A Novel Method for Soil Organic Matter Determination by Using an Artificial Olfactory System
Abstract
1. Introduction
2. Materials and Methods
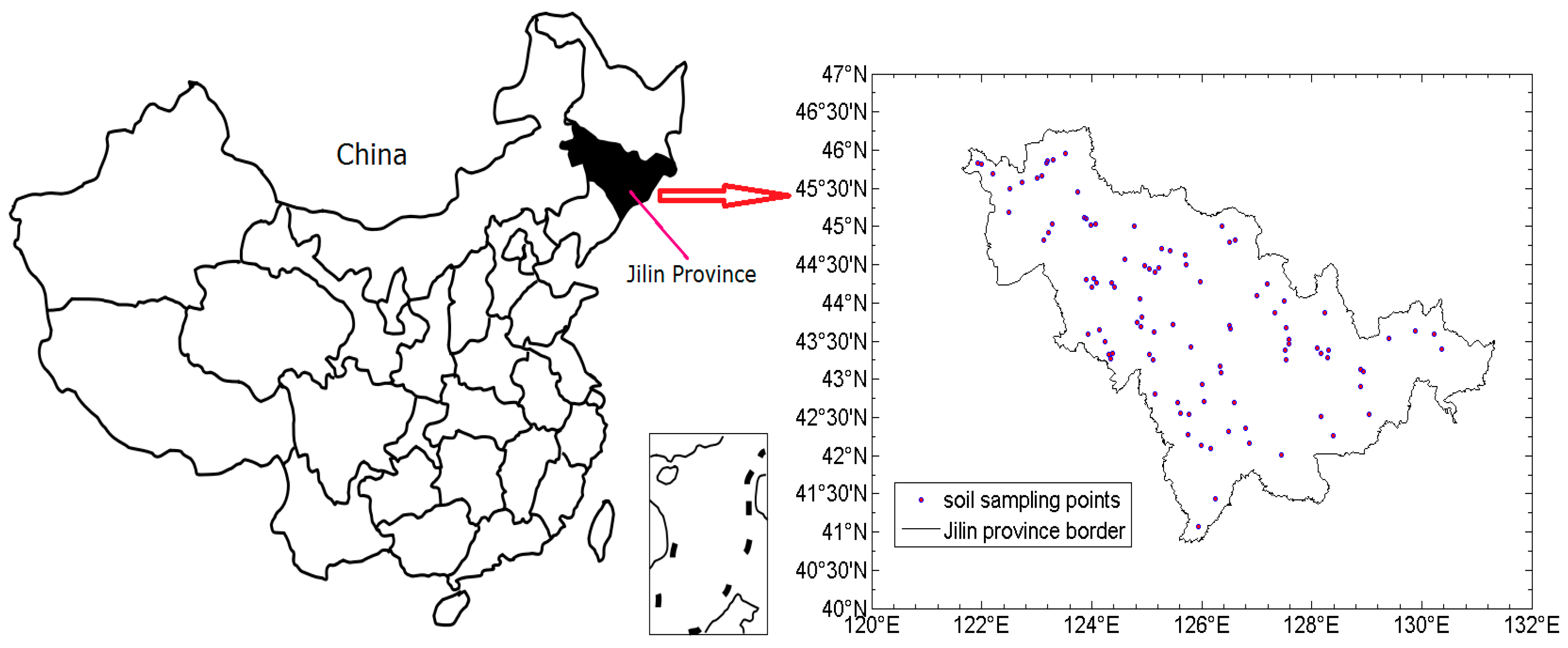
2.1. Study Area and Soil Sampling
2.2. Artificial Olfactory Measurements
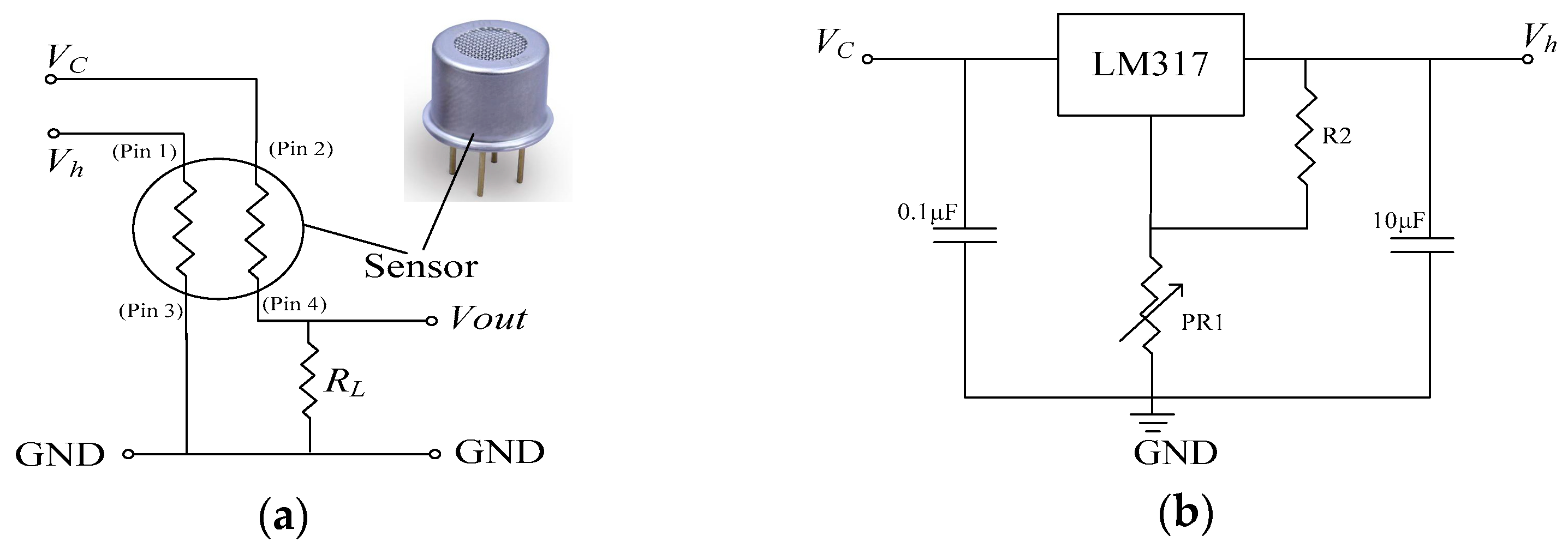
2.2.1. Measurement Setup and Data Acquisition
2.2.2. Feature Selection
2.3. Regression Calibration Models
2.3.1. BPNN
2.3.2. SVR
2.3.3. PLSR
2.4. Training Set and Validation Set
2.5. Assessment of Models
