Online Feature Selection for Robust Classification of the Microbiological Quality of Traditional Vanilla Cream by Means of Multispectral Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vanilla Cream Samples
2.2. Microbiological Analysis
2.3. Image Acquisition and Analysis
2.4. Data Labeling
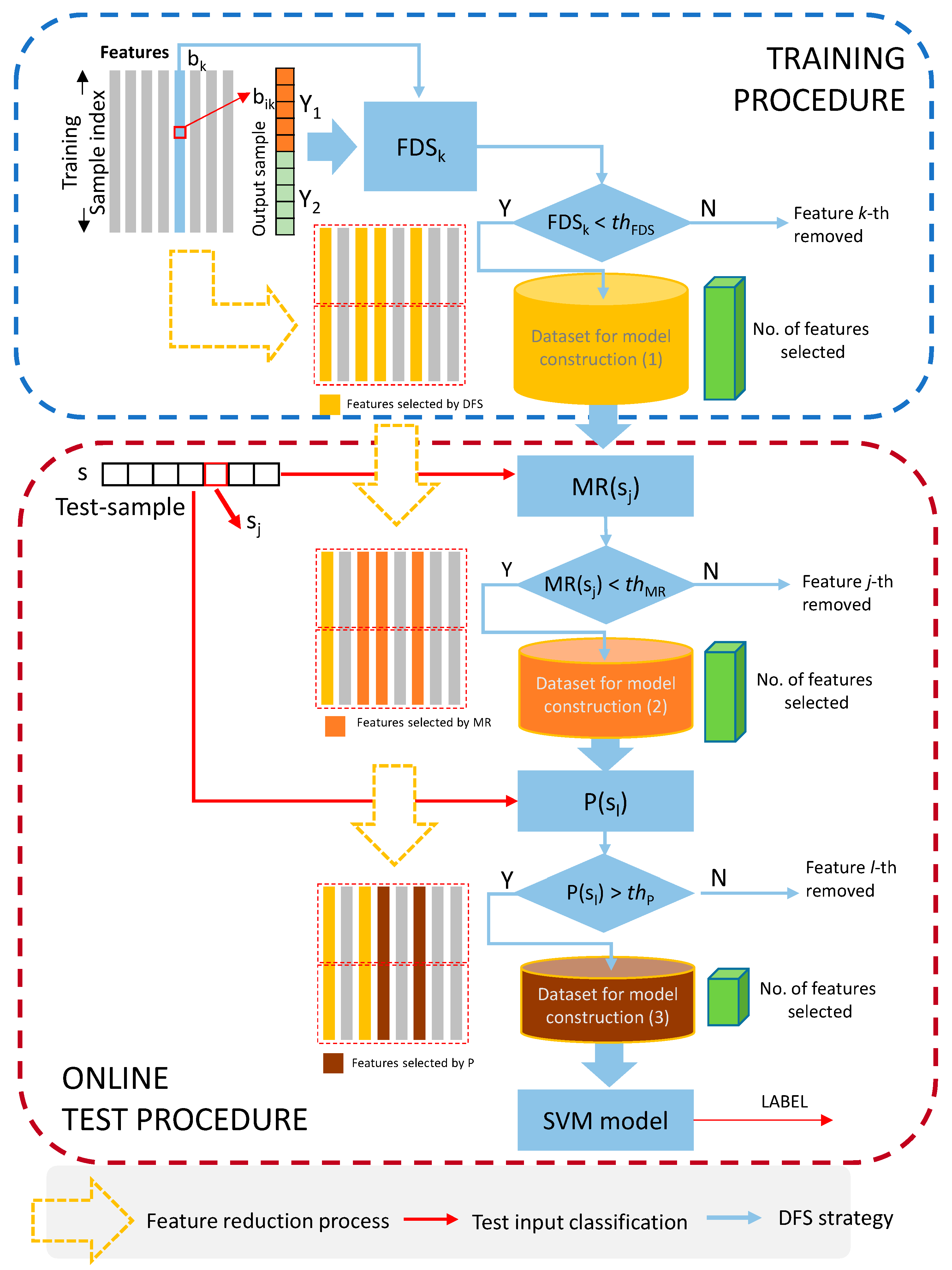
2.5. Dynamic Feature Selection (DFS) Method
2.5.1. Training-Dependent Feature Elimination Step
2.5.2. Online Test-Dependent Feature Elimination Step
3. Results
3.1. DFS Optimization Procedure
3.2. Simulation Results
3.3. Comparative Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nychas, G.-J.E.; Panagou, E.Z.; Mohareb, F. Novel approaches for food safety management and communication. Curr. Opin. Food Sci. 2016, 12, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Russ, J.C. Image analysis of foods. J. Food Sci. 2015, 80, E1974–E1987. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.; Pereira, J.A.; Silva, P.; Perestrelo, R.; Camara, J.S. Food fingerprints-A valuable tool to monitor food authenticity and safety. Food Chem. 2019, 278, 144–162. [Google Scholar] [CrossRef] [PubMed]
- ElMasry, G.; Mandour, N.; Al-Rejaie, S.; Belin, E.; Rousseau, D. Recent applications of multispectral imaging in seed phenotyping and quality monitoring—An overview. Sensors 2019, 19, 1090. [Google Scholar] [CrossRef] [PubMed]
- Su, W.H.; He, H.J.; Sun, D.W. Non-destructive and rapid evaluation of staple foods quality by using spectroscopic techniques: a review. Crit. Rev. Food Sci. Nutr. 2017, 57, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; Sun, D.W. Rapid quantification analysis and visualization of Escherichia coli loads in grass carp fish flesh by hyperspectral imaging method. Food Bioprocess Tech. 2015, 8, 951–959. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Near-infrared hyperspectral imaging in tandem with partial least squares regression and genetic algorithm for non-destructive determination and visualization of Pseudomonas loads in chicken fillets. Talanta 2013, 109, 74–83. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Sun, D.W.; Wu, D. Rapid and real-time prediction of lactic acid bacteria (LAB) in farmed salmon flesh using near-infrared (NIR) hyperspectral imaging combined with chemometric analysis. Food Res. Int. 2014, 62, 476–483. [Google Scholar] [CrossRef]
- ElMasry, G.M.; Nakauchi, S. Image analysis operations applied to hyperspectral images for non-invasive sensing of food quality—A comprehensive review. Biosyst. Eng. 2016, 142, 53–82. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review-Part II: Applications. Innov. Food Sci. Emerg. Tech. 2013, 19, 15–28. [Google Scholar] [CrossRef]
