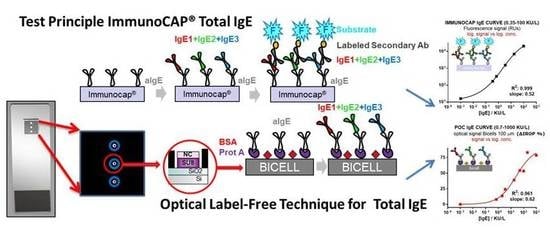
A Proof-of-Concept of Label-Free Biosensing System for Food Allergy Diagnostics in Biophotonic Sensing Cells: Performance Comparison with ImmunoCAP
Abstract
1. Introduction
2. Materials and Methods
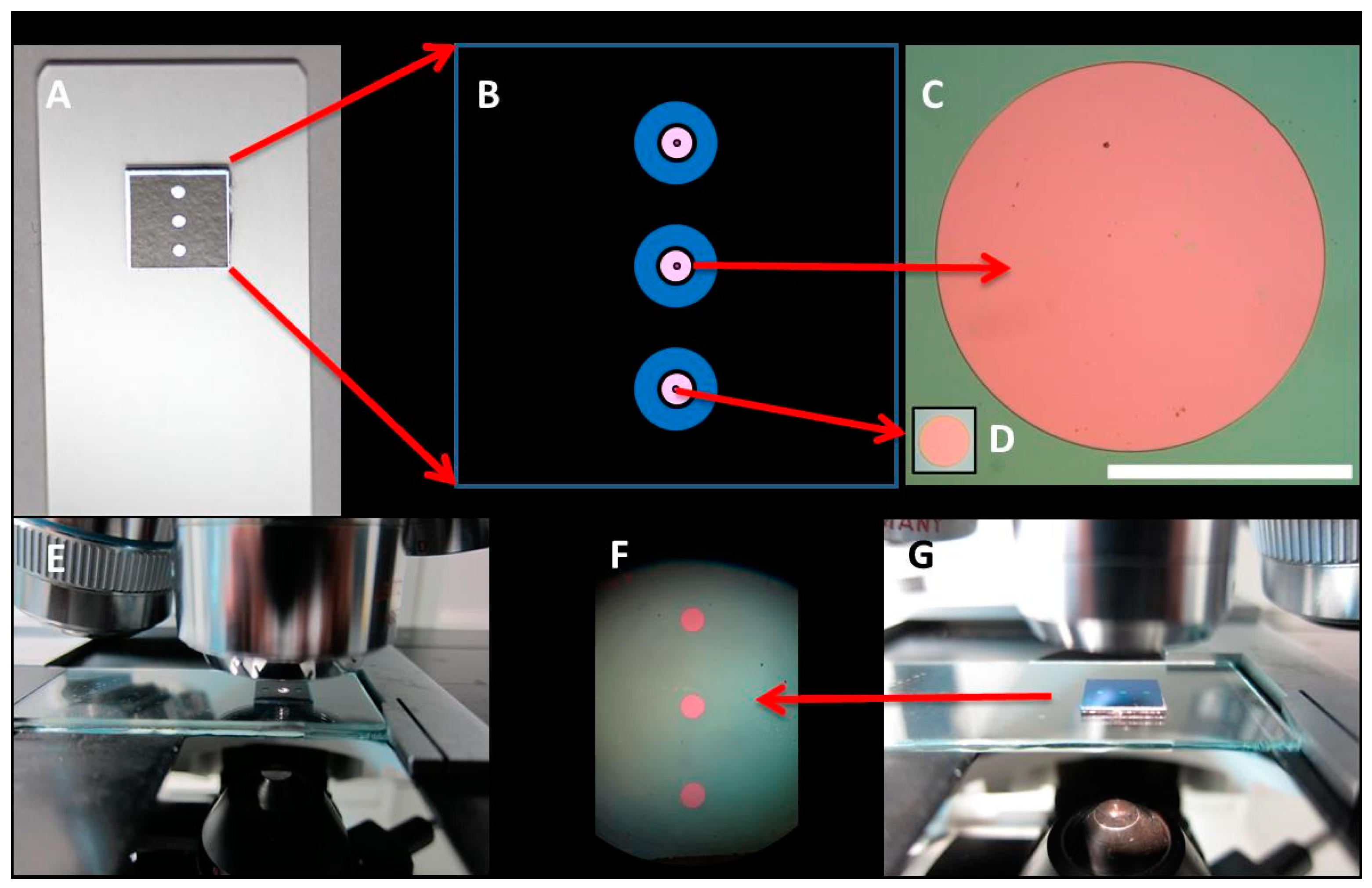
2.1. BICELLs Fabrication and Materials
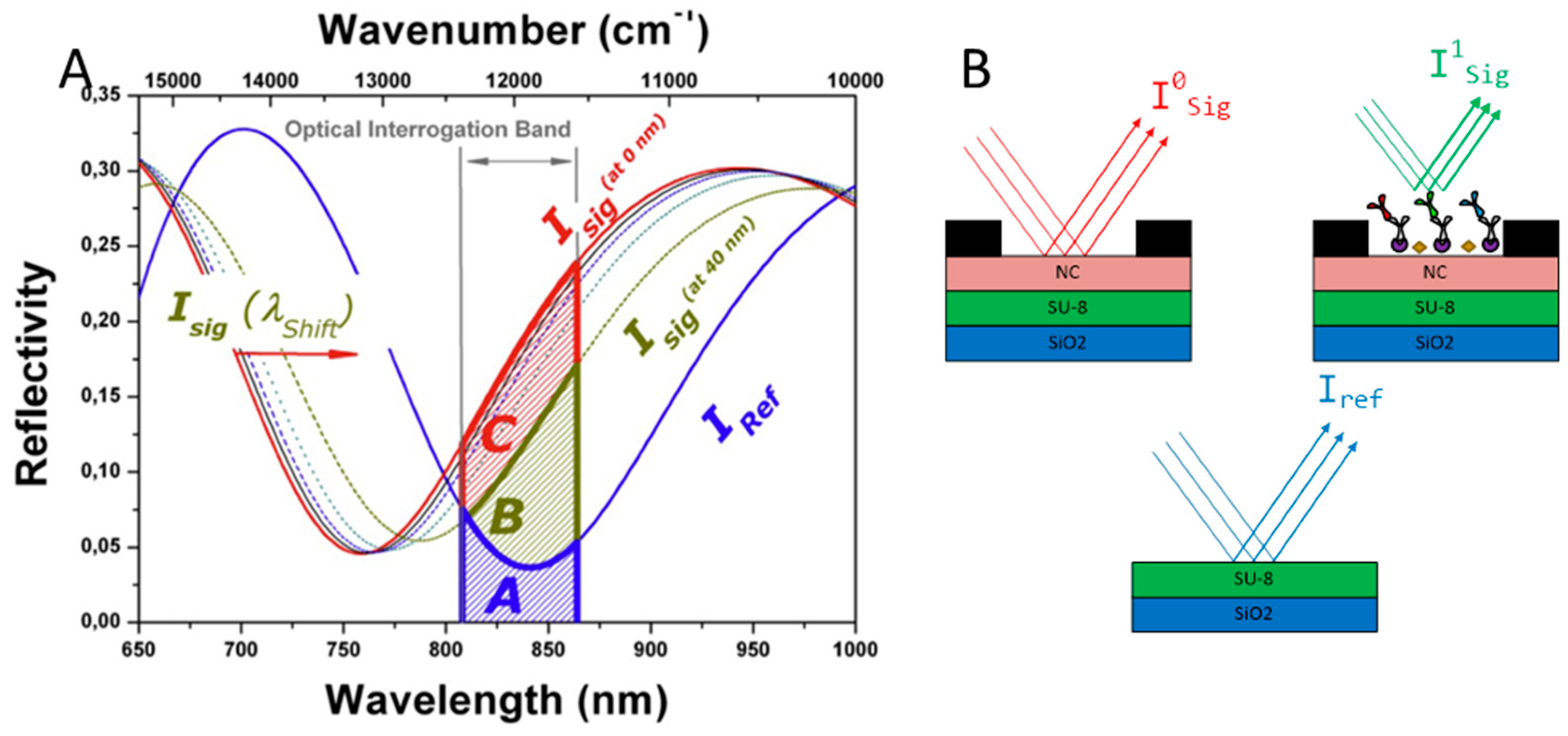
2.2. Optical Label-Free Characterization of BICELLs
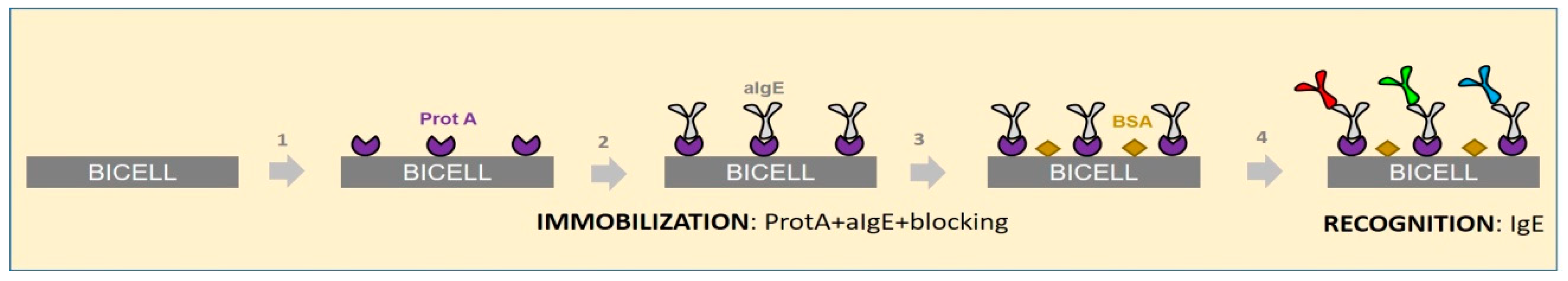
2.3. ImmunoCAP® Protocol for Obtaining of Calibration Curves Anti-IgE/IgE
2.4. Point of Care Protocol for Obtaining of Calibration Curves Anti-IgE/IgE
3. Results and Discussion
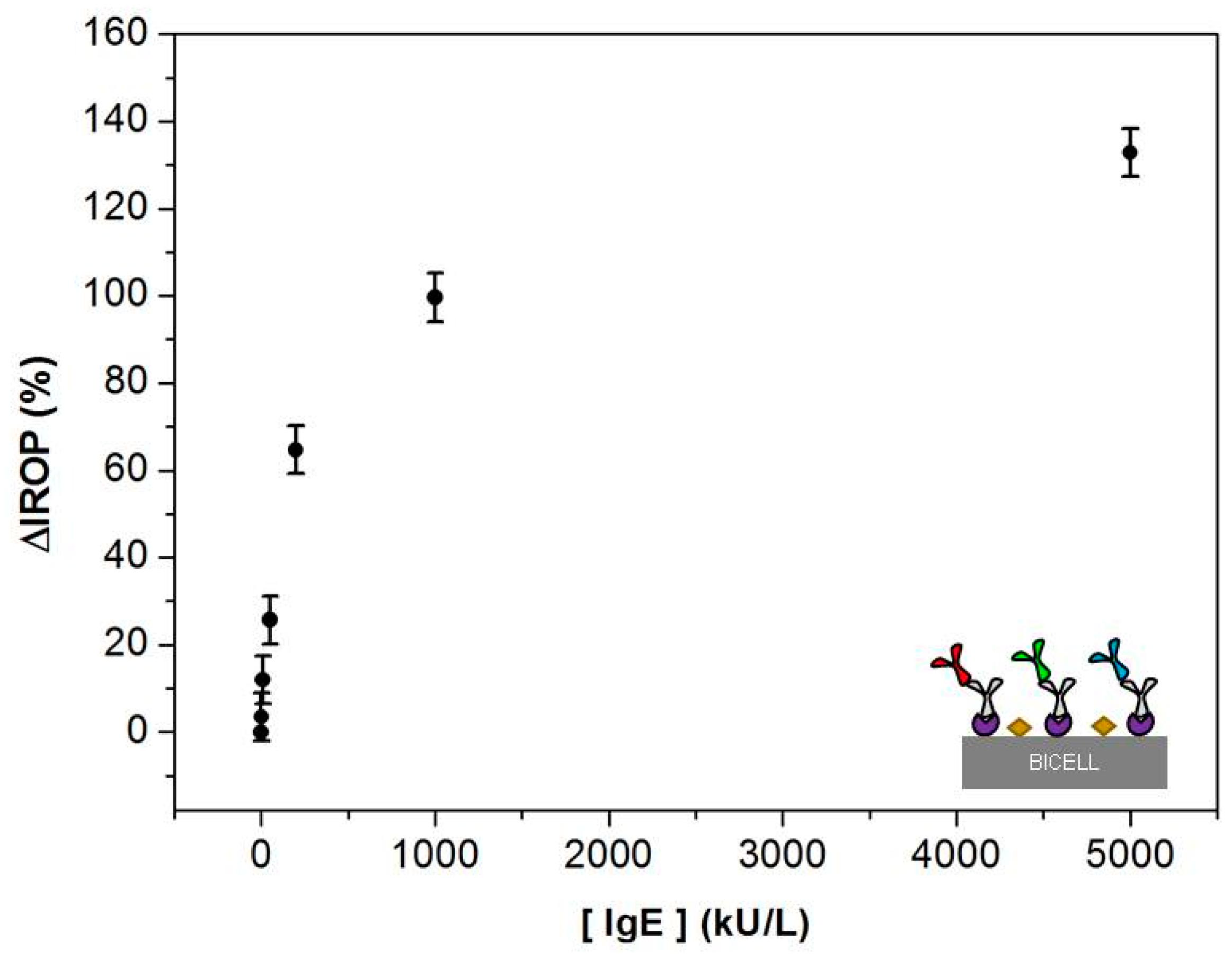
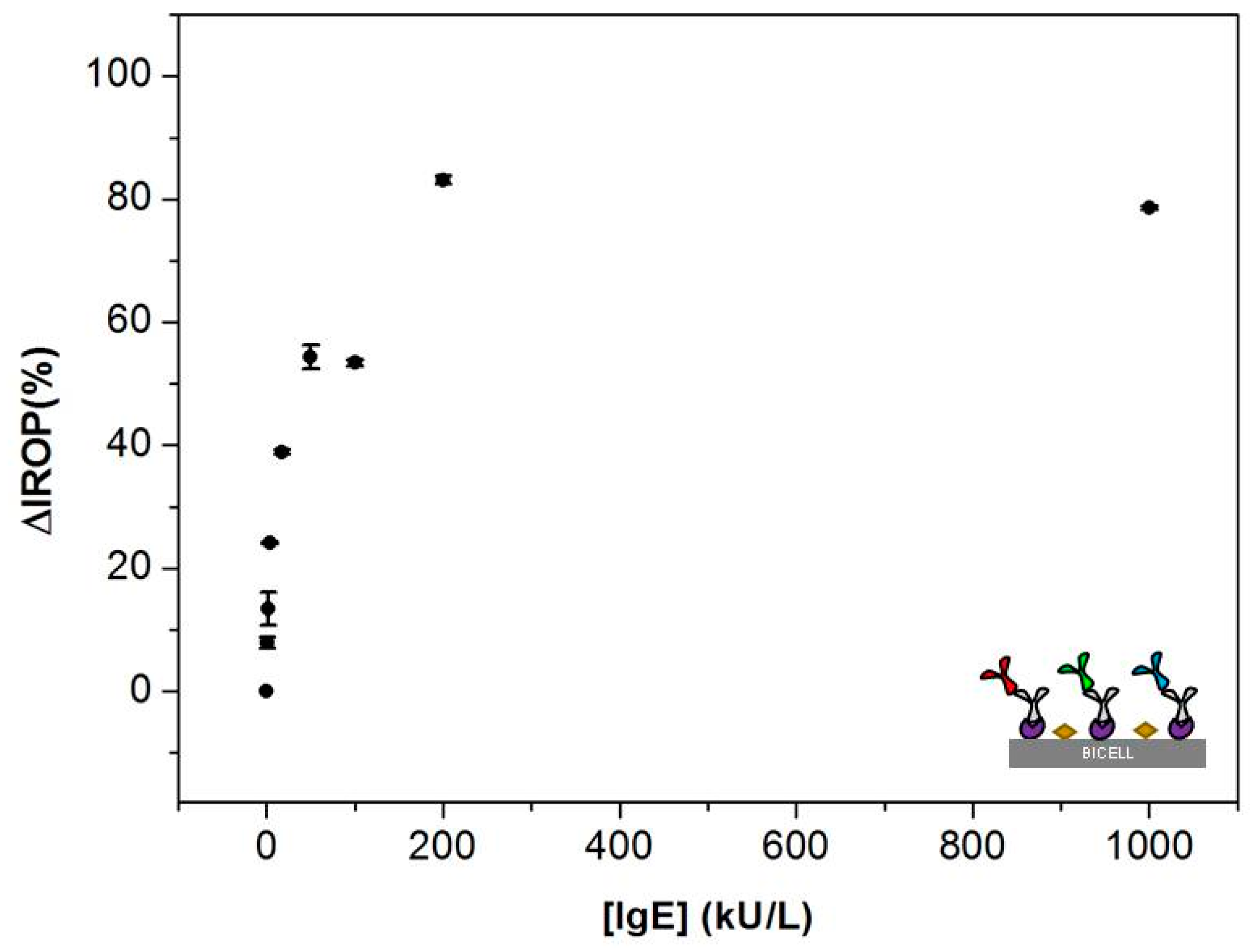
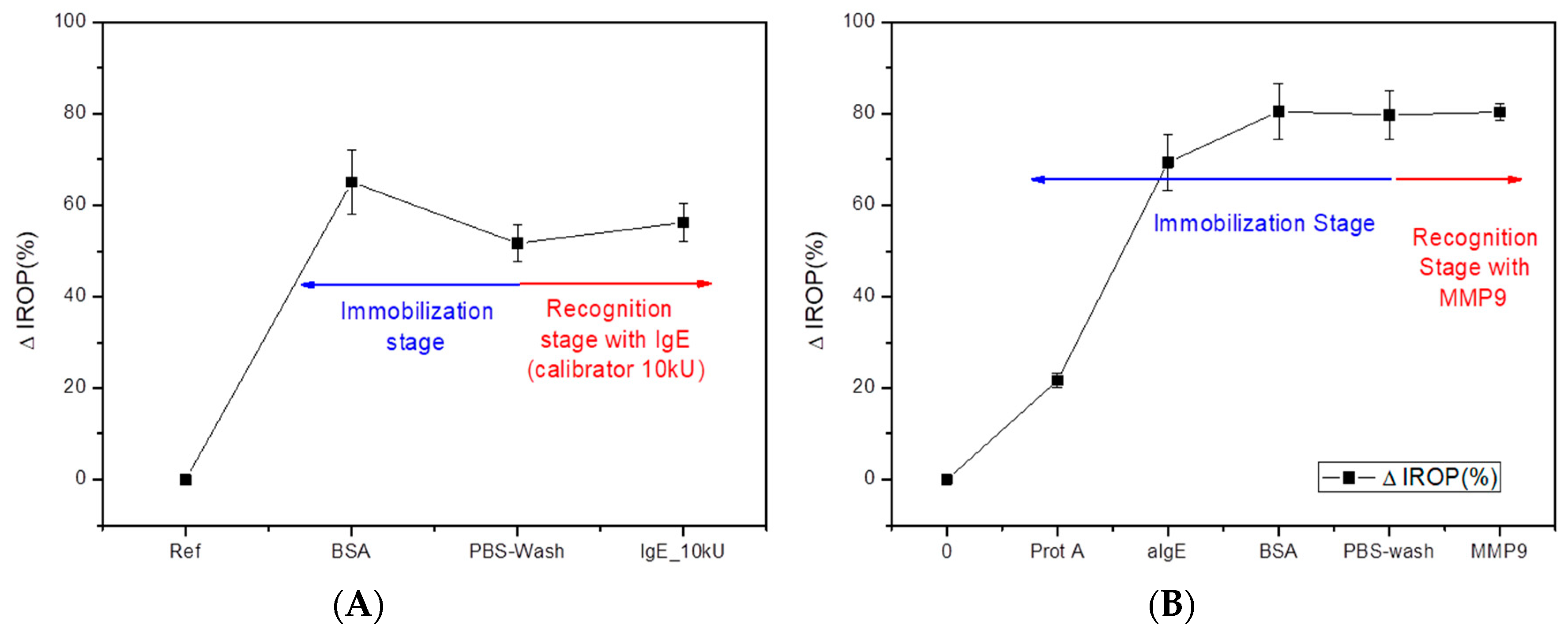
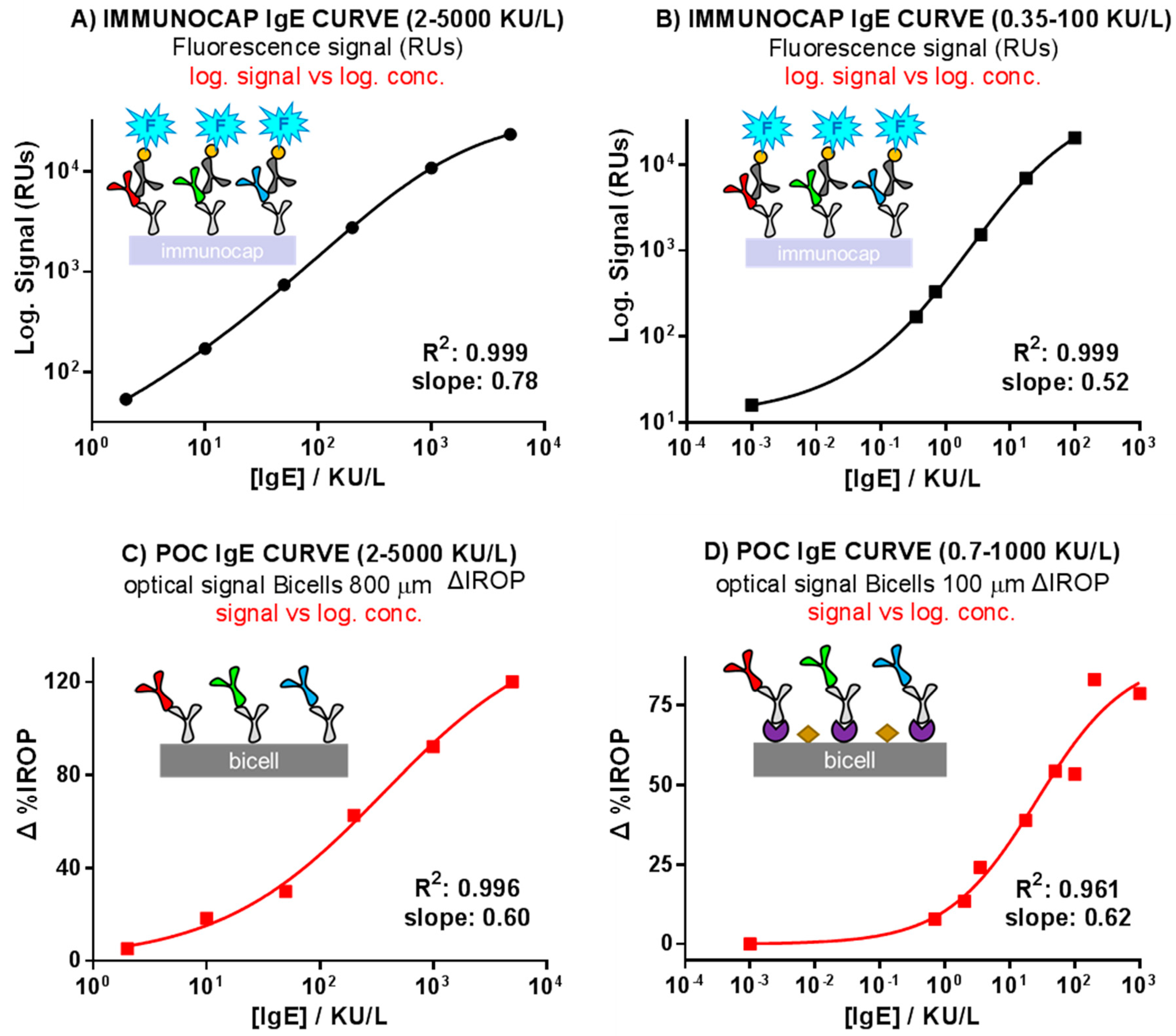
3.1. Calibration Curves Obtained by Optical Label-Free Technique
3.2. Comparison of the ImmunoCAP® Versus the Optical Label-Free Technique Calibration Curves
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muraro, A.; Werfel, T.; Hoffmann-Sommergruber, K.; Roberts, G.; Beyer, K.; Bindslev-Jensen, C.; Cardona, V.; Dubois, A.; duToit, G.; Eigenmann, P.; et