3. Results and Discussion
3.1. Chemical Analysis Results of SOM Content
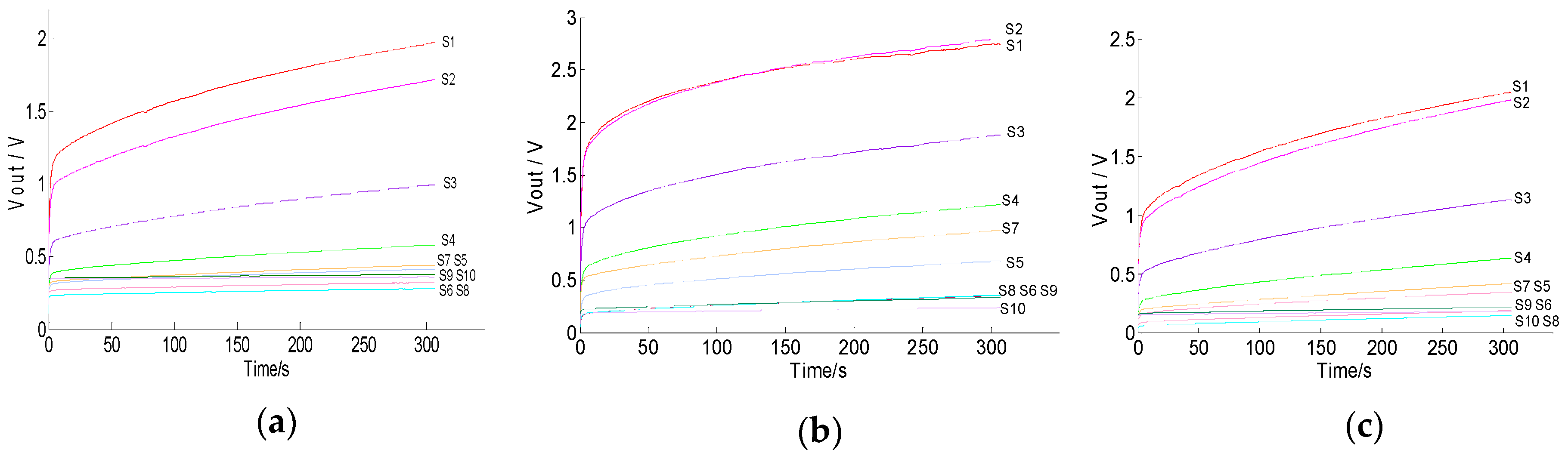
3.2. Responses of Sensors to Soil Gas
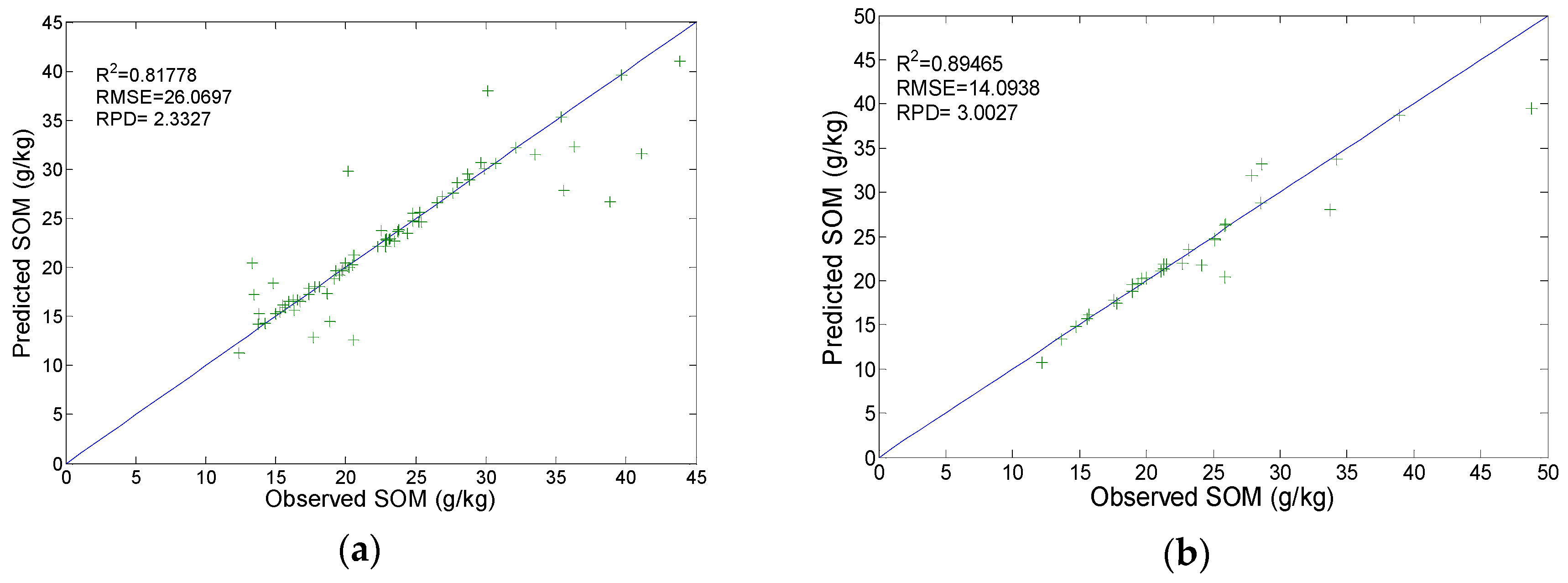
3.3. Calibration and Prediction by BPNN Model
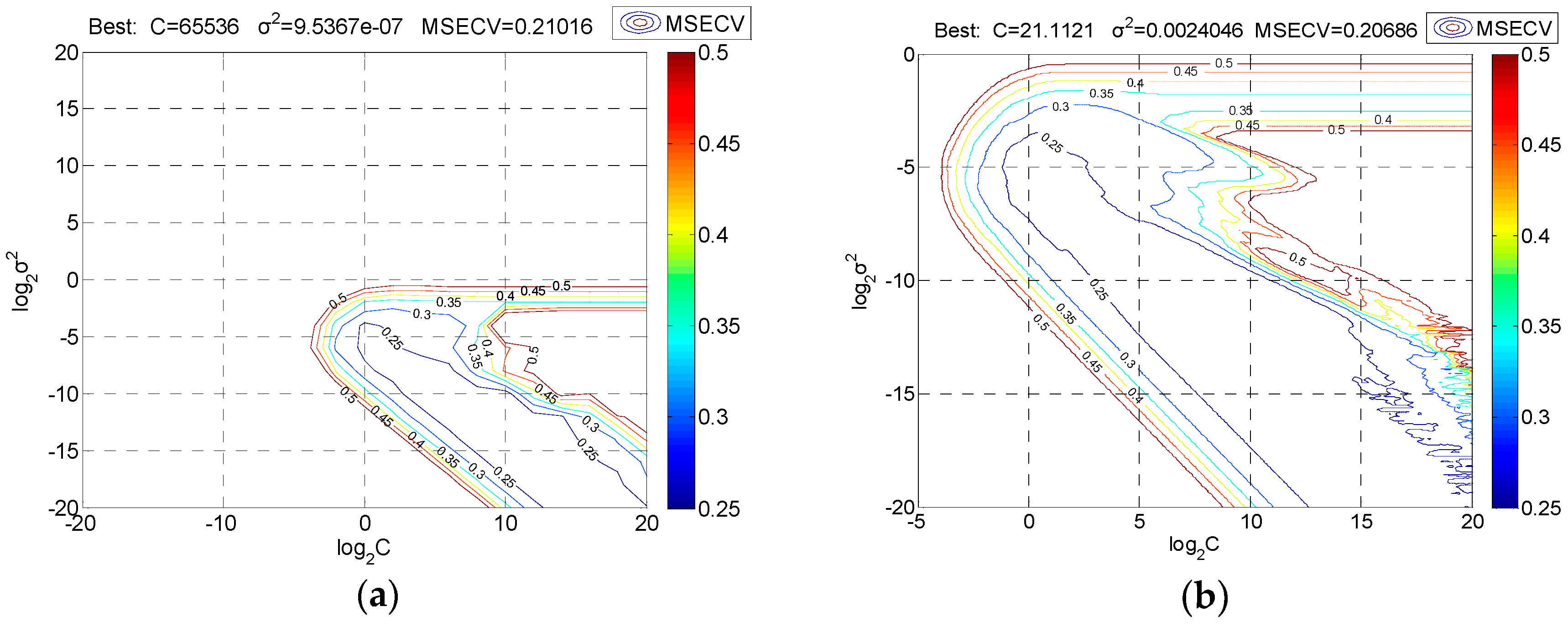
3.4. Calibration and Prediction by SVR Model
3.5. Calibration and Prediction by PLSR Model
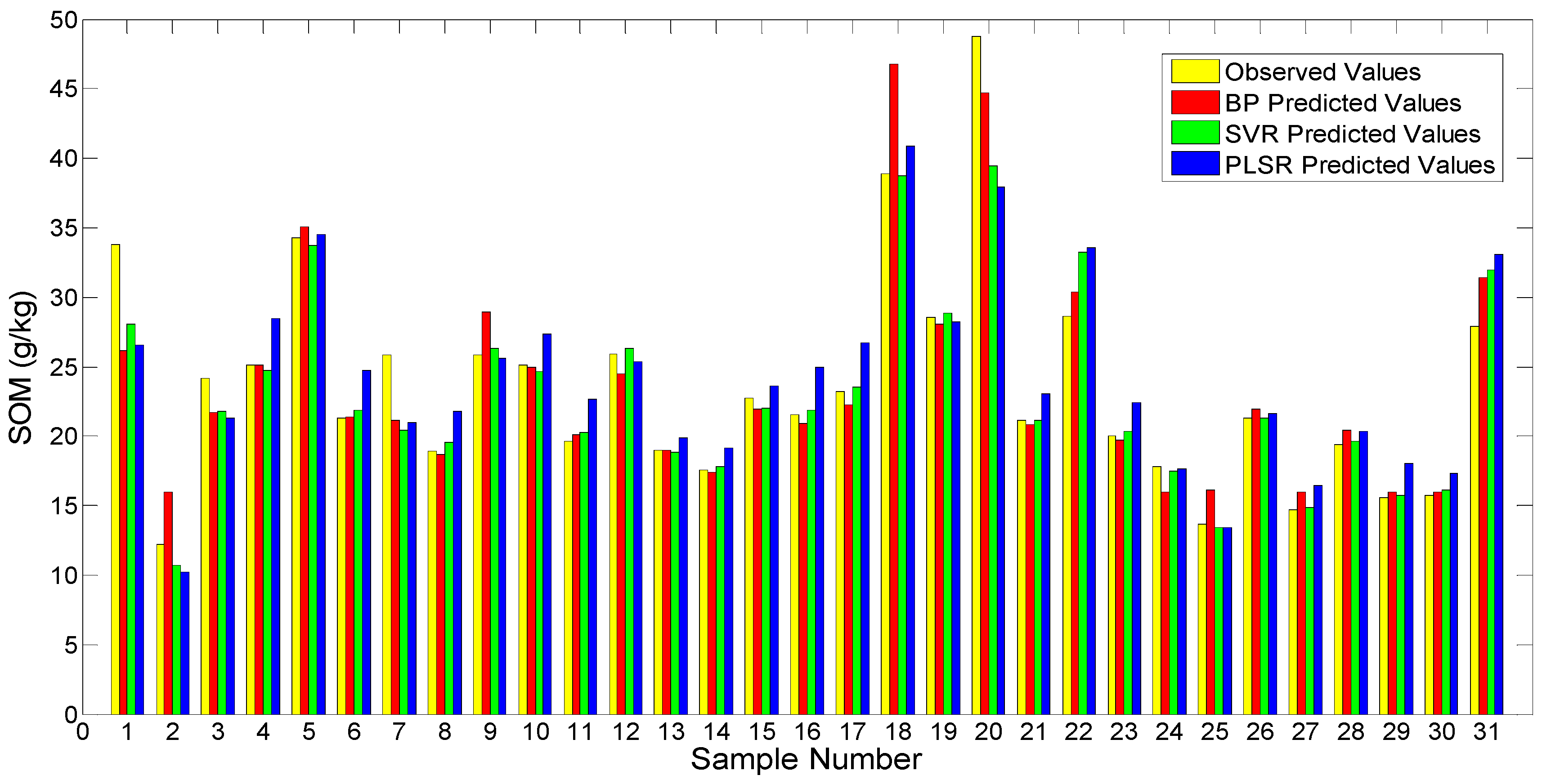
3.6. Model Comparison
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Schnitzer, M. A lifetime perspective on the chemistry of soil organic matter. Adv. Agron. 2000, 68, 1–58. [Google Scholar] [CrossRef]
- Salehi, M.H.; Beni, O.H.; Harchegani, H.B.; Borujeni, I.E.; Motaghian, H.R. Refining soil organic matter determination by loss-on-ignition. Pedosphere 2011, 21, 482. [Google Scholar] [CrossRef]
- Guan, L. Putong Turangxue, 2rd ed.; China Agricultural University Press: Beijing, China, 2016; pp. 42–45. [Google Scholar]
- Chai, X.; Shen, C.; Yuan, X.; Huang, Y. Spatial prediction of soil organic matter in the presence of different external trends with REML-EBLUP. Geoderma 2008, 148, 159–166. [Google Scholar] [CrossRef]
- Alex, M.; Damien, J.F.; Andrea, K. The dimensions of soil security. Geoderma 2014, 213, 203–213. [Google Scholar] [CrossRef]
- Johannes, L.; Markus, K. The contentious nature of soil organic matte. Nature 2015, 528, 60–68. Available online: https://www.nature.com/doifinder/10.1038/nature16069 (accessed on 5 March 2019).