- Su, W.H.; Sun, D.W. Multispectral imaging for plant food quality analysis and visualization. Compr. Rev. Food Sci. F. 2017, 17, 220–239. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Lu, X.; Ma, F.; Chen, W.; Yang, J.; Zheng, L. Application of multispectral imaging to determine quality attributes and ripeness stage in strawberry fruit. PLoS ONE 2014, 9, e87818. [Google Scholar] [CrossRef] [PubMed]
- Dissing, B.S.; Papadopoulou, O.S.; Tassou, C.C.; Ersbøll, B.K.; Carstensen, J.M.; Panagou, E.Z.; Nychas, G.-J.E. Using multispectral imaging for spoilage detection of pork meat. Food Bioprocess Tech. 2013, 6, 2268–2279. [Google Scholar] [CrossRef]
- Wang, K.; Pu, H.; Sun, D.W. Emerging spectroscopic and spectral imaging techniques for the rapid detection of microorganisms: An overview. Compr. Rev. Food Sci. F. 2018, 17, 256–273. [Google Scholar] [CrossRef]
- Carstensen, J.M.; Hansen, M.E.; Lassen, N.K.; Hansen, P.W. Creating surface chemistry maps using multispectral vision technology. In 9th Medical Image Computing and Computer Assisted Intervention (MICCAI)—Workshop on Biophotonics Imaging for Diagnostics and Treatment, Institute of Mathematical Modelling; Ersbøll, B., Jørgensen, T.M., Eds.; Technical University of Denmark: Lyngby, Denmark, 2006. [Google Scholar]
- Mehl, P.M.; Chen, Y.R.; Kim, M.S.; Chan, D.E. Development of hyperspectral imaging technique for the detection of apple surface defects and contaminations. J. Food Eng. 2004, 61, 67–81. [Google Scholar] [CrossRef]
- Brereton, R.G. Consequences of sample size, variable selection and model validation and optimisation for predicting classification ability from analytical data. Trend. Anal. Chem. 2006, 25, 1103–1111. [Google Scholar] [CrossRef]
- Dai, Q.; Cheng, J.H.; Sun, D.W.; Zeng, X.A. Advances in feature selection methods for hyperspectral image processing in food industry applications. Crit. Rev. Food Sci. Nutr. 2015, 56, 1368–1382. [Google Scholar] [CrossRef]
- Ariana, D.P.; Lu, R. Hyperspectral waveband selection for internal defect detection of pickling cucumbers and whole pickles. Comput. Electron. Agric. 2010, 74, 137–144. [Google Scholar] [CrossRef]
