Detection of Osmotic Shock-Induced Extracellular Nucleotide Release with a Genetically Encoded Fluorescent Sensor of ADP and ATP
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Protein Engineering and Library Screening
3.2. Validation with Live-Cell Time-Resolved Spectroscopy
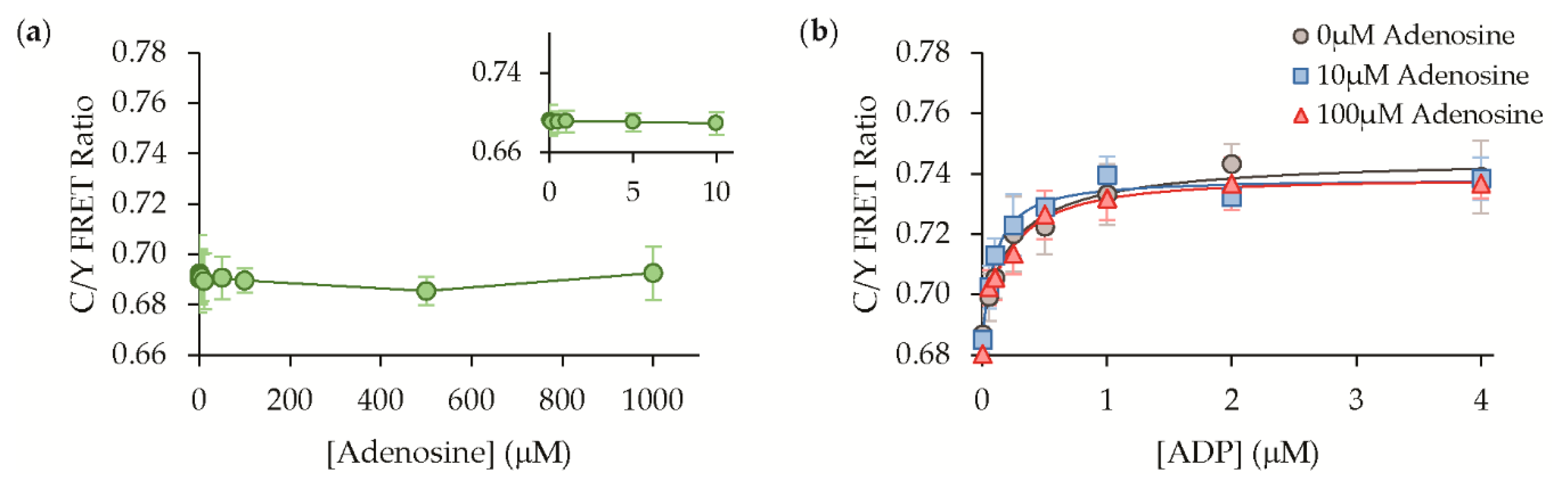
3.3. Protein Solution Characterization
3.4. Cell Surface Expression and Characterization
3.5. Detection of Physiological Nucleotide Release
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Praetorius, H.A.; Leipziger, J. ATP release from non-excitable cells. Purinergic Signal. 2009, 5, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D.; Burnstock, G. Purinergic signalling in neuron–glia interactions. Nat. Rev. Neurosci. 2006, 7, 423–436. [Google Scholar] [CrossRef]
- Junger, W.G. Immune cell regulation by autocrine purinergic signalling. Nat. Rev. Immunol. 2011, 11, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 16, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Burnstock, G.; Di Virgilio, F. Purinergic signalling and cancer. Purinergic Signal. 2013, 9, 491–540. [Google Scholar] [CrossRef]
- Robson, S.C.; Sévigny, J.; Zimmermann, H. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal. 2006, 2, 409–430. [Google Scholar] [CrossRef] [PubMed]
