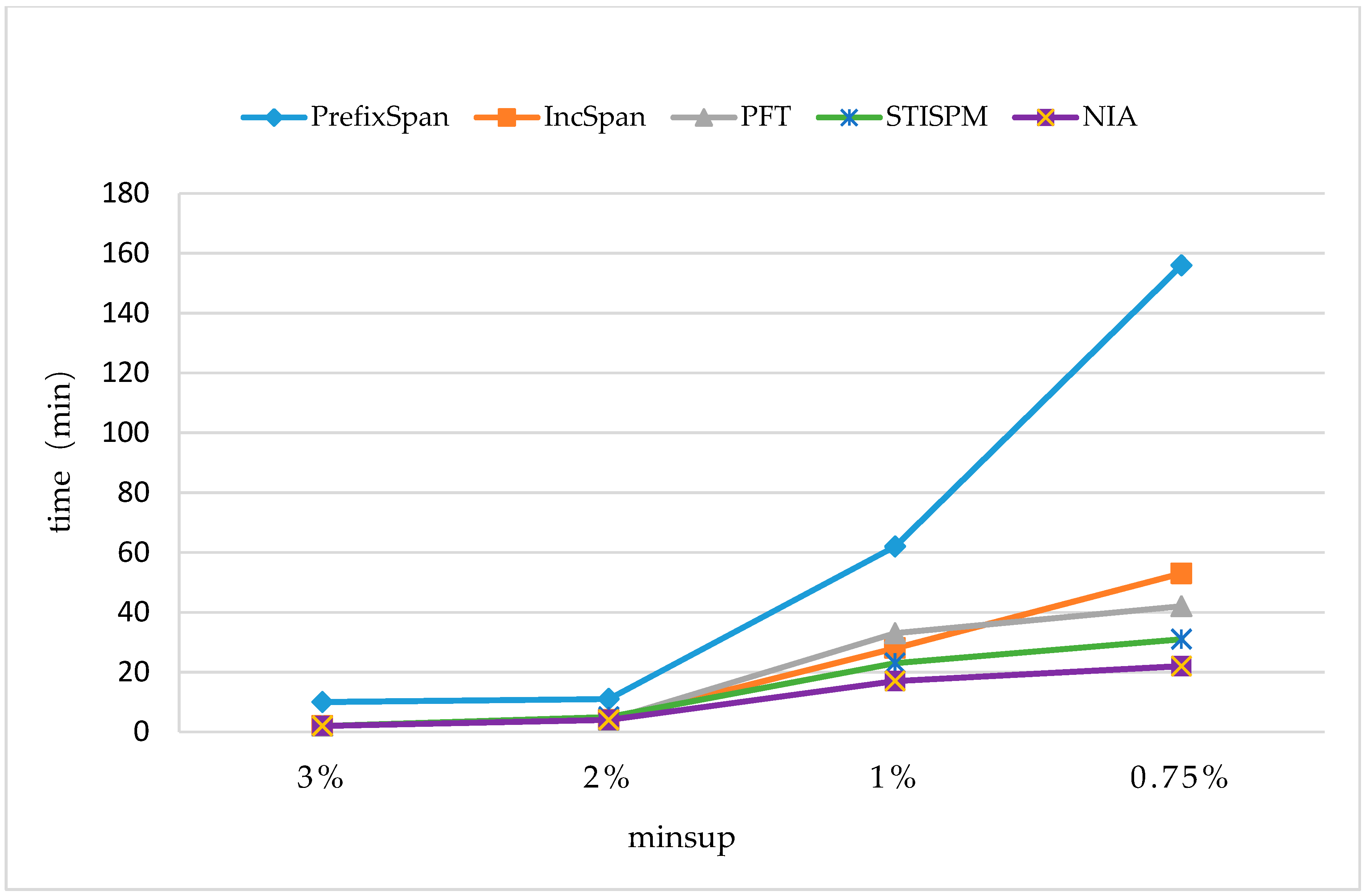
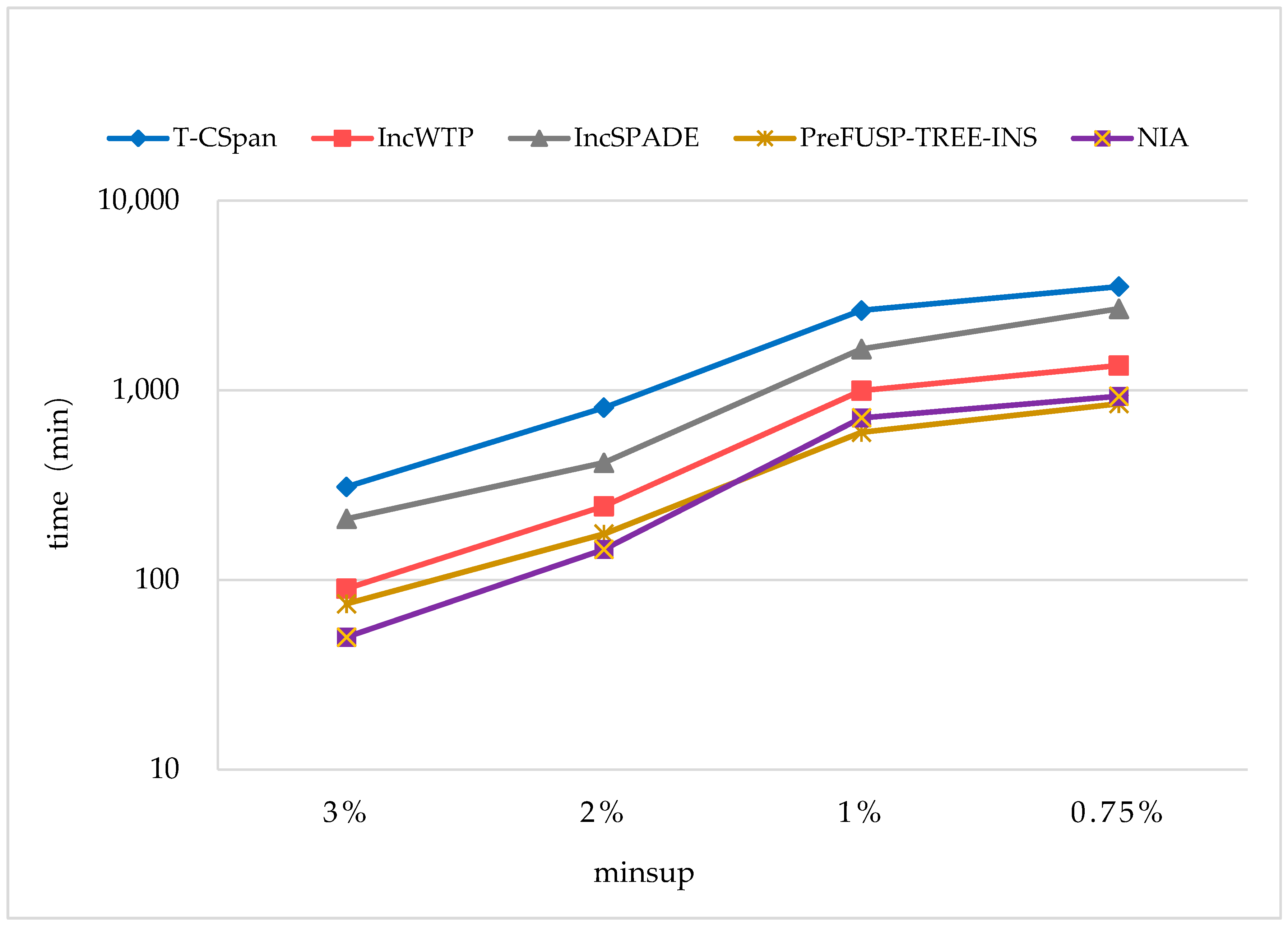
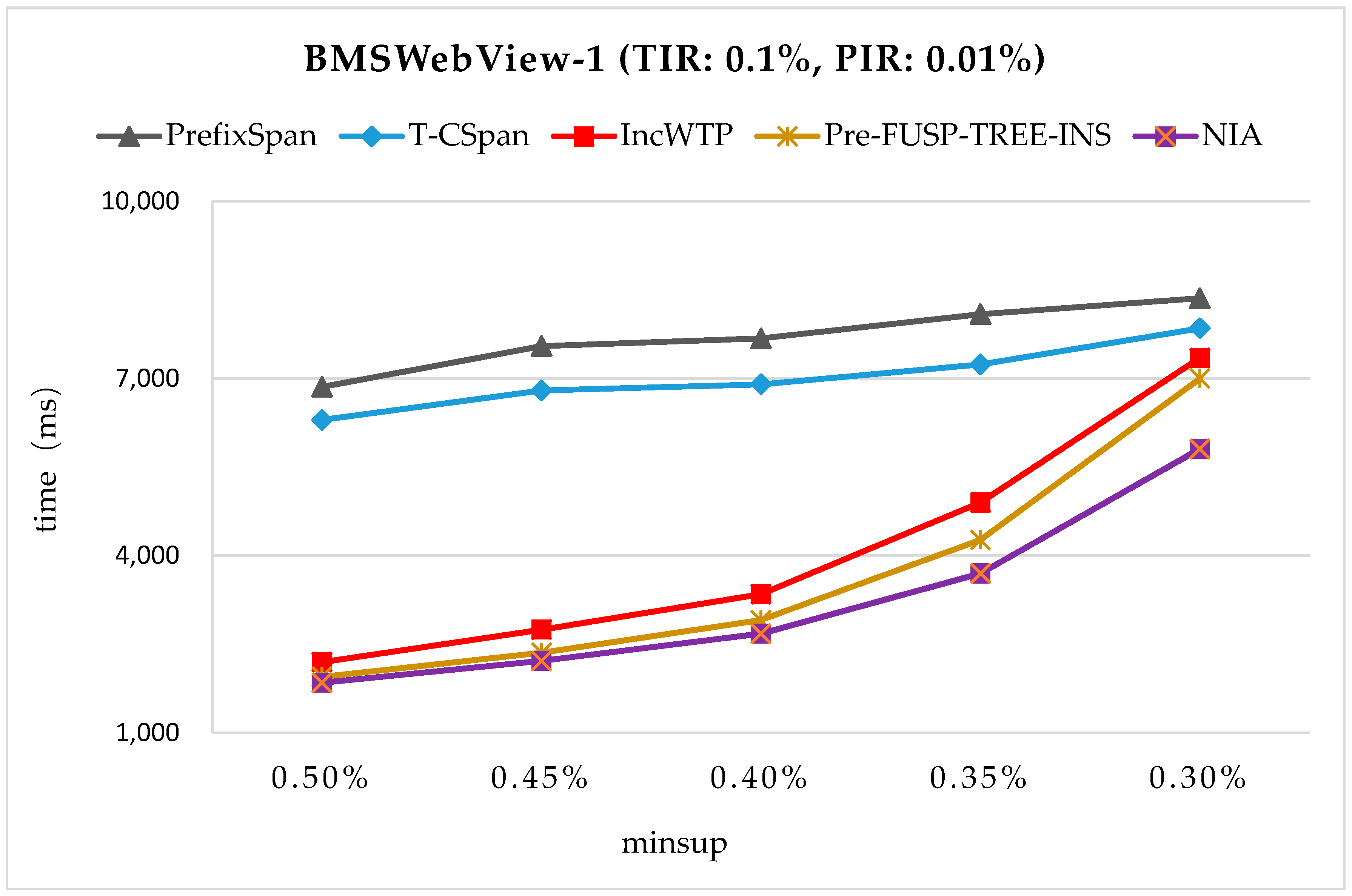
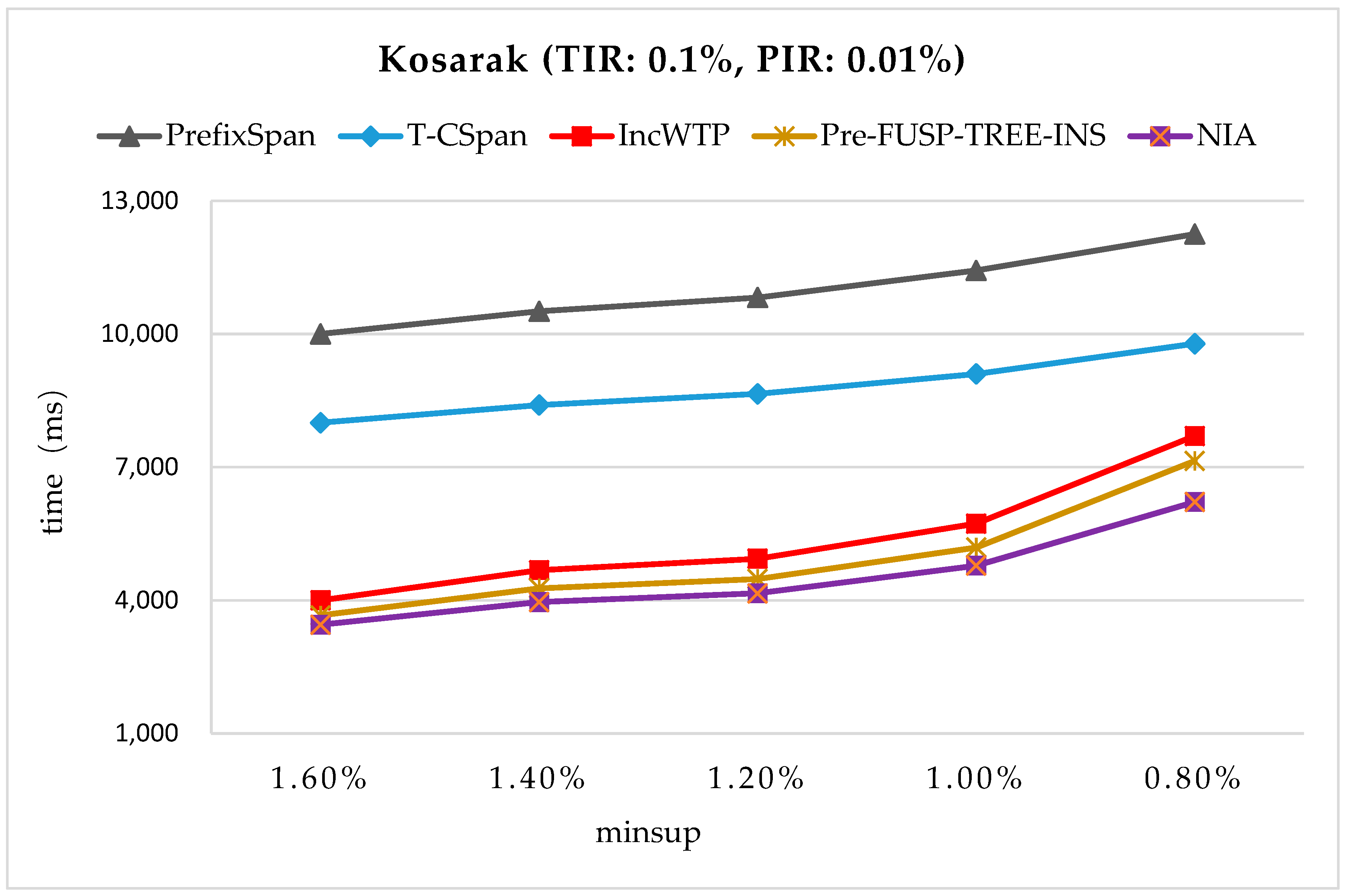
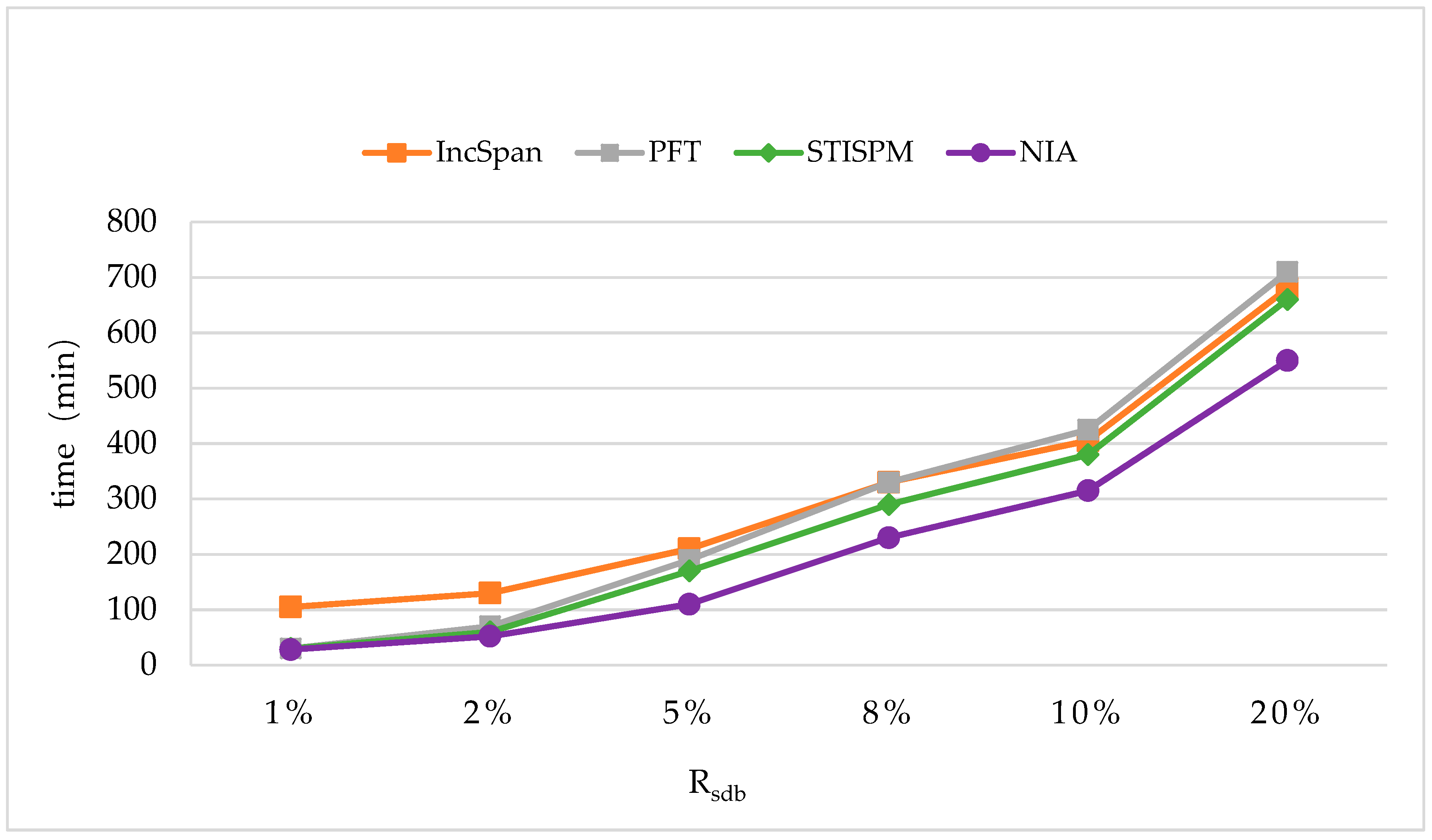
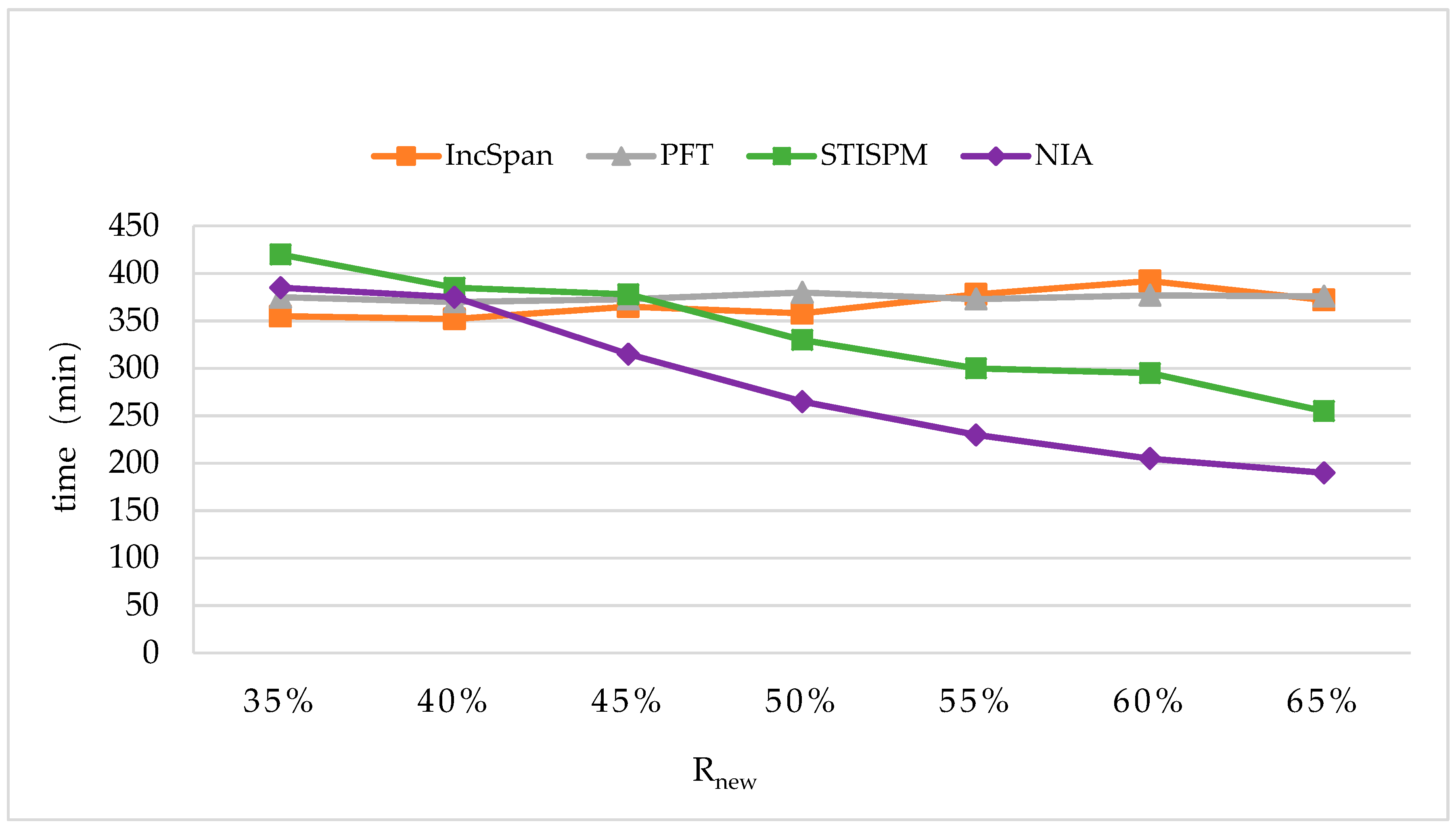
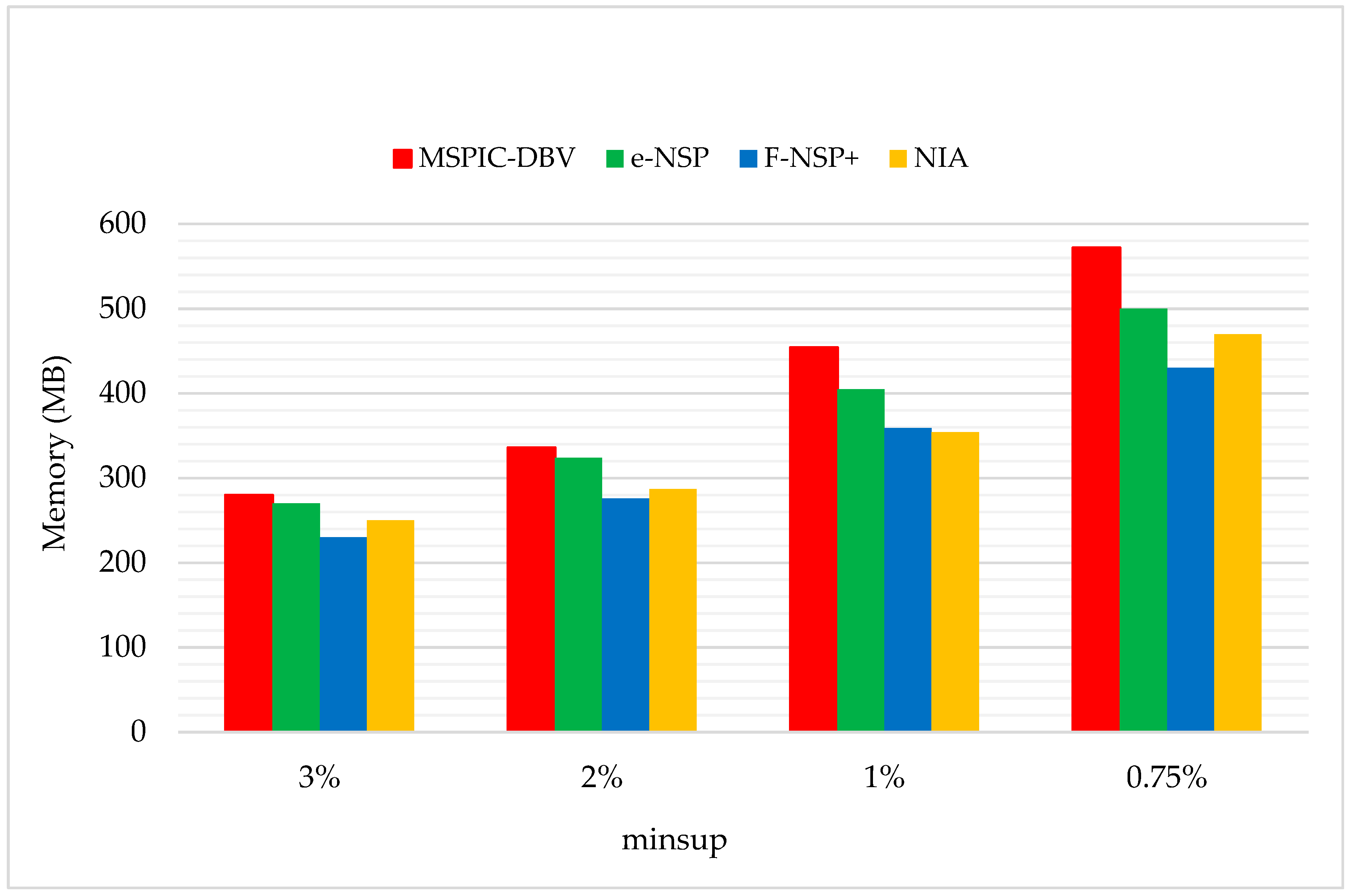
In order to maintain the set of frequent sequences and reduce the searching cost on the original database, a novel incremental mining algorithm for discovering sequential patterns (NIA) is proposed. First, the changes of the potential frequent sequence are discussed regarding when the database is updated, then an algorithm named PrefixSpan+ is suggested to mine the