- Su, W.H.; Sun, D.W. Evaluation of spectral imaging for inspection for adulterants in terms of common wheat flour, cassava flour and corn flour in organic Avatar wheat (Triticum spp.) flour. J. Food Eng. 2017, 59–69. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S.; Liu, S. Assessment of visible near-infrared hyperspectral imaging as a tool for detection of horsemeat adulteration in minced beef. Food Bioprocess Tech. 2015, 8, 1054–1062. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Rapid and non-destructive detection of chicken adulteration in minced beef using visible near-infrared hyperspectral imaging and machine learning. J. Food Eng. 2016, 170, 8–15. [Google Scholar] [CrossRef]
- Su, W.H.; Sun, D.W. Comparative assessment of feature-wavelength eligibility for measurement of water binding capacity and specific gravity of tuber using diverse spectral indices stemmed from hyperspectral images. Comput. Electron. Agric. 2016, 130, 69–82. [Google Scholar] [CrossRef]
- Su, W.H.; Bakalis, S.; Sun, D.W. Chemometrics in tandem with near infrared (NIR) hyperspectral imaging and Fourier transform mid infrared (FT-MIR) microspectroscopy for variety identification and cooking loss determination of sweet potato. Biosyst. Eng. 2019, 180, 70–86. [Google Scholar] [CrossRef]
- Pu, H.; Kamruzzaman, M.; Sun, D.-W. Selection of feature wavelengths for developing multispectral imaging systems for quality, safety and authenticity of muscle foods: A review. Trend. Food Sci. Tech. 2015, 45, 86–104. [Google Scholar] [CrossRef]
- Magna, G.; Mosciano, F.; Martinelli, E.; Di Natale, C. Unsupervised On-Line Selection of Training Features for a robust classification with drifting and faulty gas sensors. Sensor. Actuat. B: Chem. 2018, 258, 1242–1251. [Google Scholar] [CrossRef]
- Lianou, A.; Malavazos, C.; Triantafyllou, I.; Nychas, G.-J.E.; Panagou, E.Z. Rapid assessment of the microbiological quality of pasteurized vanilla cream by means of Fourier transform infrared spectroscopy in tandem with support vector machine analysis. Food Anal. Meth. 2018, 11, 840–847. [Google Scholar] [CrossRef]
- Carstensen, J.M.; Hansen, J.F. An Apparatus and a Method of Recording an Image of an Object. US Patent EP1051660, 2005. [Google Scholar]
- Ropodi, A.I.; Pavlidis, D.E.; Mohareb, F.; Panagou, E.Z.; Nychas, G.-J.E. Multispectral image analysis approach to detect adulteration of beef and pork in raw meats. Food Res. Int. 2015, 67, 12–18. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Pavlidis, D.E.; Nychas, G.-J.E. High throughput multispectral image processing with applications in food science. PLoS ONE 2015, 10, e0140122. [Google Scholar] [CrossRef]
- McLachlan, G.; Peel, D. Finite Mixture Models; Wiley Series in Probability and Statistics; John Wiley & Sons Inc.: New York, NY, USA, 2000. [Google Scholar]
