Talanta 2014, 120, 135–140. [Google Scholar] [CrossRef]
- Gunda, N.S.K.; Dasgupta, S.; Mitra, S.K. DipTest: A litmus test for E. coli detection in water. PLoS ONE 2017, 12, e0183234. [Google Scholar] [CrossRef]
- Rengaraj, S.; Cruz-Izquierdo, Á.; Scott, J.L.; Di Lorenzo, M. Impedimetric paper-based biosensor for the detection of bacterial contamination in water. Sens. Actuators B Chem. 2018, 265, 50–58. [Google Scholar] [CrossRef]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef]
- San Park, T.; Yoon, J.Y. Smartphone detection of Escherichia coli from field water samples on paper microfluidics. IEEE Sens. J. 2014, 15, 1902–1907. [Google Scholar] [CrossRef]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef]
- Kim, M.; Jung, T.; Kim, Y.; Lee, C.; Woo, K.; Seol, J.H.; Yang, S. A microfluidic device for label-free detection of Escherichia coli in drinking water using positive dielectrophoretic focusing, capturing, and impedance measurement. Biosens. Bioelectron. 2015, 74, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Lyu, J.; Shi, J.; Tan, F.; Yang, M. A polymeric microfluidic device integrated with nanoporous alumina membranes for simultaneous detection of multiple foodborne pathogens. Sens. Actuators B Chem. 2016, 225, 312–318. [Google Scholar] [CrossRef]
- Hossain, S.Z.; Ozimok, C.; Sicard, C.; Aguirre, S.D.; Ali, M.M.; Li, Y.; Brennan, J.D. Multiplexed paper test strip for quantitative bacterial detection. Anal. Bioanal. Chem. 2012, 403, 1567–1576. [Google Scholar]
- Chen, Y.; Cheng, N.; Xu, Y.; Huang, K.; Luo, Y.; Xu, W. Point-of-care and visual detection of P. aeruginosa and its toxin genes by multiple LAMP and lateral flow nucleic acid biosensor. Biosens. Bioelectron. 2016, 81, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Yang, Y.; He, R.; Park, Y.I.; Lee, A.; Bai, D.; Li, F.; Lu, T.J.; Xu, F.; Lin, M. Lateral flow aptamer assay integrated smartphone-based portable device for simultaneous detection of multiple targets using upconversion nanoparticles. Sens. Actuators B Chem. 2018, 276, 48–56. [Google Scholar] [CrossRef]
- Choi, J.R.; Yong, K.W.; Tang, R.; Gong, Y.; Wen, T.; Li, F.; Pingguan-Murphy, B.; Bai, D.; Xu, F. Advances and challenges of fully integrated paper-based point-of-care nucleic acid testing. TrAC Trends Anal. Chem. 2017, 93, 37–50. [Google Scholar] [CrossRef]
- Singh, A.; Lantigua, D.; Meka, A.; Taing, S.; Pandher, M.; Camci-Unal, G. Paper-based sensors: Emerging themes and applications. Sensors 2018, 18, 2838. [Google Scholar] [CrossRef]
- Cate, D.M.; Adkins, J.A.; Mettakoonpitak, J.; Henry, C.S. Recent developments in paper-based microfluidic devices. Anal. Chem. 2014, 87, 19–41. [Google Scholar] [CrossRef]
- Pelton, R. Bioactive paper provides a low-cost platform for diagnostics. TrAC Trends Anal. Chem. 2009, 28, 925–942. [Google Scholar] [CrossRef]
- Nery, E.W.; Kubota, L.T. Sensing approaches on paper-based devices: A review. Anal. Bioanal. Chem. 2013, 405, 7573–7595. [Google Scholar] [CrossRef]
- Alhan, S.; Nehra, M.; Dilbaghi, N.; Singhal, N.K.; Kim, K.H.; Kumar, S. Potential use of ZnO@ activated carbon nanocomposites for the adsorptive removal of Cd2+ ions in aqueous solutions. Environ. Res. 2019, 173, 411–418. [Google Scholar] [CrossRef]
- Madhura, L.; Singh, S.; Kanchi, S.; Sabela, M.; Bisetty, K. Nanotechnology-based water quality management for wastewater treatment. Environ. Chem. Lett. 2019, 17, 65–121. [Google Scholar] [CrossRef]