- Pellegatti, P.; Falzoni, S.; Pinton, P.; Rizzuto, R.; Di Virgilio, F. A Novel Recombinant Plasma Membrane-targeted Luciferase Reveals a New Pathway for ATP Secretion. Mol. Biol. Cell 2005, 16, 3659–3665. [Google Scholar] [CrossRef] [PubMed]
- Mikolajewicz, N.; Mohammed, A.; Morris, M.; Komarova, S.V. Mechanically stimulated ATP release from mammalian cells: Systematic review and meta-analysis. J. Cell Sci. 2018, 131, jcs223354. [Google Scholar] [CrossRef]
- Koyama, T.; Oike, M.; Ito, Y. Involvement of Rho-kinase and tyrosine kinase in hypotonic stress-induced ATP release in bovine aortic endothelial cells. J. Physiol. 2001, 532, 759–769. [Google Scholar] [CrossRef]
- Pedersen, S.; Pedersen, S.F.; Nilius, B.; Lambert, I.H.; Hoffmann, E.K. Mechanical stress induces release of ATP from Ehrlich ascites tumor cells. Biochim. Biophys. Acta 1999, 1416, 271–284. [Google Scholar] [CrossRef]
- Darby, M.; Kuzmiski, J.B.; Panenka, W.; Feighan, D.; MacVicar, B.A. ATP released from astrocytes during swelling activates chloride channels. J. Neurophysiol. 2003, 89, 1870–1877. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D.; Ni, Y. Nonsynaptic communication through ATP release from volume-activated anion channels in axons. Sci. Signal. 2010, 3, ra73. [Google Scholar] [CrossRef] [PubMed]