al. EAACI Food Allergy and Anaphylaxis Guidelines: Diagnosis and management of food allergy. Allergy 2014, 69, 1008–1025. [Google Scholar] [CrossRef] [PubMed]
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Roberts, G.; Muraro, A.; Sheikh, A. Prevalence of common food allergies in Europe: A systematic review and meta-analysis. Allergy 2014, 69, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Muraro, A.; Werfel, T.; Cardona, V.; Dubois, A.E.J.; Halken, S.; Hoffmann-Sommergruber, K.; Poulsen, L.K.; et al. EAACI Food Allergy and Anaphylaxis Guidelines Group The epidemiology of food allergy in Europe: A systematic review and meta-analysis. Allergy 2014, 69, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Pawankar, R.; Canonica, G.W.; Holgate, S.T.; Lockey, R.F. World Allergy Organization (WAO) White Book on Allergy; WAO: Milwaukee, WI, USA, 2011; ISBN 0615461824. [Google Scholar]
- Panesar, S.S.; Nwaru, B.I.; Hickstein, L.; Rader, T.; Hamadah, H.; Ali, D.F.I.; Patel, B.; Muraro, A.; Roberts, G.; Worm, M.; et al. The epidemiology of anaphylaxis in Europe: Protocol for a systematic review. Clin. Transl. Allergy 2013, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.A.; Assa’ad, A.; Burks, A.W.; Jones, S.M.; Sampson, H.A.; Wood, R.A.; Plaut, M.; Cooper, S.F.; Fenton, M.J.; et al. Guidelines for the diagnosis and management of food allergy in the United States: Report of the NIAID-sponsored expert panel. J. Allergy Clin. Immunol. 2010, 126, S1–S58. [Google Scholar] [CrossRef] [PubMed]
- Klimek, L.; Hoffmann, H.J.; Renz, H.; Demoly, P.; Werfel, T.; Matricardi, P.M.; Muraro, A.; Schmid-Grendelmeier, P.; Cardona, V.; Papadopoulos, N.G. Diagnostic test allergens used for in vivo diagnosis of allergic diseases are at risk: A European Perspective. Allergy Eur. J. Allergy Clin. Immunol. 2015, 70, 1329–1331. [Google Scholar] [CrossRef] [PubMed]
- Asarnoj, A.; Hamsten, C.; Lupinek, C.; Melén, E.; Andersson, N.; Anto, J.M.; Bousquet, J.; Valenta, R.; van Hage, M.; Wickman, M. Prediction of peanut allergy in adolescence by early childhood storage protein-specific IgE signatures: The BAMSE population-based birth cohort. J. Allergy Clin. Immunol. 2017, 140, 587–590.e7. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, E.; Hamsten, C.; Sibanda, E.; Ochome, M.; Focke-Tejkl, M.; Asarnoj, A.; Önell, A.; Lilja, G.; Gallerano, D.; Lupinek, C.; et al. Natural clinical tolerance to peanut in African patients is caused by poor allergenic activity of peanut IgE. Allergy Eur. J. Allergy Clin. Immunol. 2015, 70, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Van Hage, M.; Hamsten, C.; Valenta, R. ImmunoCAP assays: Pros and cons in allergology. J. Allergy Clin. Immunol. 2017, 140, 974–977. [Google Scholar] [CrossRef] [PubMed]
- Matricardi, P.M.; Kleine-Tebbe, J.; Hoffmann, H.J.; Valenta, R.; Hilger, C.; Hofmaier, S.; Aalberse, R.C.; Agache, I.; Asero, R.; Ballmer-Weber, B.; et al. EAACI Molecular Allergology User’s Guide. Pediatr. Allergy Immunol. 2016, 27, 1–250. [Google Scholar] [CrossRef] [PubMed]
- Borres, M.P.; Maruyama, N.; Sato, S.; Ebisawa, M. Recent advances in component resolved diagnosis in food allergy. Allergol. Int. 2016, 65, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.B.; Barnes, J.H.; Szeinbach, S.L.; Sullivan, T.J. Analytic precision and accuracy of commercial immunoassays for specific IgE: Establishing a standard. J. Allergy Clin. Immunol. 