- Kumar, S.; Nehra, M.; Kedia, D.; Dilbaghi, N.; Tankeshwar, K.; Kim, K.H. Nanodiamonds: Emerging face of future nanotechnology. Carbon 2019, 143, 678–699. [Google Scholar] [CrossRef]
- Vikesland, P.J. Nanosensors for water quality monitoring. Nat. Nanotechnol. 2018, 13, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Artiles, M.S.; Rout, C.S.; Fisher, T.S. Graphene-based hybrid materials and devices for biosensing. Adv. Drug Deliv. Rev. 2011, 63, 1352–1360. [Google Scholar] [CrossRef]
- Pandey, C.M.; Tiwari, I.; Singh, V.N.; Sood, K.N.; Sumana, G.; Malhotra, B.D. Highly sensitive electrochemical immunosensor based on graphene-wrapped copper oxide-cysteine hierarchical structure for detection of pathogenic bacteria. Sens. Actuators B Chem. 2017, 238, 1060–1069. [Google Scholar] [CrossRef]
- Lu, J.; Ge, S.; Ge, L.; Yan, M.; Yu, J. Electrochemical DNA sensor based on three-dimensional folding paper device for specific and sensitive point-of-care testing. Electrochim. Acta 2012, 80, 334–341. [Google Scholar] [CrossRef]
- Ali, M.M.; Brown, C.L.; Jahanshahi-Anbuhi, S.; Kannan, B.; Li, Y.; Filipe, C.D.; Brennan, J.D. A printed multicomponent paper sensor for bacterial detection. Sci. Rep. 2017, 7, 12335. [Google Scholar] [CrossRef]
- Gosselin, D.; Belgacem, M.N.; Joyard-Pitiot, B.; Baumlin, J.M.; Navarro, F.; Chaussy, D.; Berthier, J. Low-cost embossed-paper micro-channels for spontaneous capillary flow. Sens. Actuators B 2017, 248, 395–401. [Google Scholar] [CrossRef]
- Busa, L.; Mohammadi, S.; Maeki, M.; Ishida, A.; Tani, H.; Tokeshi, M. Advances in microfluidic paper-based analytical devices for food and water analysis. Micromachines 2016, 7, 86. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, F.; Cai, G.; Li, Y.; Lin, J. Rapid and sensitive detection of Escherichia coli O157: H7 using coaxial channel-based DNA extraction and microfluidic PCR. J. Dairy Sci. 2018, 101, 9736–9746. [Google Scholar] [CrossRef]
- Salman, A.; Carney, H.; Bateson, S.; Ali, Z. Shunting microfluidic PCR device for rapid bacterial detection. Talanta 2020, 207, 120303. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ju, R.; Sekine, S.; Zhang, D.; Zhuang, S.; Yamaguchi, Y. All-in-one microfluidic device for on-site diagnosis of pathogens based on integrated continuous flow PCR and electrophoresis biochip. Lab Chip 2019, 19, 2663–2668. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.; Zaferani, M.; Cheong, S.H.; Abbaspourrad, A. Pathogenic bacteria detection using RNA-based loop-mediated isothermal-amplification-assisted nucleic acid amplification via droplet microfluidics. ACS Sens. 2019, 4, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, L.; Cai, G.; Liu, N.; Liao, M.; Li, Y.; Zhang, X.; Lin, J. A microfluidic biosensor for online and sensitive detection of Salmonella typhimurium using fluorescence labeling and smartphone video processing. Biosens. Bioelectron. 2019, 140, 111333. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Yew, M.; Ren, Y.; Koh, K.S.; Sun, C.; Snape, C. A review of state-of-the-art microfluidic technologies for environmental applications: Detection and remediation. Glob. Chall. 2019, 3, 1800060. [Google Scholar] [CrossRef]
- Fernández-la-Villa, A.; Pozo-Ayuso, D.F.; Castaño-Álvarez, M. Microfluidics and electrochemistry: An emerging tandem for next-generation analytical microsystems. Curr. Opin. Electrochem. 2019, 15, 175–185. [Google Scholar] [CrossRef]
- Loo, J.F.; Ho, A.H.; Turner, A.P.; Mak, W.C. Integrated printed microfluidic biosensors. Trends Biotechnol. 2019, 37, 1104–1120. [Google Scholar] [CrossRef]