required spaces, that is, the incremental database sdb and the projection database on OD, to obtain complete potential frequent sequences. Finally, the sequence support degree is computed. A data structure, named reticular sequence tree, is introduced in this process. The sequence tree is generated using the corresponding potential frequent sequence, then each support degree can be computed from the root. This method greatly reduces the time to scan the original database during computing the sequence support.
3.1. PrefixSpan+
PrefixSpan (Prefix-Projected Pattern Growth) [
17] is a classic SPM algorithm. Its general ideal is to examine only the prefix subsequences and project only their corresponding postfix subsequences into projected databases. In each projected database, sequential patterns are grown by exploring only local frequent patterns. There is no candidate sequence produced during the mining process and the projected database keep shrinking; however, PrefixSpan still involves a high computation cost owing to constructing the projection database recursively, so it is unsuitable for large-scale sequence datasets with various items. In this section, an improvement of the original PrefixSpan algorithm, called PrefixSpan+, is suggested with the use of a structure named CMAP (Co-occurrence MAP) [
27] in order to enhance the efficiency of the recursive process mentioned above. CMAP is a structure for storing co-occurrence information, and it was originally used to candidate pruning for vertical sequential pattern mining. Here, it is used to extend the prefixes to frequent sequential patterns instead of constructing projection databases recursively. The definition of CMAP is given below.
Definition 1. An item is said to succeed by to an item in a sequence iff for an integer such that and , where means lexicographical order.
Definition 2. An item is said to succeed by to an item in a sequence iff and for some integers and such that .