- Conley, J.M.; Radhakrishnan, S.; Valentino, S.A.; Tantama, M. Imaging extracellular ATP with a genetically-encoded, ratiometric fluorescent sensor. PLoS ONE 2017, 12, e0187481. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.A.; Paoletta, S.; Katritch, V.; Wu, B.; Gao, Z.-G.; Zhao, Q.; Stevens, R.C.; Kiselev, E. Nucleotides Acting at P2Y Receptors: Connecting Structure and Function. Mol. Pharmacol. 2015, 88, 220–230. [Google Scholar] [CrossRef] [PubMed]
- van den Ent, F.; Møller-Jensen, J.; Amos, L.A.; Gerdes, K.; Löwe, J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002, 21, 6935–6943. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, S.; Webb, M.R. A Fluorescent, Reagentless Biosensor for ADP Based on Tetramethylrhodamine-Labeled ParM. ACS Chem. Biol. 2010, 5, 415–425. [Google Scholar] [CrossRef]
- Kunzelmann, S.; Webb, M.R. A biosensor for fluorescent determination of ADP with high time resolution. J. Biol. Chem. 2009, 284, 33130–33138. [Google Scholar] [CrossRef]
- Fritz, R.D.; Letzelter, M.; Reimann, A.; Martin, K.; Fusco, L.; Ritsma, L.; Ponsioen, B.; Fluri, E.; Schulte-Merker, S.; van Rheenen, J.; et al. A Versatile Toolkit to Produce Sensitive FRET Biosensors to Visualize Signaling in Time and Space. Sci. Signal. 2013, 6, rs12. [Google Scholar] [CrossRef]
- Yaginuma, H.; Kawai, S.; Tabata, K.V.; Tomiyama, K.; Kakizuka, A.; Komatsuzaki, T.; Noji, H.; Imamura, H. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci. Rep. 2015, 4, 6522. [Google Scholar] [CrossRef]
