Single Particle Differentiation through 2D Optical Fiber Trapping and Back-Scattered Signal Statistical Analysis: An Exploratory Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Optical Trapping Experiments
2.1.1. Fabrication of the Polymeric Lens for Optical Trapping
2.1.2. Optical Trapping and Particles Sensing Setup
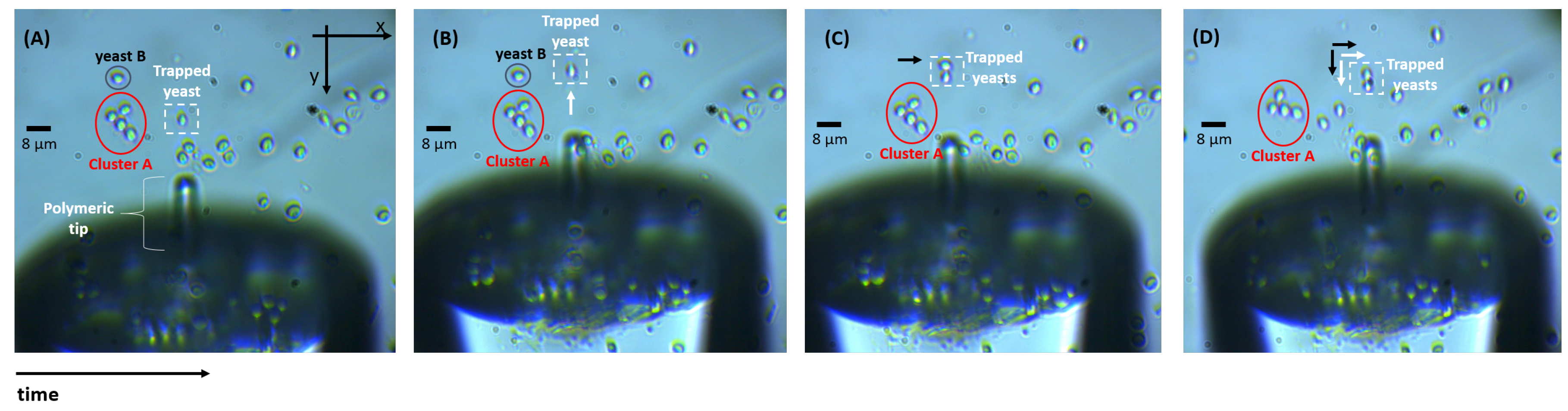
2.2. Particles Trapping and Sensing Using the Polymeric Lens
2.2.1. Back-Scattered Signal Acquisition and Processing Steps
2.2.2. Particles Differentiation Using Back-Scattered Trapping Signal
Features
Time Doman-Derived Features
Frequency Doman-Derived Features
2.2.3. Statistical Analysis
2.2.4. Features Dimensionality Reduction Using Linear Discriminant Analysis (LDA)
3. Results and Discussion
3.1. Optical Trapping
3.2. Back-Scattered Signal Analysis
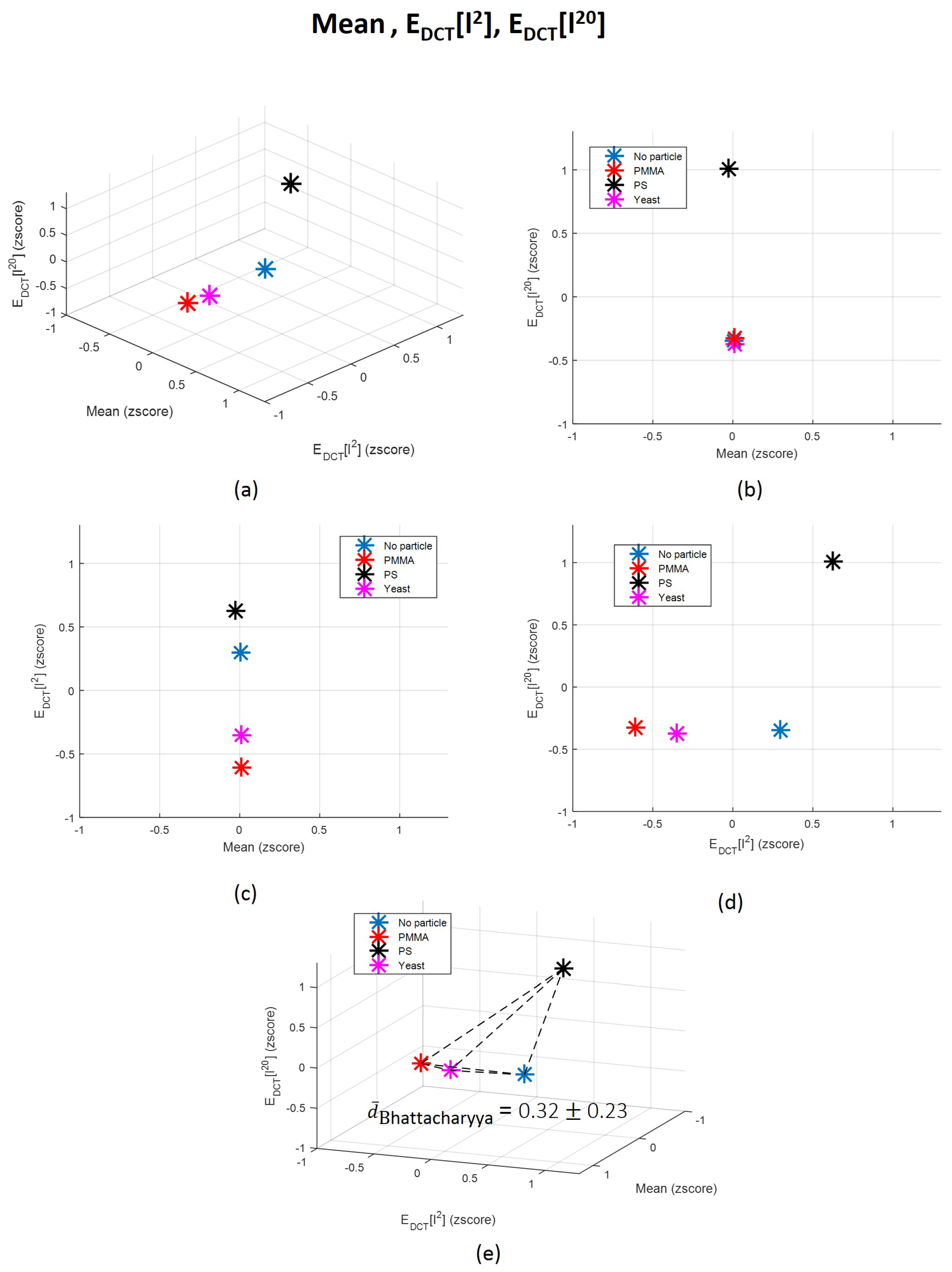
3.2.1. Time Domain Features Analysis
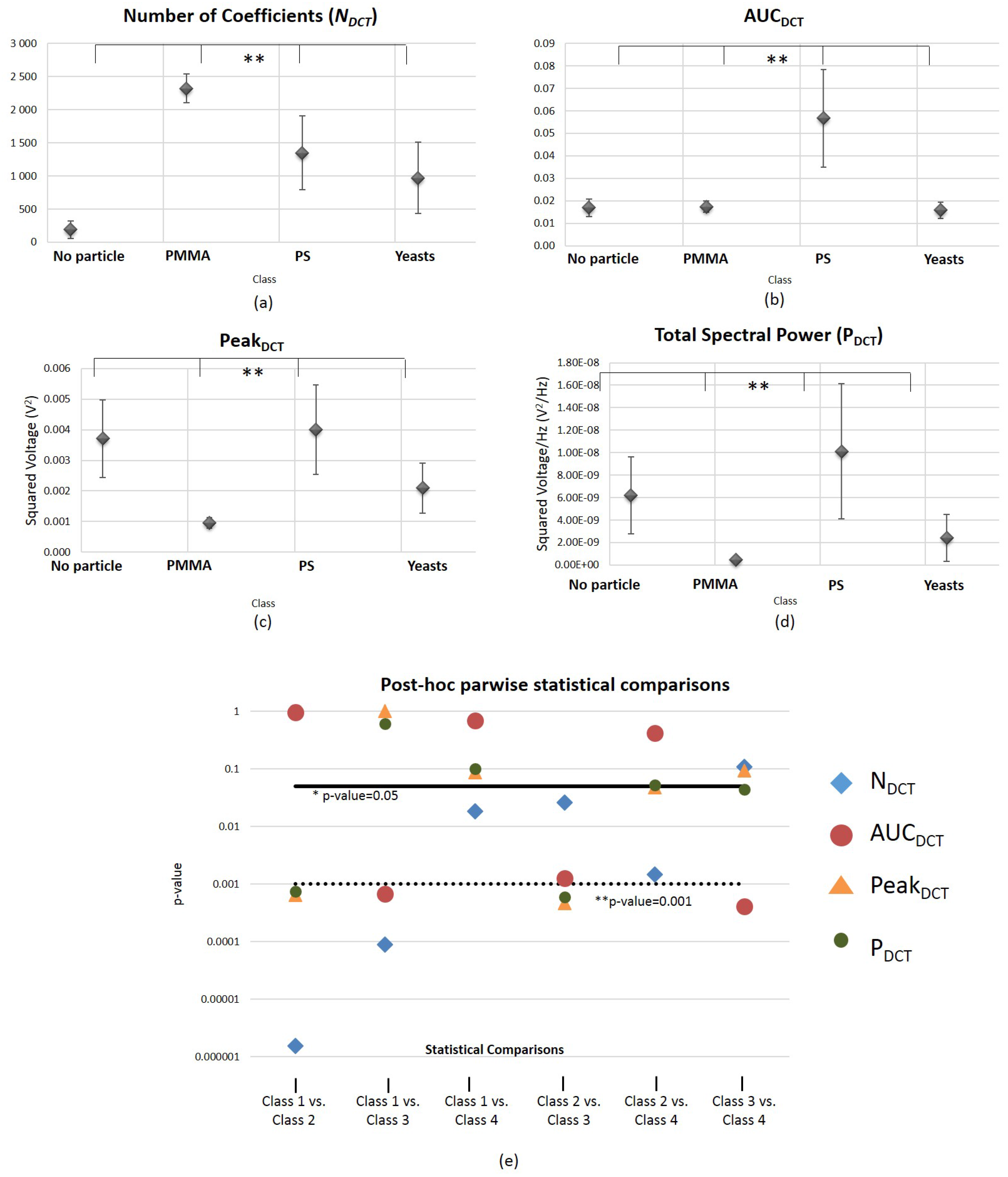
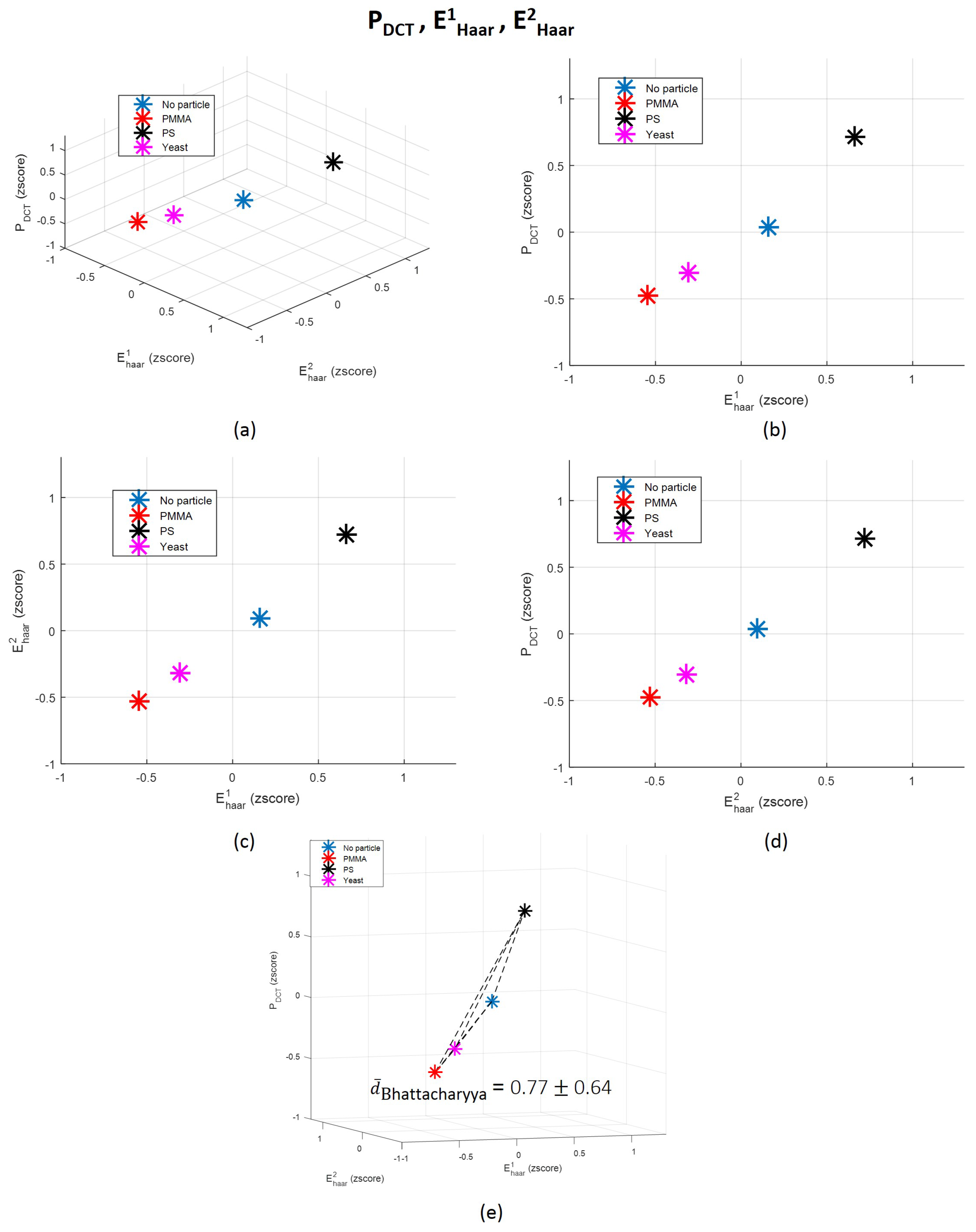
3.2.2. Frequency Domain Features Analysis