- Gram, L.; Ravn, L.; Rasch, M.; Bruch, J.B.; Christiansen, A.B.; Givskov, M. Food spoilage interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002, 87, 79–97. [Google Scholar] [CrossRef]
- Mosciano, F.; Mencattini, A.; Ringeval, F.; Schuller, B.; Martinelli, E.; Di Natale, C. An array of physical sensors and an adaptive regression strategy for emotion recognition in a noisy scenario. Sensor. Actuators A Phys. 2017, 267, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Mencattini, A.; Mosciano, F.; Comes, M.C.; Di Gregorio, T.; Raguso, G.; Daprati, E.; Ringeval, F.; Schuller, B.; Di Natale, C.; Martinelli, E. An emotional modulation model as signature for the identification of children developmental disorders. Sci. Rep. 2018, 8, 14487. [Google Scholar] [CrossRef] [PubMed]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Disc. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; John Wiley & Sons: New York, NY, USA, 2012. [Google Scholar]
- Ma, F.; Yao, J.; Xie, T.; Liu, C.; Chen, W.; Chen, C.; Zheng, L. Multispectral imaging for rapid and non-destructive determination of aerobic plate count (APC) in cooked pork sausages. Food Res. Int. 2014, 62, 902–908. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Y.; Wang, Q.; Pan, W.; Ma, F.; Liu, C.; Chen, W.; Yang, J.; Zheng, L. Rapid and non-destructive identification of water-injected beef samples using multispectral image analysis. Food Chem. 2016, 190, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Tsakanikas, P.; Pavlidis, D.; Panagou, E.; Nychas, G.-J. Exploiting multispectral imaging for non-invasive contamination assessment and mapping of meat samples. Talanta 2016, 161, 606–614. [Google Scholar] [CrossRef]
- Feng, C.H.; Makino, Y.; Oshita, S.; Garcia Martin, J.F. Hyperspectral imaging and multispectral imaging as the novel techniques for detecting defects in raw and processed meat products: Current state-of-the-art research advances. Food Control 2018, 84, 165–176. [Google Scholar] [CrossRef]
- Airado-Rodríguez, D.; Høy, M.; Skaret, J.; Wold, J.P. From multispectral imaging of autofluoresence to chemical and sensory images of lipid oxidation in cod caviar paste. Talanta 2014, 122, 70–79. [Google Scholar] [CrossRef]
- Cheng, J.H.; Sun, D.W.; Qu, J.H.; Pu, H.B.; Zhang, X.C.; Song, Z.; Chen, X.; Zhang, H. Developing a multispectral imaging for simultaneous prediction of freshness indicators during chemical spoilage of grass carp fish fillet. J. Food Eng. 2016, 182, 9–17. [Google Scholar] [CrossRef]