- Haynes, E.P.; Rajendran, M.; Henning, C.K.; Mishra, A.; Lyon, A.M.; Tantama, M. Quantifying Acute Fuel and Respiration Dependent pH Homeostasis in Live Cells Using the mCherryTYG Mutant as a Fluorescence Lifetime Sensor. Anal. Chem. 2019, 91, 8466–8475. [Google Scholar] [CrossRef]
- Marvin, J.S.; Borghuis, B.G.; Tian, L.; Cichon, J.; Harnett, M.T.; Akerboom, J.; Gordus, A.; Renninger, S.L.; Chen, T.-W.; Bargmann, C.I.; et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 2013, 10, 162–170. [Google Scholar] [CrossRef]
- Fehr, M.; Lalonde, S.; Lager, I.; Wolff, M.W.; Frommer, W.B. In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J. Biol. Chem. 2003, 278, 19127–19133. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Henderson, J.N.; Remington, S.J.; Campbell, R.E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: Structural characterization and applications in fluorescence imaging. Biochem. J. 2006, 400, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Imamura, H.; Huynh Nhat, K.P.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Clister, T.L.; Lowry, P.R.; Seldin, M.M.; Wong, G.W.; Zhang, J. Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Rep. 2015, 10, 1767–1777. [Google Scholar] [CrossRef]