Definition 3. A map (CMAP) is a structure mapping each item to a set of items succeeding it. We define two CMAPs named and . maps each item to the set of all items succeeding by in no less than minsup sequences of SDB. maps each item to the set of all items succeeding by in no less than minsup sequences of SDB.
The CMAP structure can be used for pruning candidates generated by sequential pattern mining algorithms based on the following properties.
Property 1 (pruning an i-extension). Let there be a frequent sequential pattern and an item . If there exists an item in the last itemset of such that belongs to , then the i-extension of with is frequent.
Property 2 (pruning an i-extension). Let there be a frequent sequential pattern and an item . If there exists an item such that the item belongs to , then the s-extension of with is frequent.
Property 3 (pruning a prefix). The previous properties can be generalized to prune all patterns starting with a given prefix. Let there be a frequent sequential pattern and an item . If there exists an item (equivalently in the last itemset of ) such that there is an item (equivalently in ), then all supersequences having as prefix and where succeeds by s-extension (equivalently i-extension to the last itemset) in in are frequent.
PrefixSpan+ and the original PrefixSpan are based on the same principle to obtain frequent sequential patterns. The difference is that in PrefixSpan+, all the patterns are extended by i-extension and s-extension of the prefixes, and the completeness is guaranteed by the properties stated above. Then, the whole recursive process is more efficient.
We refer readers to Reference [
27] for more details on CMAP. Now the ideal of PrefixSpan+ and the mining process are illustrated with an example. Given a sequence database (
Table 9), suppose
, then the threshold of the support number is 2. The aim of the algorithm is to find out all the frequent sequential patterns satisfied with the support requirement.
First, calculate support number of the prefixes with
. The results are listed in
Table 10. Apparently, (10), (20), (50), and (60) are not frequent sequences under the required minsup, and all the sequences prefixed with them are infrequent, therefore they are deleted from the original database (
Table 11). Meanwhile, length-1 sequential patterns are obtained, which are (30), (40), (70), (80), and (90).
Second, construct a CMAP for length-1 sequential patterns from the modified sequence database according to Definition 1–3 (see
Table 12). Item
is in the last itemset of a frequent sequential pattern (
i-extension) or it is a part of a frequent sequential pattern (
s-extension). In length-1 sequential patterns,
.
The extensions that are frequent are demonstrated in Property 1–3. We can infer from the properties that if an item belongs to a certain or , then the i-extension/s-extension of the corresponding pattern with is frequent. That is to say, all the frequent sequential patterns can be obtained through proper extensions. Taking length-1 sequential pattern (30) as example, the length-2 sequential patterns prefixed with (30) can be generated from and . The results of i-extension of (30) with the items in are (30, 70) and (30, 80), which is the extension in the itemset; the results of the s-extension of (30) with the items in are (30)(40), (30)(70), and (30)(90), which is the extension between the itemsets. In the same way, the length-2 sequential patterns prefixed with (40) and (70) can be obtained, that is, (40, 70) and (70, 80). We can notice that it just needs an i-extension since , and there is no need to carry out any extension on (80) and (90), since both of the and are empty. Then, all the length-2 sequential patterns are obtained, which are (30, 70), (30, 80), (30)(40), (30)(70), (30)(90), (40, 70), and (70, 80).
Next, by constructing the CMAP for length-2 sequential patterns, it can be found that only structure
exists on (30)(40) and (30, 70) as shown in
Table 13. Therefore, all the length-3 sequential patterns can be obtained through
i-extension, which are (30)(40, 70) and (30, 70, 80). The CMAP for length-3 sequential patterns are constructed recursively; however, there is no
or
that exists, which means no length-4 sequential patterns exist. Then, the mining process is completed, and all the frequent sequential patterns are listed in
Table 14.
Through two kinds of extensions, PrefixSpan+ can effectively discover all the frequent sequential patterns without constructing the projection database recursively. The core code of PrefixSpan+ is as shown in Algorithm 1.
| Algorithm 1 The core code of PrefixSpan+ |
| Input: Sequence dataset S, Threshold of support degree min_sup |
| Output: The set of frequent sequential pattern FS |
| 1: | Scan S, count the support number of all the items with length = 1, and delete the items from S, of which the support degree is less than min_sup, then obtain S’. The set of length-1 sequential pattern P contains all the items that are satisfied with the required support degree. |
| 2: | FS = P |
| 3: | Extension(S’, P) |
| 4: | For each , construct CMAP for from S’ |
| 5: | if |
| 6: | return FS |
| 7: | else |
| 8: | if , traverse |
| 9: | extend all the items in to by i-extension, and obtain |
| 10: | if , traverse |
| 11: | extend all the items in to by s-extension, and obtain |
| 12: | // Property 3 |
| 13: | , |
| 14: | Extension(S’, P’) |
3.2. Obtain Potential Frequent Sequence
In this section, the sequence-related properties in the updating process are analyzed in depth, then the constitution of the set of potential frequent sequence is confirmed; furthermore, an algorithm named GetPFS is designed to obtain the potential frequent sequences.
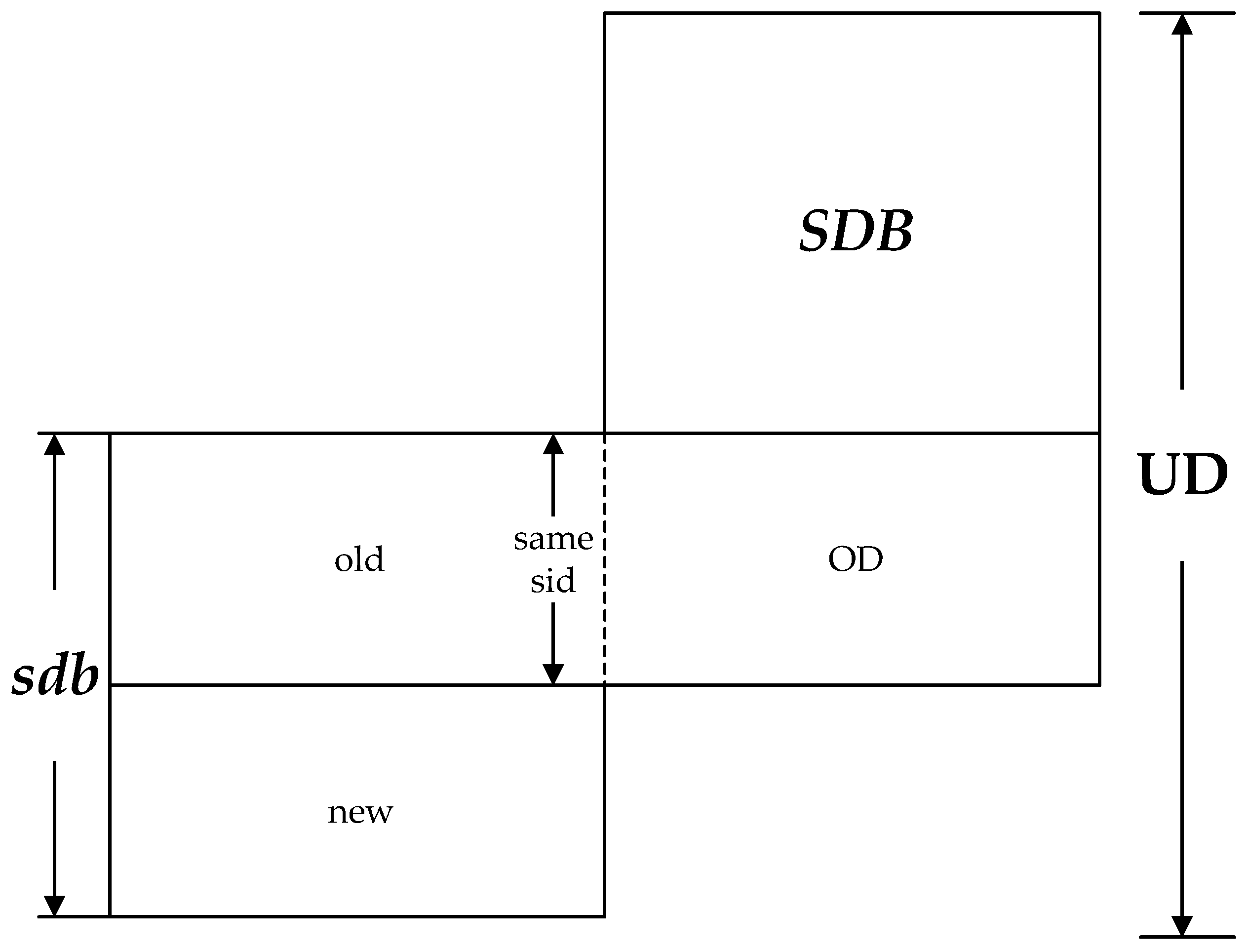
Theorem 1. The new frequent sequences in UD come from the following three parts:
- 1.