- Fengou, L.C.; Lianou, A.; Tsakanikas, P.; Gkana, E.N.; Panagou, E.Z.; Nychas, G.-J.E. Evaluation of Fourier transform infrared spectroscopy and multispectral imaging as means of estimating the microbiological spoilage of farmed sea bream. Food Microbiol. 2019, 79, 27–34. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Chen, W.; Yang, J.; Zheng, L. Feasibility in multispectral imaging for predicting the content of bioactive compounds in intact tomato fruit. Food Chem. 2015, 173, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; He, H.; Li, E.; Li, H. Multispectral imaging for predicting sugar content of ‘Fuji’ apples. Opt. Laser Techn. 2018, 106, 280–285. [Google Scholar] [CrossRef]
- Liu, C.; Hao, G.; Su, M.; Chen, Y.; Zheng, L. Potential of multispectral imaging combined with chemometric methods for rapid detection of sucrose adulteration in tomato paste. J. Food Eng. 2017, 215, 78–83. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.-J.E. Multispectral imaging (MSI): a promising method for the detection of minced beef adulteration with horsemeat. Food Control 2017, 73, 57–63. [Google Scholar] [CrossRef]
- Løkke, M.M.; Seefeldt, H.F.; Skov, T.; Edelenbos, M. Color and textural quality of packaged wild rocket measured by multispectral imaging. Postharvest Biol. Tech. 2013, 75, 86–95. [Google Scholar] [CrossRef]
- Huang, W.; Li, J.; Wang, Q.; Chen, L. Development of a multispectral imaging system for online detection of bruises on apples. J. Food Eng. 2015, 146, 62–71. [Google Scholar] [CrossRef]
- Khodabakhshian, R.; Emadi, B.; Khojastehpour, M.; Golzarian, M.R. Determining quality and maturity of pomegranates using multispectral imaging. J. Saudi Soc. Agric. Sci. 2017, 16, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Kalkan, H.; Beriat, P.; Yardimci, Y.; Pearson, T.C. Detection of contaminated hazelnuts and ground red chilli pepper flakes by multispectral imaging. Comput. Electron. Agric. 2011, 77, 28–34. [Google Scholar] [CrossRef]
- Sendin, K.; Manley, M.; Williams, P.J. Classification of white maize defects with multispectral imaging. Food Chem. 2018, 243, 311–318. [Google Scholar] [CrossRef]
- Clemmensen, L.H.; Hansen, M.E.; Ersbøll, B.K.; Frisvad, J.C. A method for comparison of growth media in objective identification of Penicillium based on multispectral imaging. J. Microbiol. Meth. 2007, 69, 249–255. [Google Scholar] [CrossRef]
- Ebrahimi, P.; van den Berg, F.; Aunsbjerg, S.D.; Honoré, A.; Benfeldt, C.; Jensen, H.M.; Engelsen, S.B. Quantitative determination of mold growth and inhibition by multispectral imaging. Food Control 2015, 55, 82–89. [Google Scholar] [CrossRef]