2000, 105, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Goikoetxea, M.J.; Sanz, M.L.; García, B.E.; Mayorga, C.; Longo, N.; Gamboa, P.M.; Barber, D.; Caballero Molina, T.; de la Calle Toral, A.; et al. Recommendations for the use of in vitro methods to detect specific immunoglobulin E: Are they comparable? J. Investig. Allergol. Clin. Immunol. 2013, 23, 448–454. [Google Scholar] [PubMed]
- Lee, Y.W.; Sohn, J.H.; Lee, J.-H.; Hong, C.-S.; Park, J.-W. Allergen-specific IgE measurement with the IMMULITE 2000 system: Intermethod comparison of detection performance for allergen-specific IgE antibodies from Korean allergic patients. Clin. Chim. Acta 2009, 401, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Von Lode, P. Point-of-care immunotesting: Approaching the analytical performance of central laboratory methods. Clin. Biochem. 2005, 38, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Holgado Bolaños, M.; Casquel del Campo, R.; Molpeceres Álvarez, C.; Ocaña Moreno, J.L.; Laguna Heras, M.F.; Morales Furio, M. Optical Detection System for Labelling-Free High Sensitivity Bioassays. European Patent EP20090811139, 16 March 2010. [Google Scholar]
- Holgado, M.; Maigler, M.V.; Santamaría, B.; Hernandez, A.L.; Lavín, A.; Laguna, M.F.; Sanza, F.J.; Granados, D.; Casquel, R.; Portilla, J.; et al. Towards reliable optical label-free point-of-care (PoC) biosensing devices. Sens. Actuators B Chem. 2016, 236, 765–772. [Google Scholar] [CrossRef]
- Holgado Bolaños, M.; Sanza Gutierrez, F.J.; Laguna Heras, M.F.; Lavin Hueros, A.; Casquel del Campo, R. Interferometric Detection Method. European Patent EP2013/066298, 2 May 2014. [Google Scholar]
- Laguna, M.F.; Sanza, F.J.; Soria, J.; Jara, M.; Lavín, Á.; Casquel, R.; López, A.; Suarez, T.; Holgado, M. Label-free biosensing by means of BICELLs for dry eye. Sens. Actuators B Chem. 2014, 203, 209–212. [Google Scholar] [CrossRef]
- Laguna, M.; Holgado, M.; Hernandez, A.; Santamaría, B.; Lavín, A.; Soria, J.; Suarez, T.; Bardina, C.; Jara, M.; Sanza, F.; Casquel, R. Antigen-Antibody Affinity for Dry Eye Biomarkers by Label Free Biosensing. Comparison with the ELISA Technique. Sensors 2015, 15, 19819–19829. [Google Scholar] [CrossRef] [PubMed]