- Ramalingam, N.; Rui, Z.; Liu, H.B.; Dai, C.C.; Kaushik, R.; Ratnaharika, B.; Gong, H.Q. Real-time PCR-based microfluidic array chip for simultaneous detection of multiple waterborne pathogens. Sens. Actuators B Chem. 2010, 145, 543–552. [Google Scholar] [CrossRef]
- Ishii, S.; Kitamura, G.; Segawa, T.; Kobayashi, A.; Miura, T.; Sano, D.; Okabe, S. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl. Environ. Microbiol. 2014, 80, 7505–7511. [Google Scholar] [CrossRef]
- Ishii, S.; Segawa, T.; Okabe, S. Simultaneous quantification of multiple food-and waterborne pathogens by use of microfluidic quantitative PCR. Appl. Environ. Microbiol. 2013, 79, 2891–2898. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Song, J.; Cho, B.; Hong, S.; Hoxha, O.; Kang, T.; Kim, D.; Lee, L.P. Bubble-free rapid microfluidic PCR. Biosens. Bioelectron. 2019, 126, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, C. A novel paper-based microfluidic enhanced chemiluminescence biosensor for facile, reliable and highly-sensitive gene detection of Listeria monocytogenes. Sens. Actuators B Chem. 2015, 209, 399–406. [Google Scholar] [CrossRef]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of pathogenic microorganisms by microfluidics based analytical methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef]
- Safavieh, M.; Ahmed, M.U.; Tolba, M.; Zourob, M. Microfluidic electrochemical assay for rapid detection and quantification of Escherichia coli. Biosens. Bioelectron. 2012, 31, 523–528. [Google Scholar] [CrossRef]
- San Park, T.; Li, W.; McCracken, K.E.; Yoon, J.Y. Smartphone quantifies Salmonella from paper microfluidics. Lab Chip 2013, 13, 4832–4840. [Google Scholar] [CrossRef]
- Sun, L.; Jiang, Y.; Pan, R.; Li, M.; Wang, R.; Chen, S.; Fu, S.; Man, C. A novel, simple and low-cost paper-based analytical device for colorimetric detection of Cronobacter spp. Anal. Chim. Acta 2018, 1036, 80–88. [Google Scholar] [CrossRef]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; DeRosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics integrated biosensors: A leading technology towards lab-on-a-chip and sensing applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef]
- Mirasoli, M.; Guardigli, M.; Michelini, E.; Roda, A. Recent advancements in chemical luminescence-based lab-on-chip and microfluidic platforms for bioanalysis. J. Pharm. Biomed. Anal. 2014, 87, 36–52. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. Electrochemical biosensors for rapid detection of Escherichia coli O157: H7. Talanta 2017, 162, 511–522. [Google Scholar] [CrossRef]
- Yao, L.; Wang, L.; Huang, F.; Cai, G.; Xi, X.; Lin, J. A microfluidic impedance biosensor based on immunomagnetic separation and urease catalysis for continuous-flow detection of E. coli O157: H7. Sens. Actuators B Chem. 2018, 259, 1013–1021. [Google Scholar] [CrossRef]
- Banerjee, R.; Jaiswal, A. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst 2018, 143, 1970–1996. [Google Scholar] [CrossRef] [PubMed]
- Dalirirad, S.; Steckl, A.J. Aptamer-based lateral flow assay for point of care cortisol detection in sweat. Sens. Actuators B Chem. 2019, 283, 79–86. [Google Scholar] [CrossRef]
- Ma, Q.; Yao, J.; Yuan, S.; Liu, H.; Wei, N.; Zhang, J.; Shan, W. Development of a lateral flow recombinase polymerase amplification assay for rapid and visual detection of Cryptococcus neoformans/C. gattii in cerebral spinal fluid. BMC Infect. Dis. 2019, 19, 108. [Google Scholar] [CrossRef] [PubMed]