- Tziachris, P.; Aschonitis, V.; Chatzistathis, T.; Papadopoulou, M. Assessment of spatial hybrid methods for predicting soil organic matter using DEM derivatives and soil parameters. Catena 2019, 174, 206–216. [Google Scholar] [CrossRef]
- Mallah, N.S.; Akbar, N.A.; Homaee, M. Estimating soil organic matter content from Hyperion reflectance images using PLSR, PCR, MinR and SWR models in semi-arid regions of Iran. Environ. Dev. 2018, 25, 23–32. [Google Scholar] [CrossRef]
- Mats, S.; Gustav, S.; Lars, R.; Kristin, P. Adaptation of regional digital soil mapping for precision agriculture. Precis. Agric. 2016, 17, 588–607. [Google Scholar] [CrossRef]
- Andre, C.A.; Ricardo, S.D.D.; Alexandre, T.C.; Sabine, G. A systematic study on the application of scatter-corrective and spectral-derivative preprocessing for multivariate prediction of soil organic carbon by Vis-NIR spectra. Geoderma 2018, 314, 262–274. [Google Scholar] [CrossRef]
- Cecile, G.; Raphael, A.V.R.; Alex, B.M. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Zhou, P.; Zhang, Y.; Yang, W.; Li, M.; Liu, Z.; Liu, X. Development and performance test of an in-situ soil total nitrogen-soil moisture detector based on near-infrared spectroscopy. Comput. Electron. Agric. 2019, 160, 51–58. [Google Scholar] [CrossRef]
- Christy, C.D. Real-time measurement of soil attributes using on-the-go near infrared reflectance spectroscopy. Comput. Electron. Agric. 2008, 61, 10–19. [Google Scholar] [CrossRef]
- Hummel, J.W.; Sudduth, K.A.; Hollinger, S.E. Soil moisture and organic matter prediction of surface and subsurface soils using an NIR soil sensor. Comput. Electron. Agric. 2001, 32, 149–165. [Google Scholar] [CrossRef]
- Croft, H.; Kuhn, N.J.; Anderson, K. On the use of remote sensing techniques for monitoring spatio-temporal soil organic carbon dynamics in agricultural systems. Catena 2012, 94, 64–74. [Google Scholar] [CrossRef]
- Condit, H.R. The spectral reflectance of American soils. Photogramm. Eng. 1970, 36, 955–966. [Google Scholar]
- Krishnan, P.; Alexander, J.D.; Butler, B.J.; Hummel, J.W. Reflectance Technique for Predicting Soil Organic Matter. Soil Sci. Soc. Am. J. 1980, 44, 1282–1285. [Google Scholar] [CrossRef]
- Liu, W.D.; Baret, F.; Gu, X.F.; Tong, Q.X.; Zheng, L.F.; Zhang, B. Relating soil surface moisture to reflectance. Remote. Sens. Environ. 2002, 81, 238–246. [Google Scholar] [CrossRef]
- Whiting, M.L.; Li, L.; Ustin, S.L. Predicting water content using Gaussian model on soil spectra. Remote Sens. Environ. 2004, 89, 535–552. [Google Scholar] [CrossRef]
- Bowers, S.A.; Hanks, R.J. Reflection of radiant energy from soil. Soil Sci. 1965, 100, 130–138. [Google Scholar] [CrossRef]
- Stoner, E.R.; Baumgardner, M.F. Characteristic variations in reflectance of surface soils. Soil Sci. Soc. Am. J. 1982, 45, 1161–1165. [Google Scholar] [CrossRef]
- Goutal, N.; Renault, P.; Ranger, J. Forwarder traffic impacted over at least four years soil air composition of two forest soils in northeast France. Geoderma 2013, 193, 29–40. [Google Scholar] [CrossRef]
- Vermoesen, A.; Ramon, H.; Cleemput, O.V. Composition of the soil gas phase. Permanent gases and hydrocarbons. Pedologie 1991, 41, 119–132. [Google Scholar]
- Goodlass, G.; Smith, K.A. Effect of PH, organic matter content and nitrate on the evolution of ethylene from soils. Soil Biol. Biochem. 1978, 10, 193–199. [Google Scholar] [CrossRef]
- Zhang, G.; Xie, C. A novel method in the gas identification by using WO3 gas sensor based on the temperature-programmed technique. Sens. Actuators B Chem. 2015, 206, 220–229. [Google Scholar] [CrossRef]
- Ding, H.; Ge, H.; Liu, J. High performance of gas identification by wavelet transform-based fast feature extraction from temperature modulated semiconductor gas sensors. Sens. Actuators B Chem. 2005, 107, 749–755. [Google Scholar] [CrossRef]
- Ngo, K.A.; Lauque, P.; Aguir, K. High performance of a gas identification system using sensor array and temperature modulation. Sens. Actuators B Chem. 2007, 124, 209–216. [Google Scholar] [CrossRef]
- Penza, M.; Cassano, G. Application of principal component analysis and artificial neural networks to recognize the individual VOCs of methanol/2-propanol in a binary mixture by SAW multi-sensor array. Sens. Actuators B Chem. 2003, 89, 269–284. [Google Scholar] [CrossRef]