Discrete Cosine Transform (DCT)-Derived Features
Wavelet Features
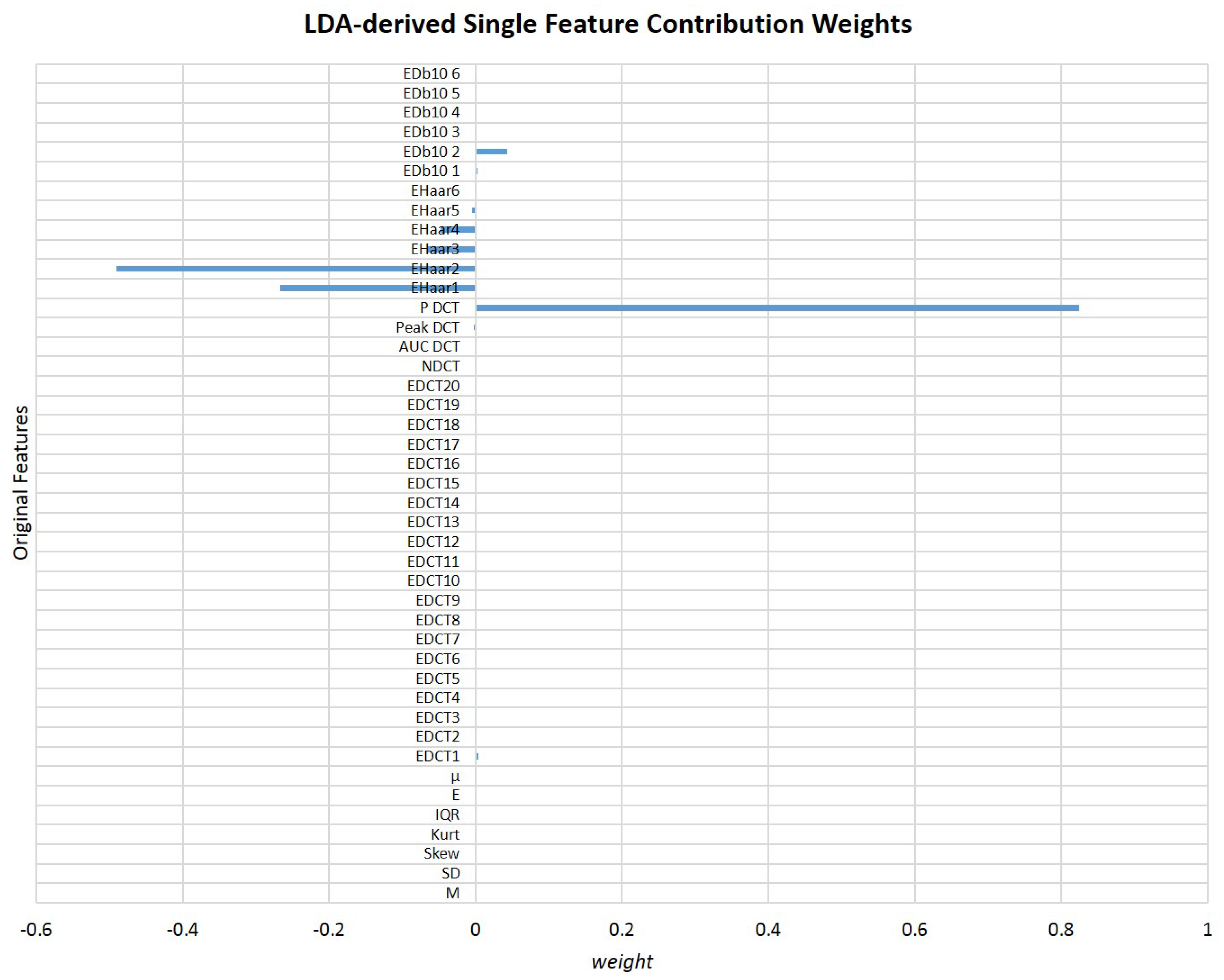
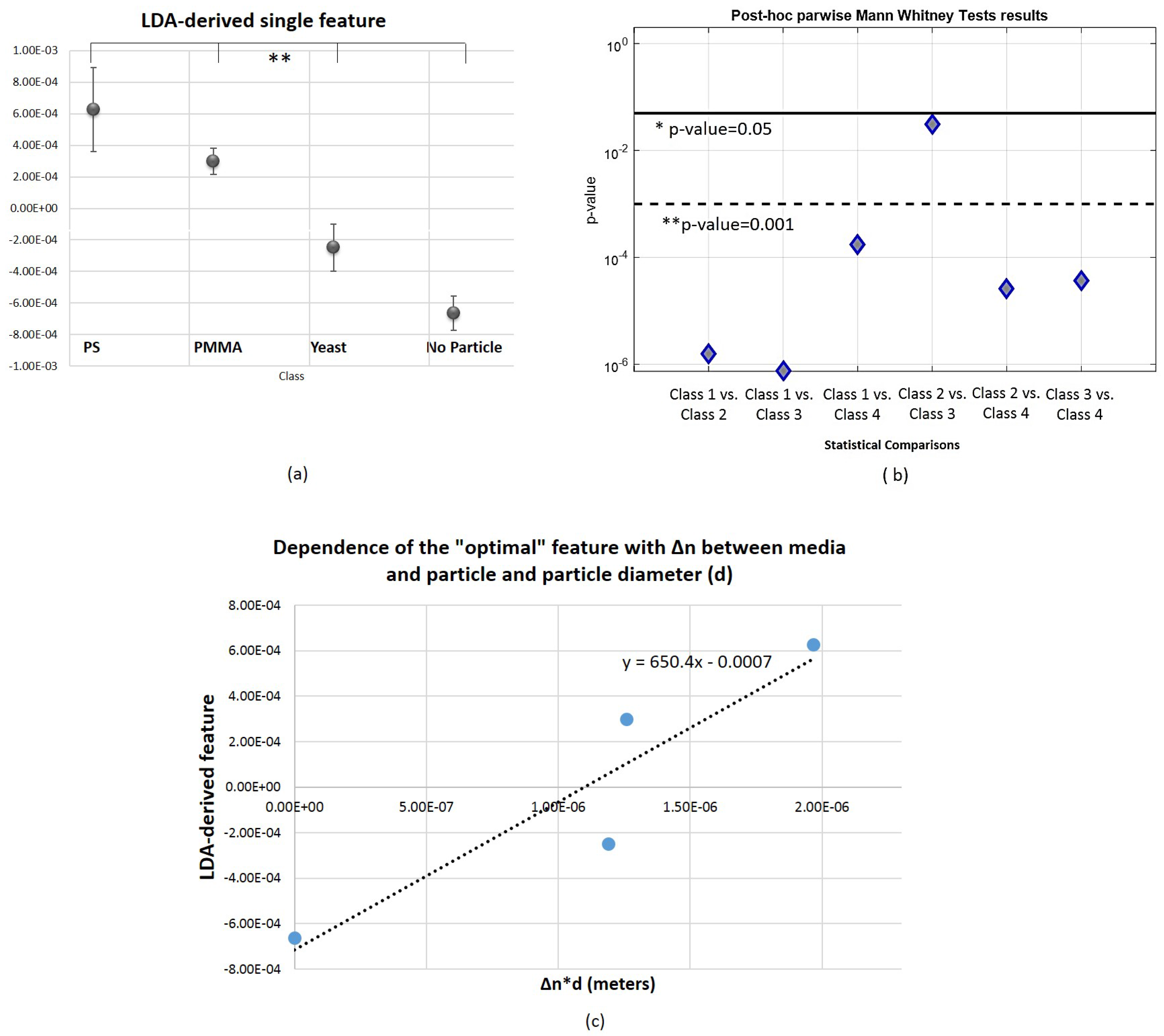
3.3. Towards a Single Feature for Particles Class Differentiation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A. Time Domain Features Analysis-Parwise Comparisons

Appendix B. Frequency Domain Features Analysis-Discrete Cosine Transform (DCT) Coefficients

Abbreviations
| AUC | Area Under the Curve |
| CMOS | Complementary Metal-Oxide-Semiconductor |
| COTs | Conventional Optical Tweezers |
| DAQ | Data Acquisition Board |
| DCT | Discrete Cosine Transform |
| E | Entropy |
| FFT | Fast-Fourier Transform |
| IQR | Interquartile Range |
| IoT | Internet of Things |
| Kurt | Kurtosis |
| LDA | Linear Discriminant Analysis |
| M | Mean |
| OFT | Optical Fiber Tweezers |
| OT | Optical Tweezers |
| PBS | Phosphate Buffered Saline |
| PCA | Principal Component Analysis |
| Probability Density Function | |
| PMMA | Poly(methyl methacrylate) |