Frequent sequences in the original SDB;
- 2.
The sequences in sdb of which the support number are no less than ;
- 3.
The sequences discovered from the prefix projection database of potential frequent sequence of sdb in DD, and .
The proof of theorem 1 is based on the following eight lemmas.
Lemma 1. The frequent sequences of original database must be contained in the set of potential frequent sequences.
Proof. To the frequent sequences in original database, the support degree of these sequences in UD cannot be decreased. Owing to , the value of is probably more than such that even the degree of the sequences is not increased in the updated database. Therefore, the potential sequences must contain the frequent sequences in the original database. □
Lemma 2. Only the sequences, of which the support number is increased after updating of database, can be the frequent sequences.
Proof. For the infrequent sequences in the original database, because of , cannot be more than if the support degree is not increased. □
Lemma 3. The change of support number will only happen in two kinds of sequences as follows after updating the database.
- 1.
The sequences emerged in sdb, including the sequences existing in SDB and the new sequences produced from sdb;
- 2.
The new sequences produced from the combination of OD and old.
Proof. Once the updating of SDB is completed, the sequence, of which the support degree has been changed, must be related with sdb; that is, either it is from database new or from database old. If the sequence is related with database new, then it is condition 1; if the sequence is related with database old, then it is entirely or partly from old, which fall into condition 1 and 2 respectively. □
Lemma 4. For each sequence in UD, the inequality holds.
Proof. For some sequences in UD, the first half part emerges in SDB, and the latter part emerges in sdb. Besides, some certain sequences may appear in both the SDB part and the sdb part of a certain sequence. However, the sequences in UD can contribute one to the support number at most, therefore, the inequality holds. □
Lemma 5. For the sequences in sdb that are infrequent in SDB, the inequation holds if the sequences are frequent in UD.
Proof. Suppose
is frequent in UD, then
. Therefore:
and
is infrequent in
SDB, then
, therefore:
□
Therefore, if the support number of sequence in sdb is no less than , the sequence may become frequent; thus, it should be included in the set of potential frequent sequence.
Lemma 6. If the new sequences produced from the combination of OD and old are frequent, then, for the part from old, it must be included in the set of potential frequent sequences of sdb derived from Lemma 5.
Proof. If sequence is frequent, the part from old is also frequent, according to Lemma 5, where the part of that exists in old is contained in potential frequent sequences. □
Lemma 7. For the new sequences generated from the combination of OD and old, the support number is.
Proof. There are new sequences that emerge after combination, and the new ones may emerge in new or the original database SDB. Owing to the support number of a same sequence, from DD and OD, it may calculate twice or even more times, for instance, in the case of ; thus, the contribution from OD should be subtracted from the sum of the support number in SDB, DD, and new during the calculation. □
Definition 4. Suppose sequences , with . is a postfix projection of to if the following requirements are satisfied: (1) is the postfix of ; (2) there does not exist a sequence that satisfies , and is the postfix of .
The attribute of postfix projection is described in Definition 4, which means a postfix projection to is the maximum subsequence of postfixed with . For instance, given a sequence , if , then the postfix projection ; if , then .
Definition 5. Suppose there exist sequences , with , then the postfix projection is according to Definition 4. Sequence which is the rest of except , and is called a prefix of to , and denoted as . Let SDB be a sequence database, and is a sequence in SDB, then the set of prefixes from projections of all the sequences in SDB to , is called a prefix database of the projection of SDB to , and denoted as .
For instance, there are two sequences in SDB, which are and , and . For , and ; for , and . Then, the is .
Based on the ideal of PrefixSpan+, a “PostfixSpan+” algorithm can be obtained through reversing the direction of i-extension and s-extension during the extending process; for instance, for a sequence , the i-extension of item 40 is 70, and the s-extension is in PrefixSpan+. In PostfixSpan+, the i-extension of item 40 is , and the s-extension is 30. In the following, the Postfix algorithm is utilized to process the new sequences produced from the combination of OD and old.
Lemma 8. Construct to the potential frequent sequences of sdb in DD. If the new sequences produced from the combination of OD and old are frequent, then they must be included in the sequences generated from the ; also, the inequation holds.
Explanation. All the sequences from old are already involved in potential frequent sequences of sdb, therefore they do not need to be the elements of the prefix projection database. For instance, sequence and potential frequent sequence ; then, the prefix of projection is not , but .
Proof. The sequences generated from the combination of OD and old are frequent, thus the part of them, which are from
sdb, must be potential sequences; meanwhile, to the prefix projection database constructed by all the potential frequent sequences, the frequent sequences generated from the combination of OD and old are involved in it. Then, the following inequation holds:
□
Therefore, if the sum of the support numbers, in DD and new, of the sequences produced by the combination OD and old is no less than , they might become frequent sequences, which means they should be involved in the set of potential frequent sequence.