- Suman, M.; Riani, G.; Dalcanale, E. Mos-based artificial olfactory system for the assessment of egg products freshness. Sens. Actuators B Chem. 2007, 125, 40–47. [Google Scholar] [CrossRef]
- Liu, Y.; Meng, Q.; Qi, P.; Sun, B.; Zhu, X. Using spike-based bio-inspired olfactory model for data processing in electronic noses. IEEE Sens. J. 2018, 18, 692–702. [Google Scholar] [CrossRef]
- Lotfivand, N.; Abdolzadeh, V.; Hamidon, M.N. Artificial olfactory system with fault-tolerant sensor array. Isa Trans. 2016, 63, 425–435. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J. Internal quality detection of Chinese pecans (Carya cathayensis) during storage using electronic nose responses combined with physicochemical methods. Postharvest Biol. Technol. 2016, 118, 17–25. [Google Scholar] [CrossRef]
- Chatterjee, D.; Bhattacharjee, P.; Bhattacharyya, N. Development of methodology for assessment of shelf-life of fried potato wedges using electronic noses: Sensor screening by fuzzy logic analysis. J. Food Eng. 2014, 133, 23–29. [Google Scholar] [CrossRef]
- Chen, Q.; Hui, Z.; Zhao, J.; Ouyang, Q. Evaluation of chicken freshness using a low-cost colorimetric sensor array with AdaBoost-OLDA classification algorithm. LWT Food Sci. Technol. 2014, 57, 502–507. [Google Scholar] [CrossRef]
- Shih, C.H.; Lin, Y.J.; Lee, K.F.; Chien, P.Y.; Drake, P. Real-time electronic nose based pathogen detection for respiratory intensive care patients. Sens. Actuators B Chem. 2010, 148, 153–157. [Google Scholar] [CrossRef]
- Hong, X.; Wang, J. Detection of adulteration in cherry tomato juices based on electronic nose and tongue: Comparison of different data fusion approaches. J. Food Eng. 2014, 126, 89–97. [Google Scholar] [CrossRef]
- Huo, D.; Wu, Y.; Yang, M.; Fa, H.; Luo, X.; Hou, C. Discrimination of Chinese green tea according to varieties and grade levels using artificial nose and tongue based on colorimetric sensor arrays. Food Chem. 2014, 145, 639–645. [Google Scholar] [CrossRef]
- Deshmukh, S.; Jana, A.; Bhattacharyya, N.; Bandyopadhyay, R.; Pandey, R.A. Quantitative determination of pulp and paper industry emissions and associated odor intensity in methyl mercaptan equivalent using electronic nose. Atmos. Environ. 2014, 82, 401–409. [Google Scholar] [CrossRef]
- Andrzej, B.; Katarzyna, J.G.; Łukasz, G.; Łagód, G.; Grzegorz, J.; Wojciech, F.; Zbigniew, S.; Henryk, S. Evaluating soil moisture status using an e-nose. Sensors 2016, 16, 886. [Google Scholar] [CrossRef]
- Cesare, F.D.; Mattia, E.D.; Pantalei, S.; Zampetti, E.; Vinciguerra, V.; Canganella, F.; Macagnano, A. Use of electronic nose technology to measure soil microbial activity through biogenic volatile organic compounds and gases release. Soil Biol. Biochem. 2011, 43, 2094–2107. [Google Scholar] [CrossRef]
- Pobkrut, T.; Kerdcharoen, T. Soil sensing survey robots based on electronic nose. In Proceedings of the 2014 14th International Conference on Control, Automation and Systems (ICCAS), Seoul, Korea, 22–25 October 2014; IEEE: Piscataway, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Lavanya, S.; Deepika, B.; Narayanan, S.; Krishna Murthy, V.; Uma, M.V. Indicative extent of humic and fulvic acids in soils determined by electronic nose. Comput. Electron. Agric. 2017, 139, 198–203. [Google Scholar] [CrossRef]
- Bieganowski, A.; Jozefaciuk, G.; Bandura, L.; Guz, L.M.; Lagod, G.; Franus, W. Evaluation of hydrocarbon soil pollution using e-nose. Sensors 2018, 18, 2463. [Google Scholar] [CrossRef]
- Arief, S.; Akio, K. Application of temperature modulation-SDP on MOS gas sensors: Capturing soil gaseous profile for discrimination of soil under different nutrient addition. J. Sens. 2016, 2016, 1–11. [Google Scholar] [CrossRef]
- Lu, P.; Wang, L.; Niu, Z.; Li, L.; Zhang, W. Prediction of soil properties using laboratory VIS–NIR spectroscopy and Hyperion imagery. J. Geochem. Explor. 2013, 132, 26–33. [Google Scholar] [CrossRef]
- Li, G.; Feng, X.; Qiu, G.; Bi, X.; Li, Z.; Zhang, C.; Wang, D.; Shang, L.; Guo, Y. Environmental mercury contamination of an artisanal zinc smelting area in Weining County, Guizhou, China. Environ. Pollut. 2008, 154, 21–31. [Google Scholar] [CrossRef]
- SGAS707—Industrial Organic Chemical Sensor | IDT. Available online: https://www.idt.com/products/sensor-products/gas-sensors/sgas707-industrial-organic-chemical-sensor (accessed on 5 March 2019).