- Pöhlmann, C.; Dieser, I.; Sprinzl, M. A lateral flow assay for identification of Escherichia coli by ribosomal RNA hybridisation. Analyst 2014, 139, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Raeisossadati, M.J.; Danesh, N.M.; Borna, F.; Gholamzad, M.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Lateral flow based immunobiosensors for detection of food contaminants. Biosens. Bioelectron. 2016, 86, 235–246. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, H.; Zhang, P.; Sun, C.; Wang, X.; Wang, X.; Yang, R.; Wang, C.; Zhou, L. Rapid multiplex detection of 10 foodborne pathogens with an up-converting phosphor technology-based 10−channel lateral flow assay. Sci. Rep. 2016, 6, 21342. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]

| S. No. | Type | Analyte | Substrate | Transduction Platform | Detection Limit (CFU/mL) | Analysis Time | Cost | Lifetime | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Paper-based | E. coli | Filter paper | Fluorescence | 10 | 5.5 h | Moderate | Single use | [54] |
| 2 | Salmonella spp. | CdSe@ZnS QDs decorated paper strips | Fluorescence | 3.8 | - | Moderate | Single use | [55] | |
| 3 | E. coli | AuNP decorated PDMS paper chips | Optical immunoassay | 57 | - | High | Reusable | [56] | |
| 4 | E. coli | Litmus paper | Colorimetry DipTest | 2 × 105 to 4 × 104 | - | Very low | Single use | [57] | |
| 5 | Bacterial Contaminant | Screen printed carbon electrode | Electrochemical impedance | 2 × 103 | Moderate | Reusable | [58] | ||
| 6 | Microfluidic | L. monocytogenes, E. coli, S. enteric | Color-producing compounds deposited on µPAD | Colorimetry | 10 | 4−12 h | Moderate | Reusable | [59] |
| 7 | E. coli | Paper fibers | Gyroscope installed in smartphone | 10 | 90 s | Moderate | Reusable | [60] | |
| 8 | E. coli | AuNP-coated biochips | Cyclic voltammetry and amperometry | 50 | 8 min | High | Reusable | [61] | |
| 9 | E. coli | Dieletrophoretic microfluidic chip | Electrochemical impedance | 300 | < 1 min | High | Reusable | [62] | |
| 10 | E. coli | Nanoporous alumina membrane | Electrochemical impedance | 100 | - | Moderate | Reusable | [63] | |
| 11 | Lateral flow | E. coli | Sol-gel-derived silica ink-coated test strips | Colorimetry | 5 | 30 min | Moderate | Reusable | [64] |
| 12 | Psuedomonas aeruginosa | AuNP-conjugated nitrocellulose membrane | Visual detection | 20 | 50 min | Low | Single use | [65] | |
| 13 | Salmonella | Upconverting nanoparticles-coated paper strips | Colorimetry | 85 | 30 min | High | Single use | [66] | |
| 14 | E. coli | Flinders Technology Associates (FTA) cards and glass fibers | Colorimetry | 10−100 | - | High | Reusable | [67] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Nehra, M.; Mehta, J.; Dilbaghi, N.; Marrazza, G.; Kaushik, A. Point-of-Care Strategies for Detection of Waterborne Pathogens. Sensors 2019, 19, 4476. https://doi.org/10.3390/s19204476
Kumar S, Nehra M, Mehta J, Dilbaghi N, Marrazza G, Kaushik A. Point-of-Care Strategies for Detection of Waterborne Pathogens. Sensors. 2019; 19(20):4476. https://doi.org/10.3390/s19204476
Chicago/Turabian StyleKumar, Sandeep, Monika Nehra, Jyotsana Mehta, Neeraj Dilbaghi, Giovanna Marrazza, and Ajeet Kaushik. 2019. "Point-of-Care Strategies for Detection of Waterborne Pathogens" Sensors 19, no. 20: 4476. https://doi.org/10.3390/s19204476
APA StyleKumar, S., Nehra, M., Mehta, J., Dilbaghi, N., Marrazza, G., & Kaushik, A. (2019). Point-of-Care Strategies for Detection of Waterborne Pathogens. Sensors, 19(20), 4476. https://doi.org/10.3390/s19204476