- Thestrup, T.; Litzlbauer, J.; Bartholomäus, I.; Mues, M.; Russo, L.; Dana, H.; Kovalchuk, Y.; Liang, Y.; Kalamakis, G.; Laukat, Y.; et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods 2014, 11, 175–182. [Google Scholar] [CrossRef]
- Hum, J.; Siegel, A.; Pavalko, F.; Day, R. Monitoring Biosensor Activity in Living Cells with Fluorescence Lifetime Imaging Microscopy. Int. J. Mol. Sci. 2012, 13, 14385–14400. [Google Scholar] [CrossRef]
- San Martín, A.; Ceballo, S.; Ruminot, I.; Lerchundi, R.; Frommer, W.B.; Barros, L.F. A Genetically Encoded FRET Lactate Sensor and Its Use to Detect the Warburg Effect in Single Cancer Cells. PLoS ONE 2013, 8, e57712. [Google Scholar] [CrossRef]
- San Martín, A.; Ceballo, S.; Baeza-Lehnert, F.; Lerchundi, R.; Valdebenito, R.; Contreras-Baeza, Y.; Alegría, K.; Barros, L.F. Imaging Mitochondrial Flux in Single Cells with a FRET Sensor for Pyruvate. PLoS ONE 2014, 9, e85780. [Google Scholar] [CrossRef]
- Baeza-Lehnert, F.; Saab, A.S.; Gutiérrez, R.; Larenas, V.; Díaz, E.; Horn, M.; Vargas, M.; Hösli, L.; Stobart, J.; Hirrlinger, J.; et al. Non-Canonical Control of Neuronal Energy Status by the Na+ Pump. Cell Metab. 2019, 29, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Mächler, P.; Wyss, M.T.; Elsayed, M.; Stobart, J.; Gutierrez, R.; von Faber-Castell, A.; Kaelin, V.; Zuend, M.; San Martín, A.; Romero-Gómez, I.; et al. In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab. 2016, 23, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Mastop, M.; Bindels, D.S.; Shaner, N.C.; Postma, M.; Gadella, T.W.J.; Goedhart, J. Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2. Sci. Rep. 2017, 7, 11999. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, E.C.; Mehta, S.; Zhang, J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118, 11707–11794. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yasuda, R. Imaging ERK and PKA Activation in Single Dendritic Spines during Structural Plasticity. Neuron 2017, 93, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive red protein calcium indicators for imaging neural activity. Elife 2016, 5, e12727. [Google Scholar] [CrossRef]
- Harada, K.; Ito, M.; Wang, X.; Tanaka, M.; Wongso, D.; Konno, A.; Hirai, H.; Hirase, H.; Tsuboi, T.; Kitaguchi, T. Red fluorescent protein-based cAMP indicator applicable to optogenetics and in vivo imaging. Sci. Rep. 2017, 7, 7351. [Google Scholar] [CrossRef] [PubMed]
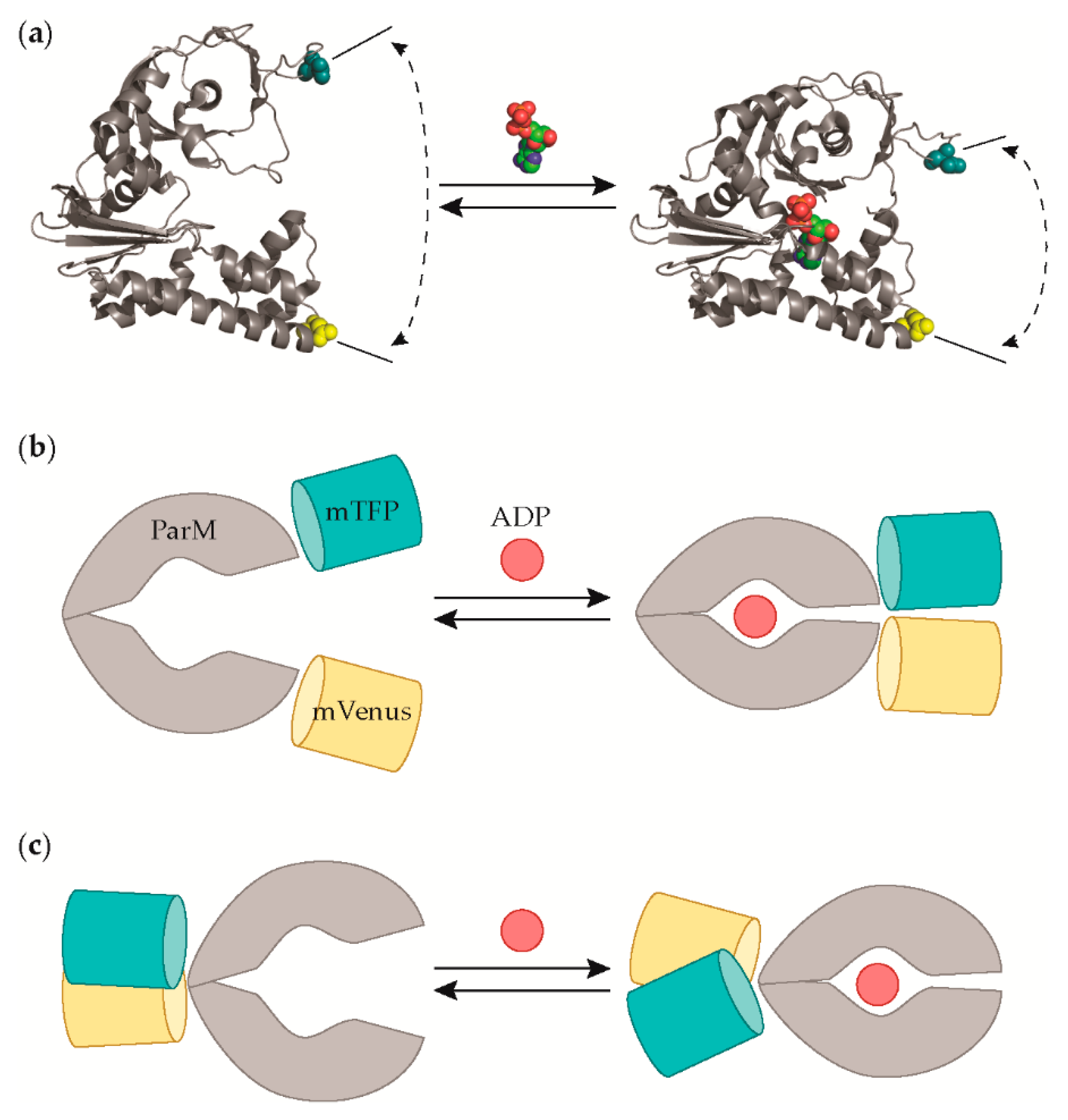

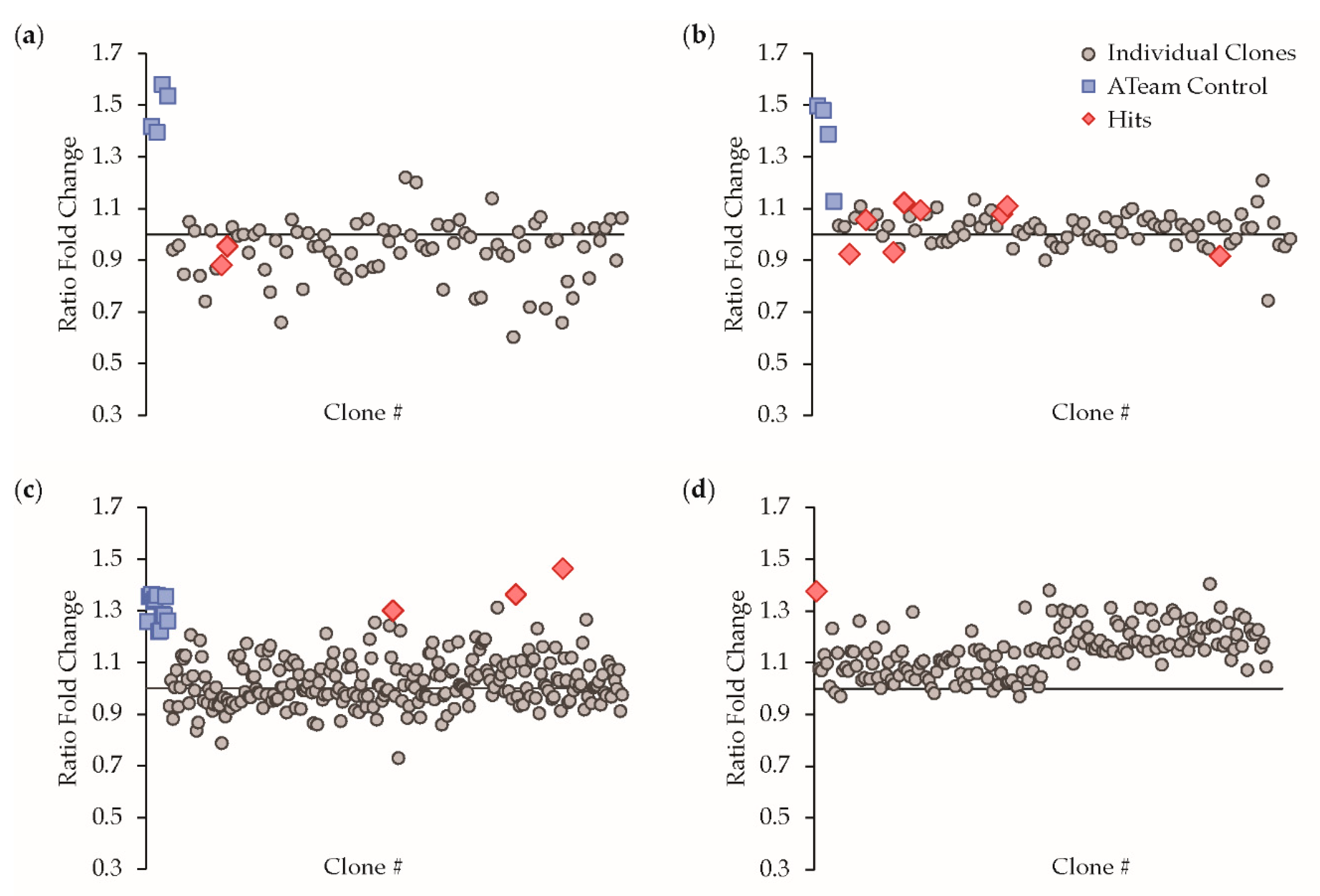
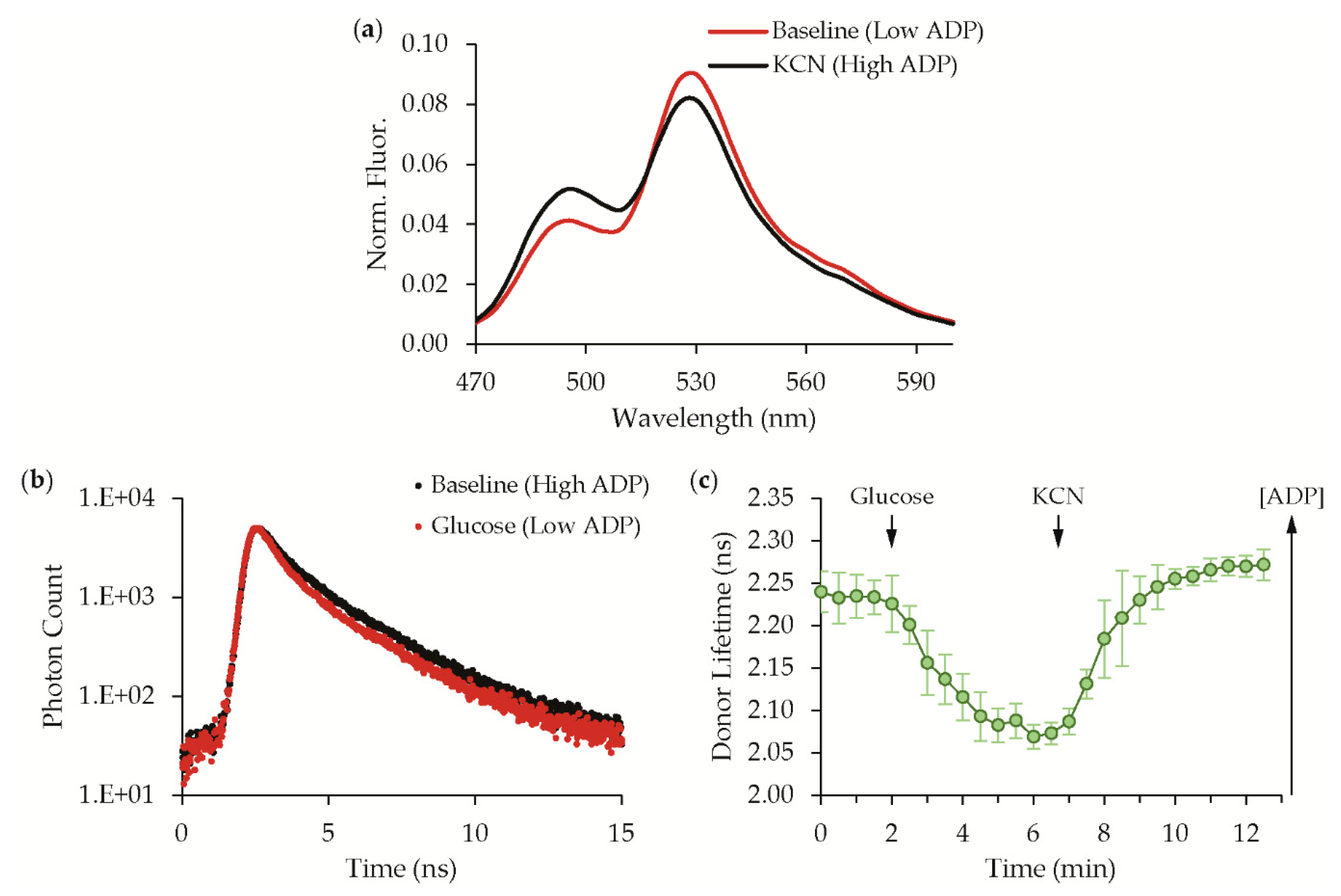

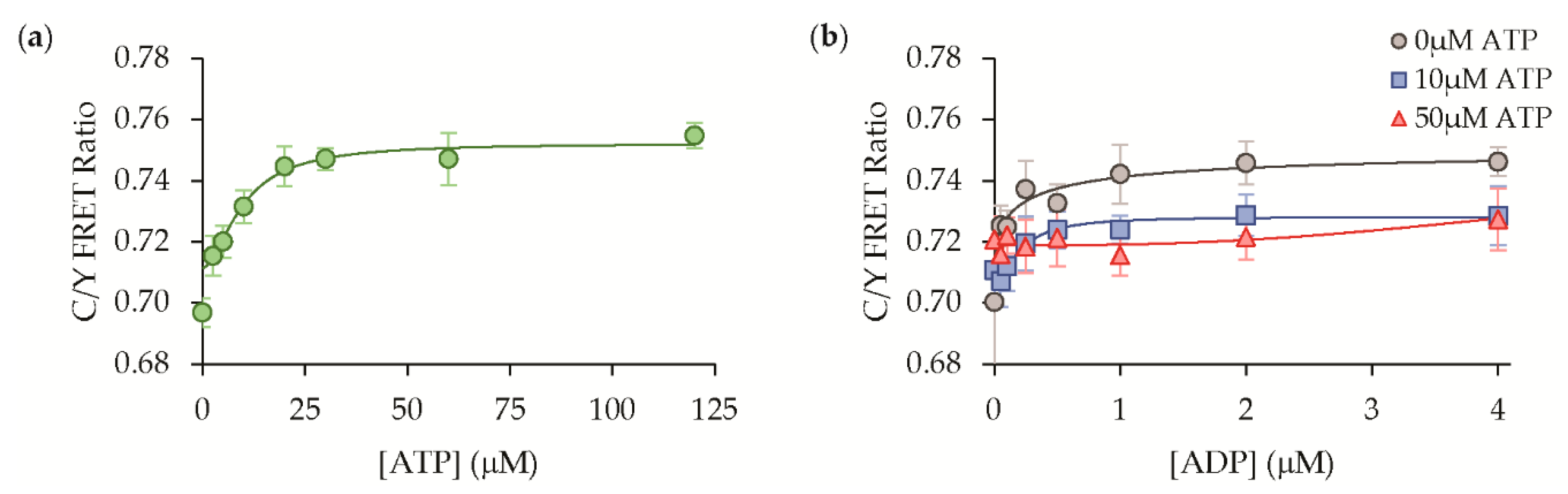



| Hit ID | mTFP1 Variant | mVenus Variant |
|---|---|---|
| Insertion Clone 1 | WT | cp157 |
| Insertion Clone 2 | cp227 | cp157 |
| Terminal Clone 1 | cp105 | cp173 |
| Terminal Clone 2 | WT | cp157 |
| Terminal Clone 3 | WT | cp173 |