- Llobet, E.; Brezmes, J.; Vilanova, X.; Sueiras, J.E.; Correig, X. Qualitative and quantitative analysis of volatile organic compounds using transient and steady-state responses of a thick-film tin oxide gas sensor array. Sens. Actuators B Chem. 1997, 41, 13–21. [Google Scholar] [CrossRef]
- Fu, J.; Li, G.; Qin, Y.; Freeman, W.J. A pattern recognition method for electronic noses based on an olfactory neural network. Sens. Actuators B Chem. 2007, 125, 489–497. [Google Scholar] [CrossRef]
- Zoest, V.; Osei, F.B.; Stein, A.; Hoek, G. Calibration of low-cost NO2 sensors in an urban air quality network. Atmos. Environ. 2019, 210, 66–75. [Google Scholar] [CrossRef]
- Osowski, S.; Brudzewski, K.; Markiewicz, T.; Ulaczyk, J. Neural methods of calibration of sensors for gas measurements and aroma identification system. J. Sens. Stud. 2008, 23, 533–557. [Google Scholar] [CrossRef]
- Cai, F.; Cui, J.; Dong, B.; Li, J.; Li, X. Training back-propagation neural network using hybrid fruit fly optimization algorithm. J. Comput. Theo. Nanos. 2016, 13, 3212–3221. [Google Scholar] [CrossRef]
- Hanafizadeh, P.; Ravasan, A.Z.; Khaki, H.R. An expert system for perfume selection using artificial neural network. Expert Syst. Appl. 2010, 37, 8879–8887. [Google Scholar] [CrossRef]
- Kaastra, I.; Boyd, M. Designing a neural network for forecasting financial and economic time series. Neurocomputing 1996, 10, 215–236. [Google Scholar] [CrossRef]
- Kolmogorov, A.N. On the representation of continuous functions of many variables by superposition of continuous functions of one variable and addition. In Doklady Akademii Nauk; Russian Academy of Sciences: Saint Petersburg, Russia, 1957; Volume 114, pp. 953–956. [Google Scholar]
- Vapnik, V.N. The Nature of Statistical Learning Theory, 2rd ed.; Springer: New York, NY, USA, 2000; pp. 138–167. [Google Scholar]
- Farquad, M.A.H.; Ravi, V.; Raju, S.B. Support vector regression based hybrid rule extraction methods for forecasting. Expert Syst. Appl. 2010, 37, 5577–5589. [Google Scholar] [CrossRef]
- Ortiz-García, E.G.; Salcedo-Sanz, S.; Pérez-Bellido, M.Á.; Portilla-Figueras, J.A.; Prieto, L. Prediction of hourly o3 concentrations using support vector regression algorithms. Atmos. Environ. 2010, 44, 4481–4488. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Zhao, M.; Ren, J.; Ji, L.; Fu, C.; Li, J.; Zhou, M. Parameter selection of support vector machines and genetic algorithm based on change area search. Neural Comput. Appl. 2012, 21, 1–8. [Google Scholar] [CrossRef]
- Qi, H.; Tarin, P.K.; Arnon, K.; Jin, X.; Li, S. Evaluating calibration methods for predicting soil available nutrients using hyperspectral VNIR data. Soil Till. Res. 2018, 175, 267–275. [Google Scholar] [CrossRef]
- Wold, S.; Ruhe, A.; Wold, H.; Dunn, I.W.J. The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J. Sci. Stat. Comput. 1984, 5, 735–743. [Google Scholar] [CrossRef]
- Rossel, R.A.V.; Walvoort, D.J.J.; Mcbratney, A.B.; Janik, L.J.; Skjemstad, J.O. Visible, near infrared, mid infrared or combined diffuse reflectance spectroscopy for simultaneous assessment of various soil properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Ji, W.; Shi, Z.; Huang, J.; Li, S. In situ measurement of some soil properties in paddy soil using visible and near-infrared spectroscopy. PLoS ONE 2014, 9, e105708. [Google Scholar] [CrossRef]
- Li, B.; Morris, J.; Martin, E.B. Model selection for partial least squares regression. Chemometr. Intell. Lab. 2002, 64, 79–89. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Chang, C.W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-infrared reflectance spectroscopy–principal components regression analyses of soil properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
- LIBSVM—A Library for Support Vector Machines. Software. Available online: http://www.csie.ntu.edu.tw/~cjlin/libsvm (accessed on 5 March 2019).