- Holgado, M.; Barrios, C.A.; Ortega, F.J.; Sanza, F.J.; Casquel, R.; Laguna, M.F.; Bañuls, M.J.; López-Romero, D.; Puchades, R.; Maquieira, A. Label-free biosensing by means of periodic lattices of high aspect ratio SU-8 nano-pillars. Biosens. Bioelectron. 2010, 25, 2553–2558. [Google Scholar] [CrossRef] [PubMed]
- Sanza, F.J.; Holgado, M.; Ortega, F.J.; Casquel, R.; López-Romero, D.; Bañuls, M.J.; Laguna, M.F.; Barrios, C.A.; Puchades, R.; Maquieira, A. Bio-Photonic Sensing Cells over transparent substrates for anti-gestrinone antibodies biosensing. Biosens. Bioelectron. 2011, 26, 4842–4847. [Google Scholar] [CrossRef] [PubMed]
- Laguna, M.; Holgado, M.; Sanza, F.; Lavín, A.; López, A.; Casquel, R. Optimization of Dengue Immunoassay by Label-Free Interferometric Optical Detection Method. Sensors 2014, 14, 6695–6700. [Google Scholar] [CrossRef] [PubMed]
- SU-8 2000 Permanent Epoxy Negative Photoresist, Processing Guidelines. Available online: www.microchem.com/pdf/SU-82000DataSheet2000_5thru2015Ver4.pdf (accessed on 14 August 2018).