| PS | Polystyrene |
| RBC | Red Blood Cell |
| RI | Refractive Index |
| RMS | Root Mean Square |
| SD | Standard Deviation |
| Skew | Skewness |
| SMF | Single Mode Fiber |
| SNR | Signal-to-noise Ratio |
References
- Ribeiro, R.; Dahal, P.; Guerreiro, A.; Jorge, P.; Viegas, J. Fabrication of Fresnel plates on optical fibres by FIB milling for optical trapping, manipulation and detection of single cells. Sci. Rep. 2017, 7. [Google Scholar]
- Genuer, V.; Gal, O.; Méteau, J.; Marcoux, P.; Schultz, E.; Lacot, É.; Maurin, M.; Dinten, J. Optical elastic scattering for early label-free identification of clinical pathogens. arXiv, 2016; arXiv:1610.02980. [Google Scholar]
- Greiner, C.; Hunter, M.; Huang, P.; Rius, F.; Georgakoudi, I. Confocal backscattering spectroscopy for leukemic and normal blood cell discrimination. Cytom. Part A 2011, 79, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Ullah, R.; Khan, A.; Wahab, N.; Bilal, M.; Ahmed, M. Analysis of dengue infection based on Raman spectroscopy and support vector machine (SVM). Biomed. Opt. Express 2016, 7, 2249–2256. [Google Scholar] [CrossRef] [PubMed]