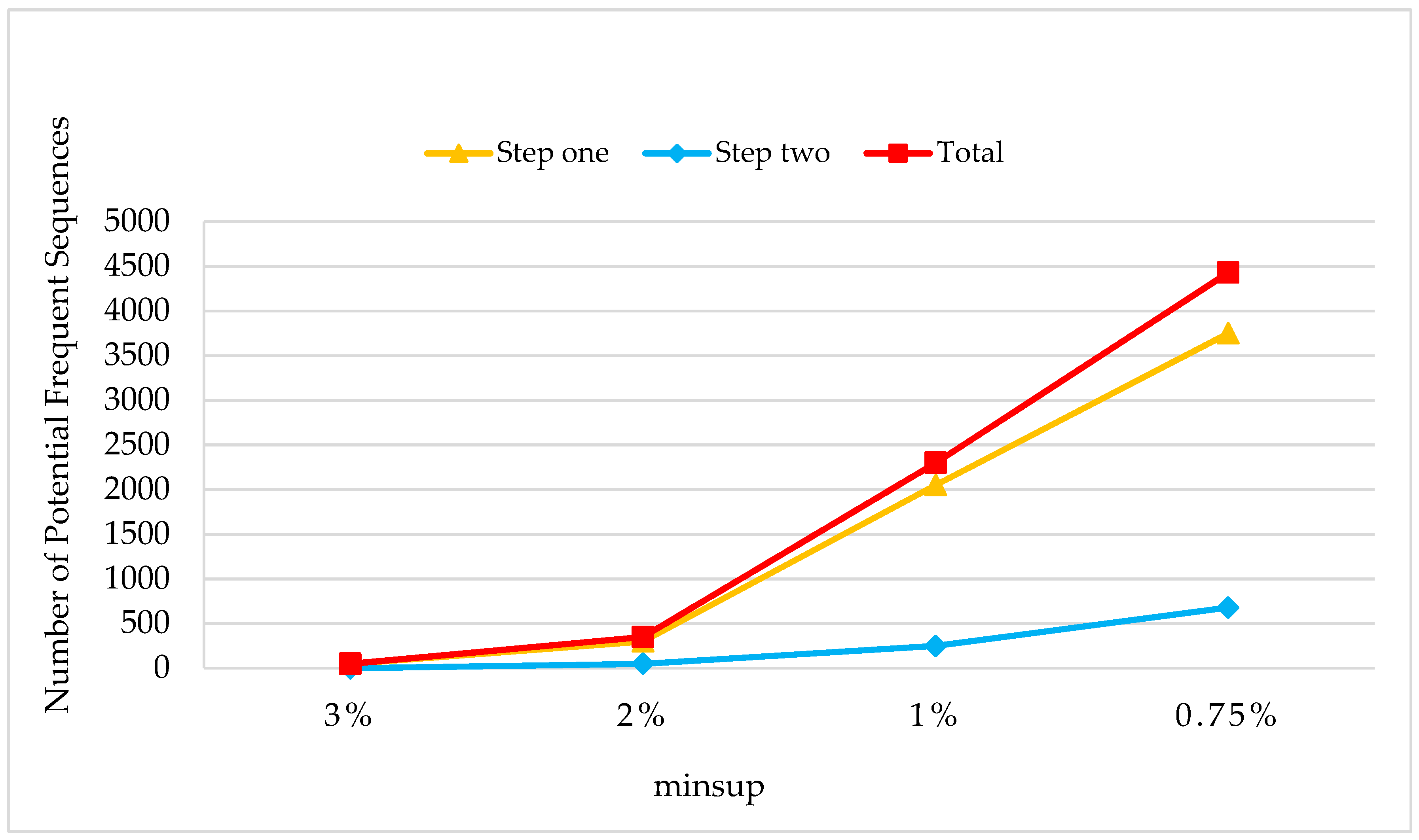
Therefore, we conclude from the above lemmas that if a sequence is frequent in UD, then it must be satisfied by one of the conditions mentioned above, which means it will be added into the set of potential frequent sequence. It just needs two steps to obtain a potential frequent sequence:
Step one: Implement sequence mining on sdb by using PrefixSpan+, setting the threshold of the support number as ;
Step two: Construct the prefix projection database under DD to potential frequent sequences in sdb, then implement sequence mining on the constructed databases by using PostfixSpan+, setting the threshold of the support number as .
Suppose the FS (frequent sequence) is the set of frequent sequences in SDB, PFS (potential frequent sequence) as the set of potential frequent sequence, and NFS (new frequent sequence) is the set of frequent sequence in updated database. The core code of algorithm GetPFS, which is used to obtain the potential frequent sequences, is described in Algorithm 2:
| Algorithm 2 GetPFS (OD, DD, FS, old, new) |
| Input: OD, DD, FS, old, new |
| Output: PFS |
| 1: | Put FS into PFS |
| 2: | Set |
| 3: | Putinto PFS |
| 4: | Forin PFS |
| 5: | Construct projection database of DD to the part of that belongs to OD |
| 6: | Set |
| 7: | Put into PFS |
3.3. Fast Support Number-Counting Algorithm
In order to calculate the support degree once the potential sets of frequent sequence (PFS) are obtained, a fast counting algorithm is proposed in this section. To level up the efficiency of calculating, a reticular structure is designed to store the PFS, making the speed of counting faster than the traditional methods such as a hash function-based algorithm.
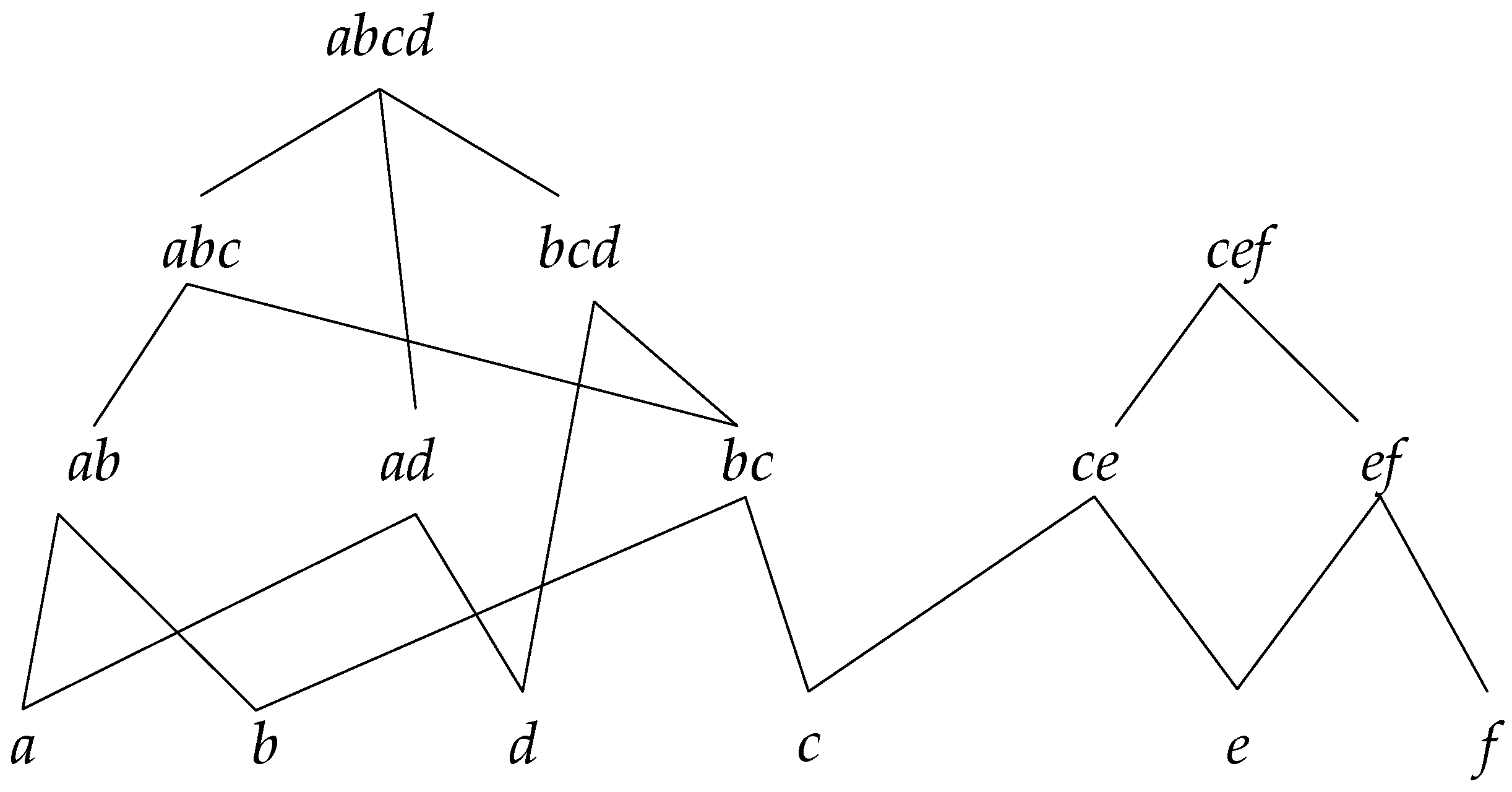
Figure 2 shows a reticular sequence tree structure, with
.