- López-Romero, D.; Barrios, C.A.; Holgado, M.; Laguna, M.F.; Casquel, R. High aspect-ratio SU-8 resist nano-pillar lattice by e-beam direct writing and its application for liquid trapping. Microelectron. Eng. 2010, 87, 663–667. [Google Scholar] [CrossRef]
- Santamaría, B.; Laguna, M.; López-Romero, D.; Hernandez, A.; Sanza, F.; Lavín, Á.; Casquel, R.; Maigler, M.; Espinosa, R.; Holgado, M. Development towards Compact Nitrocellulose-Based Interferometric Biochips for Dry Eye MMP9 Label-Free In-Situ Diagnosis. Sensors 2017, 17, 1158. [Google Scholar] [CrossRef] [PubMed]
- Holgado, M.; Sanza, F.; López, A.; Lavín, Á.; Casquel, R.; Laguna, M. Description of an Advantageous Optical Label-Free Biosensing Interferometric Read-Out Method to Measure Biological Species. Sensors 2014, 14, 3675–3689. [Google Scholar] [CrossRef] [PubMed]
- Lavín, Á.; Casquel, R.; Sanza, F.J.; Laguna, M.F.; Holgado, M. Efficient design and optimization of bio-photonic sensing cells (BICELLs) for label free biosensing. Sens. Actuators B Chem. 2013, 176, 753–760. [Google Scholar] [CrossRef]
- Winter, W.E.; Hardt, N.S.; Fuhrman, S. Immunoglobulin E: Importance in parasitic infections and hypersensitivity responses. Arch. Pathol. Lab. Med. 2000, 124, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Qi, Y.; Zhao, Y.; Xiang, Y.; Chen, Q.; Fu, W. Aptamer-based piezoelectric quartz crystal microbalance biosensor array for the quantification of IgE. Biosens. Bioelectron. 2009, 24, 2499–2503. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.; Mesa-Antunez, P.; Estevez, M.C.; Ruiz-Sanchez, A.J.; Otte, M.A.; Sepulveda, B.; Collado, D.; Mayorga, C.; Torres, M.J.; Perez-Inestrosa, E.; et al. Highly sensitive dendrimer-based nanoplasmonic biosensor for drug allergy diagnosis. Biosens. Bioelectron. 2015, 66, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Johansson, S. ImmunoCAP® Specific IgE test: An objective tool for research and routine allergy diagnosis. Expert Rev. Mol. Diagn. 2004, 4, 273–279. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa, R.L.; Laguna, M.F.; Fernández, F.; Santamaria, B.; Sanza, F.J.; Maigler, M.V.; Álvarez-Millán, J.J.; Canalejas-Tejero, V.; Holgado, M. A Proof-of-Concept of Label-Free Biosensing System for Food Allergy Diagnostics in Biophotonic Sensing Cells: Performance Comparison with ImmunoCAP. Sensors 2018, 18, 2686. https://doi.org/10.3390/s18082686
Espinosa RL, Laguna MF, Fernández F, Santamaria B, Sanza FJ, Maigler MV, Álvarez-Millán JJ, Canalejas-Tejero V, Holgado M. A Proof-of-Concept of Label-Free Biosensing System for Food Allergy Diagnostics in Biophotonic Sensing Cells: Performance Comparison with ImmunoCAP. Sensors. 2018; 18(8):2686. https://doi.org/10.3390/s18082686
Chicago/Turabian StyleEspinosa, Rocio L., María Fe Laguna, Fátima Fernández, Beatriz Santamaria, Francisco Javier Sanza, Maria Victoria Maigler, Juan J. Álvarez-Millán, Víctor Canalejas-Tejero, and Miguel Holgado. 2018. "A Proof-of-Concept of Label-Free Biosensing System for Food Allergy Diagnostics in Biophotonic Sensing Cells: Performance Comparison with ImmunoCAP" Sensors 18, no. 8: 2686. https://doi.org/10.3390/s18082686
APA StyleEspinosa, R. L., Laguna, M. F., Fernández, F., Santamaria, B., Sanza, F. J., Maigler, M. V., Álvarez-Millán, J. J., Canalejas-Tejero, V., & Holgado, M. (2018). A Proof-of-Concept of Label-Free Biosensing System for Food Allergy Diagnostics in Biophotonic Sensing Cells: Performance Comparison with ImmunoCAP. Sensors, 18(8), 2686. https://doi.org/10.3390/s18082686