| Terminal Clone 4 | cp159 | WT |
| Terminal Clone 5 | WT | cp195 |
| Terminal Clone 6 | cp227 | WT |
| Terminal Clone 7 | cp227 | cp195 |
| Terminal Clone 8 | cp227 | cp173 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trull, K.J.; Miller, P.; Tat, K.; Varney, S.A.; Conley, J.M.; Tantama, M. Detection of Osmotic Shock-Induced Extracellular Nucleotide Release with a Genetically Encoded Fluorescent Sensor of ADP and ATP. Sensors 2019, 19, 3253. https://doi.org/10.3390/s19153253
Trull KJ, Miller P, Tat K, Varney SA, Conley JM, Tantama M. Detection of Osmotic Shock-Induced Extracellular Nucleotide Release with a Genetically Encoded Fluorescent Sensor of ADP and ATP. Sensors. 2019; 19(15):3253. https://doi.org/10.3390/s19153253
Chicago/Turabian StyleTrull, Keelan J., Piper Miller, Kiet Tat, S. Ashley Varney, Jason M. Conley, and Mathew Tantama. 2019. "Detection of Osmotic Shock-Induced Extracellular Nucleotide Release with a Genetically Encoded Fluorescent Sensor of ADP and ATP" Sensors 19, no. 15: 3253. https://doi.org/10.3390/s19153253
APA StyleTrull, K. J., Miller, P., Tat, K., Varney, S. A., Conley, J. M., & Tantama, M. (2019). Detection of Osmotic Shock-Induced Extracellular Nucleotide Release with a Genetically Encoded Fluorescent Sensor of ADP and ATP. Sensors, 19(15), 3253. https://doi.org/10.3390/s19153253