In this tree structure, and are the root nodes, and each child node is the subsequence of the parent node; also, if a sequence is the subsequence of another, then it must be in the reticular sequence tree with the parent sequence as root node.
The concrete process for constructing a reticular sequence tree is as follows. Each layer is comprised of the sequences with the same length. The first layer is the set of the longest sequences, and each node is a root node. Then, check the rest of the sequences in decreasing order to determine if they are the subsequence of the node in the higher layer; if so, add them as the corresponding child node, and if not, set them as the root node.
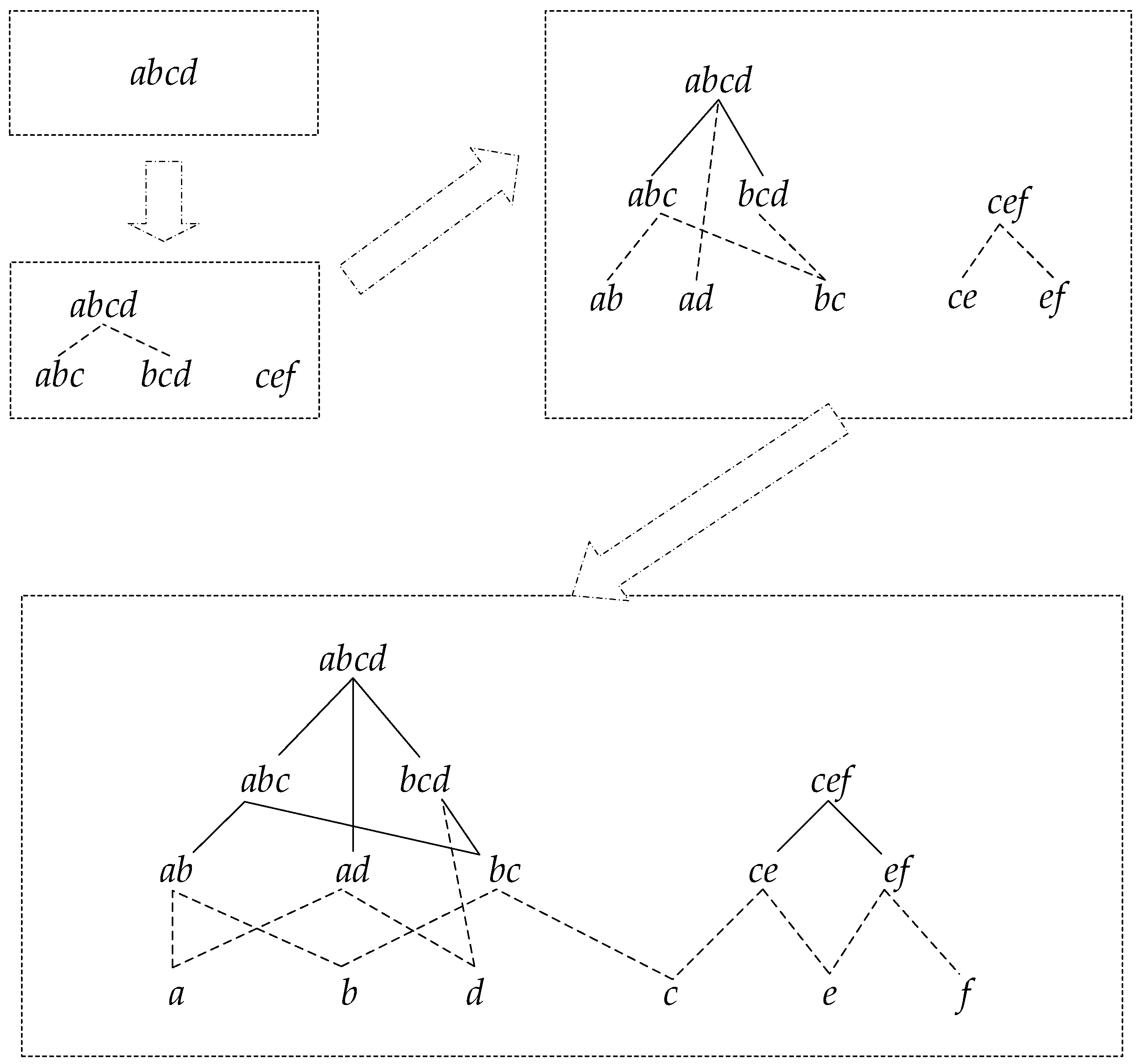
Figure 3 shows the detailed construction process.
The support number of each node can be obtained through two steps. First, count the elementary support number. Check whether the sequence contains a root node traversing all the sequences in the updated database; if so, the support number of the root node adds one. If not, check if it contains the sequence of the child node; if so, the support number of the child node adds one, and if not, check recursively. For instance, if there is a sequence
in the database, for root node
, which is contained in the sequence, then its support number adds one; if the sequence is
, it does not contain root node
, but for its child node
, the support number should add one. We can notice that the child nodes do not need to be checked if there is any father node included in the sequence. Supposing there is a set of results counted from step one, as shown in
Figure 4, the number on the right side of each node is the elementary support number.
In step two, the real support number of each node is obtained by calculating the sum of elementary support number of itself and all the father nodes. For instance, as shown in
Figure 4, the elementary support number of node
is 2, and the numbers of its father nodes
and
are 2 and 3, respectively; therefore, the real support number of
is 7. However, for node
, it has three father nodes—
, and
—where
is the common father node of
and
; then, its elementary support number can just be counted once, so the real support number of
is 9. That is to say, in a diamond structure like
Figure 5, the elementary support number of all the common father nodes can only be added one time when counting the real support number.
In the real programming process, counting the real support number and determining whether the sequence is frequent or not could be carried out simultaneously. First, the support number is calculated from a leaf node: if it is equal or greater than the minimum support number, then determine whether its father node is frequent; if the support number is less than the minimum support number, it can be confirmed that all the father nodes are not in the frequent sequence.
To sum up, the fast support number-counting algorithm is comprised of three steps:
Construct a reticular sequence tree;
Count the support number on the constructed reticular sequence tree by traversing the updated sequence database;
Count the real support number and determine the frequent sequences.