- Paiva, J.; Ribeiro, R.; Jorge, P.; Rosa, C.; Guerreiro, A.; Cunha, J. 2D Computational Modeling of Optical Trapping Effects on Malaria-Infected Red Blood Cells; OSA Frontiers in Optics; OSA: Washington DC, USA, 2017. [Google Scholar]
- Vaiano, P.; Carotenuto, B.; Pisco, M.; Ricciardi, A.; Quero, G.; Consales, M.; Crescitelli, A.; Esposito, E.; Cusano, A. Lab on Fiber Technology for biological sensing applications. Laser Photonics Rev. 2016, 10, 922–961. [Google Scholar] [CrossRef]
- Ribeiro, R.; Soppera, O.; Oliva, A.; Guerreiro, A.; Jorge, P. New Trends on Optical Fiber Tweezers. J. Lightw. Technol. 2015, 33, 3394–3405. [Google Scholar] [CrossRef]
- Ashkin, A. Acceleration and trapping of particles by radiation pressure. Phys. Rev. Lett. 1970, 24, 156. [Google Scholar] [CrossRef]
- Novotny, L.; Hecht, B. Principles of Nano-Optics; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Liberale, C.; Minzioni, P.; Bragheri, F.; De Angelis, F.; Di Fabrizio, E.; Cristiani, I. Miniaturized all-fibre probe for three-dimensional optical trapping and manipulation. Nat. Photonics 2007, 1, 723–727. [Google Scholar] [CrossRef]
- Paiva, J.; Ribeiro, R.; Jorge, P.; Rosa, C.; Cunha, J. Computational modeling of red blood cells trapping using Optical Fiber Tweezers. In Proceedings of the 2017 IEEE 5th Portuguese Meeting on Bioengineering (ENBENG), Coimbra, Portugal, 16–18 February 2017; pp. 1–4. [Google Scholar]
- Ribeiro, R.; Soppera, O.; Guerreiro, A.; Jorge, P. Polymeric optical fiber tweezers as a tool for single cell micro manipulation and sensing. In Proceedings of the 24th International Conference on Optical Fibre Sensors (OFS24), International Society for Optics and Photonics, Curitiba, Brazil, 28 September–2 October 2015. [Google Scholar]
- Abedin, K.; Kerbage, C.; Fernandez-Nieves, A.; Weitz, D. Optical manipulation of liquid crystal drops: Application towards all-optical tunable photonic devices. In