- Kuang, B.; Mouazen, A. Calibration of visible and near infrared spectroscopy for soil analysis at the field scale on three European farms. Eur. J. Soil Sci. 2011, 62, 629–636. [Google Scholar] [CrossRef]





| Sensor Number | Vh (V) | Working Temperature (°C) | Sensor Number | Vh (V) | Working Temperature (°C) |
|---|---|---|---|---|---|
| S1 | 1.25 | 34.4 | S6 | 2.50 | 48.1 |
| S2 | 1.50 | 36.0 | S7 | 2.75 | 52.5 |
| S3 | 1.75 | 37.8 | S8 | 3.00 | 60.0 |
| S4 | 2.00 | 40.4 | S9 | 3.25 | 65.7 |
| S5 | 2.25 | 43.0 | S10 | 3.50 | 74.3 |
| Dataset | SOM (g·kg–1) | Max (g·kg–1) | Min (g·kg–1) | Mean (g·kg–1) | SD (g·kg–1) | CV (%) |
|---|---|---|---|---|---|---|
| Training set | 20.51; 27.62; 33.50; 20.23; 23.11; 24.43; 28.71; 26.53; 18.88; 26.92; 14.97; 20.48; 17.69; 13.76; 17.38; 19.97; 32.13; 29.87; 28.85; 39.64; 12.37; 17.33; 14.22; 22.85; 15.49; 22.85; 25.27; 22.55; 18.13; 20.52; 25.20; 23.72; 13.44; 16.24; 15.67; 41.10; 22.31; 20.17; 13.29; 19.54; 35.55; 36.28; 43.85; 19.14; 25.42; 19.79; 13.79; 15.90; 30.71; 19.27; 23.16; 30.14; 24.76; 23.80; 27.95; 20.60; 22.88; 24.75; 23.46; 18.67; 35.38; 16.53; 15.32; 16.31; 16.74; 17.78; 22.89; 14.80; 29.65; 38.86; 19.750 | 43.85 | 12.37 | 22.98 | 7.27 | 31.64 |
| Validation set | 33.77; 12.19; 24.15; 25.11; 34.24; 21.32; 25.86; 18.94; 25.85; 25.10; 19.64; 25.94; 18.96; 17.58; 22.71; 21.50; 23.18; 38.92; 28.58; 48.79; 21.13; 28.62; 20.01; 17.78; 13.64; 21.28; 14.72; 19.37; 15.59; 15.71; 27.89 | 48.79 | 12.19 | 23.49 | 7.73 | 32.90 |
| Neuron Number | R2T | RMSET | ||||
|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | |
| 6 | 0.627 | 0.906 | 0.793 | 15.794 | 26.600 | 21.351 |
| 7 | 0.382 | 0.810 | 0.678 | 18.841 | 37.911 | 25.515 |
| 8 | 0.450 | 0.845 | 0.630 | 18.955 | 31.600 | 26.440 |
| 9 | 0.280 | 0.824 | 0.690 | 18.190 | 40.136 | 25.424 |
| 10 | 0.512 | 0.832 | 0.650 | 17.906 | 36.760 | 27.188 |
| 11 | 0.503 | 0.804 | 0.716 | 28.880 | 34.671 | 24.384 |
| 12 | 0.568 | 0.832 | 0.704 | 18.252 | 30.010 | 24.299 |
| 13 | 0.391 | 0.867 | 0.726 | 17.611 | 33.872 | 23.202 |
| 14 | 0.127 | 0.848 | 0.599 | 16.768 | 55.106 | 32.411 |
| 15 | 0.561 | 0.812 | 0.681 | 18.927 | 40.192 | 27.247 |
| 16 | 0.300 | 0.857 | 0.672 | 17.897 | 37.628 | 25.187 |
| Models | R2 | RMSE | RPD | Category |
|---|---|---|---|---|
| BPNN | 0.880 | 14.916 | 2.837 | A |
| SVR | 0.895 | 14.094 | 3.003 | A |
| PLRS | 0.808 | 18.890 | 2.240 | A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Jia, H.; Chen, Y.; Wang, Q.; Li, M.; Huang, D.; Bai, Y. A Novel Method for Soil Organic Matter Determination by Using an Artificial Olfactory System. Sensors 2019, 19, 3417. https://doi.org/10.3390/s19153417
Zhu L, Jia H, Chen Y, Wang Q, Li M, Huang D, Bai Y. A Novel Method for Soil Organic Matter Determination by Using an Artificial Olfactory System. Sensors. 2019; 19(15):3417. https://doi.org/10.3390/s19153417
Chicago/Turabian StyleZhu, Longtu, Honglei Jia, Yibing Chen, Qi Wang, Mingwei Li, Dongyan Huang, and Yunlong Bai. 2019. "A Novel Method for Soil Organic Matter Determination by Using an Artificial Olfactory System" Sensors 19, no. 15: 3417. https://doi.org/10.3390/s19153417
APA StyleZhu, L., Jia, H., Chen, Y., Wang, Q., Li, M., Huang, D., & Bai, Y. (2019). A Novel Method for Soil Organic Matter Determination by Using an Artificial Olfactory System. Sensors, 19(15), 3417. https://doi.org/10.3390/s19153417