- Magna, G.; Di Natale, C.; Martinelli, E. Self-repairing classification algorithms for chemical sensor array. Sensor. Actuat. B Chem. 2019, 297, 126721. [Google Scholar] [CrossRef]
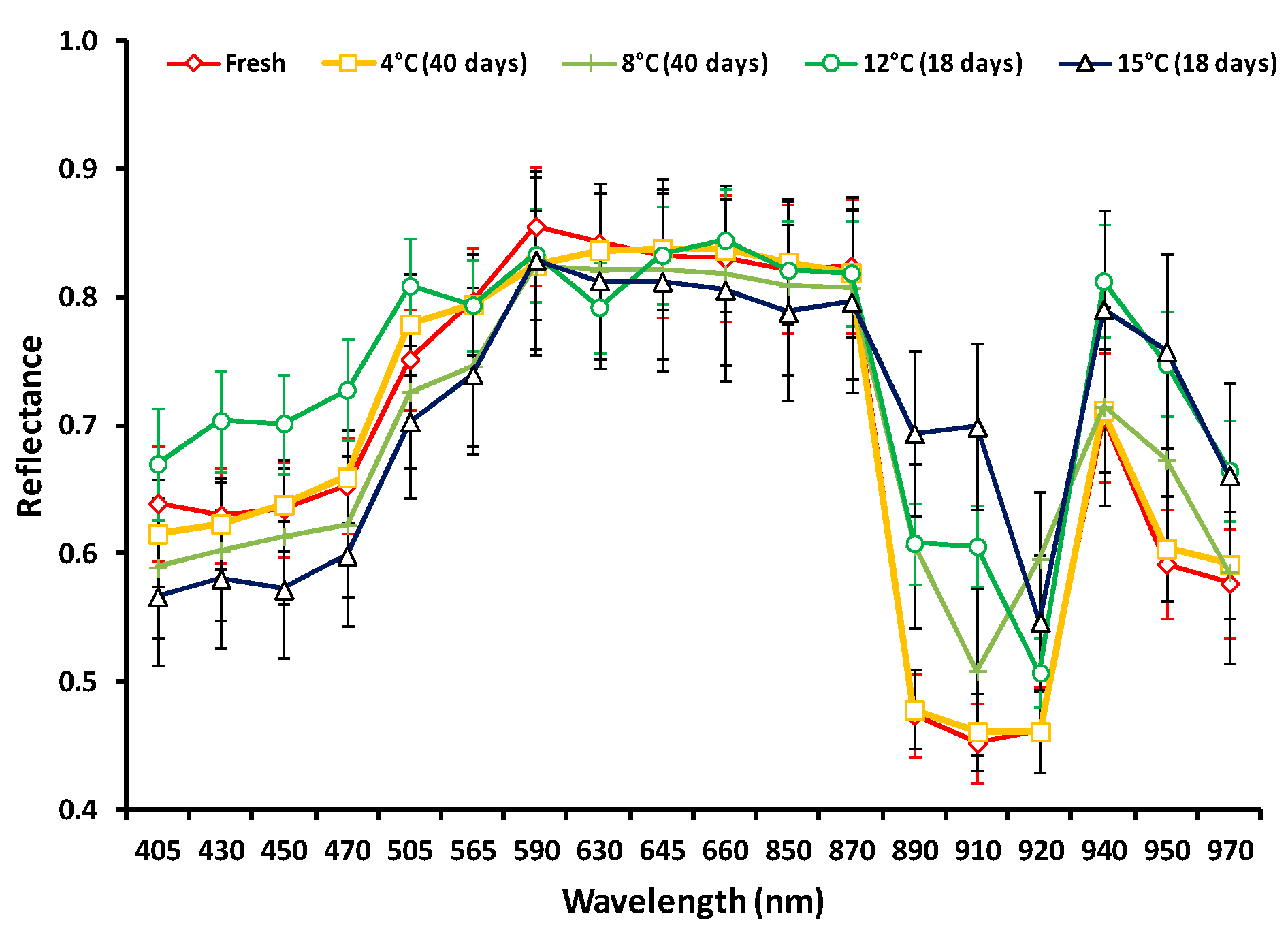



| Training Data | Validation Data | Test Data | ||||
|---|---|---|---|---|---|---|
| TVC ≤ 2 | TVC ≥ 6 | TVC ≤ 2 | TVC ≥ 6 | TVC ≤ 2 | TVC ∈ (2,6) | TVC ≥ 6 |
| 29/65 | 36/65 | 7/48 | 41/48 | 106/132 | 18/132 | 8/132 |
| Positive Assignment | Negative Assignment | |
|---|---|---|
| TVC <2 | 4/103 (4%) | 99/103 (96%) |
| TVC ∈ [2,3) | 0/7 (0%) | 7/7 (100%) |
| TVC ∈ [3,4) | 0/2 (0%) | 2/2 (100%) |
| TVC ∈ [4,5) | 0/2 (0%) | 2/2 (100%) |
| TVC ∈ [5,6) | 6/10 (60%) | 4/10 (40%) |
| TVC ≥ 6 | 7/8 (87.5%) | 1/8 (12.5%) |
| Comparative Analysis | |||
|---|---|---|---|
| Method | Accuracy | Sensitivity Per Subcategory | |
| TVC < 6 vs. TVC ≥ 6 | TVC ∈ (2 ÷ 5) | TVC ∈ (5 ÷ 6) | |
| SVM + DFS | 91.7% | 0% to SPOILED | 60% to SPOILED |
| LDA + DFS | 84.9% | 57% to SPOILED | 70% to SPOILED |
| QDA + DFS | 85.7% | 14% to SPOILED | 80% to SPOILED |
| SVM + SE | 70.2% | 40% to SPOILED | 80% to SPOILED |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lianou, A.; Mencattini, A.; Catini, A.; Di Natale, C.; Nychas, G.-J.E.; Martinelli, E.; Panagou, E.Z. Online Feature Selection for Robust Classification of the Microbiological Quality of Traditional Vanilla Cream by Means of Multispectral Imaging. Sensors 2019, 19, 4071. https://doi.org/10.3390/s19194071
Lianou A, Mencattini A, Catini A, Di Natale C, Nychas G-JE, Martinelli E, Panagou EZ. Online Feature Selection for Robust Classification of the Microbiological Quality of Traditional Vanilla Cream by Means of Multispectral Imaging. Sensors. 2019; 19(19):4071. https://doi.org/10.3390/s19194071
Chicago/Turabian StyleLianou, Alexandra, Arianna Mencattini, Alexandro Catini, Corrado Di Natale, George-John E. Nychas, Eugenio Martinelli, and Efstathios Z. Panagou. 2019. "Online Feature Selection for Robust Classification of the Microbiological Quality of Traditional Vanilla Cream by Means of Multispectral Imaging" Sensors 19, no. 19: 4071. https://doi.org/10.3390/s19194071
APA StyleLianou, A., Mencattini, A., Catini, A., Di Natale, C., Nychas, G.-J. E., Martinelli, E., & Panagou, E. Z. (2019). Online Feature Selection for Robust Classification of the Microbiological Quality of Traditional Vanilla Cream by Means of Multispectral Imaging. Sensors, 19(19), 4071. https://doi.org/10.3390/s19194071