Proceedings of the 2006 Quantum Electronics and Laser Science Conference CLEO/QELS 2006 Conference on Lasers and Electro-Optics, Long Beach, CA, USA, 21–26 May 2006; pp. 1–2. [Google Scholar]
- Minzioni, P.; Bragheri, F.; Liberale, C.; Di Fabrizio, E.; Cristiani, I. A novel approach to fiber-optic tweezers: Numerical analysis of the trapping efficiency. IEEE J. Sel. Top. Quantum Electron. 2008, 14, 151–157. [Google Scholar] [CrossRef]
- Zhou, H.; Feng, G.; Yang, H.; Deng, G.; Ding, H. Fiber Optical Tweezers for the Operation of Particles. In Proceedings of the 2010 Symposium on Photonics and Optoelectronic (SOPO), Chengdu, China, 19–21 June 2010; pp. 1–4. [Google Scholar]
- Zhang, Y.; Yuan, L.; Liu, Z.; Yang, J. Dual optical tweezers integrated in a four-core fiber: Design and simulation. In Proceedings of the 4th Asia Pacific Optical Sensors Conference, International Society for Optics and Photonics, Wuhan, China, 1 October 2013. [Google Scholar]
- Decombe, J.; Valdivia-Valero, F.; Dantelle, G.; Leménager, G.; Gacoin, T.; Des Francs, G.; Huant, S.; Fick, J. Luminescent nanoparticle trapping with far-field optical fiber-tip tweezers. Nanoscale 2016, 8, 5334–5342. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, Z.; Yang, J.; Guan, C. Twin-core fiber optical tweezers. Opt. Express 2008, 16, 4559–4566. [Google Scholar] [CrossRef] [PubMed]
- Juan, M.; Righini, M.; Quidant, R. Plasmon nano-optical tweezers. Nat. Photonics 2011, 5, 349–356. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, L.; Santschi, C.; Martin, O. Trapping and sensing 10 nm metal nanoparticles using plasmonic dipole antennas. Nano Lett. 2010, 10, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, Y. Origin and future of plasmonic optical tweezers. Nanomaterials 2015, 5, 1048–1065. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Moore, S.; Schmidt, B.; Klug, M.; Lipson, M.; Erickson, D. Optical manipulation of nanoparticles and biomolecules in sub-wavelength slot waveguides. Nature 2009, 457, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Sainidou, R.; De Abajo, F. Optically tunable surfaces with trapped particles in microcavities. Phys. Rev. Lett. 2008, 101, 136802. [Google Scholar] [CrossRef] [PubMed]
- Grigorenko, A.; Roberts, N.; Dickinson, M.; Zhang, Y. Nanometric optical tweezers based on nanostructured substrates. Nat. Photonics 2008, 2, 365–370. [Google Scholar] [CrossRef]
- Harvey, T.; Faria, E.; Henderson, A.; Gazi, E.; Ward, A.; Clarke, N.; Brown, M.; Snook, R.; Gardner, P. Spectral discrimination of live prostate and bladder cancer cell lines using Raman optical tweezers. J. Biomed. Opt. 2008, 13. [Google Scholar] [CrossRef] [PubMed]
- Jess, P.; Garcés-Chávez, V.; Smith, D.; Mazilu, M.; Paterson, L.; Riches, A.; Herrington, C.; Sibbett, W.; Dholakia, K. Dual beam fibre trap for Raman microspectroscopy of single cells. Opt. Express 2006, 14, 5779–5791. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.; Lin, Y.; Wei, J.; Bono, T.; Lindquist, R. An enhanced LSPR fiber-optic nanoprobe for ultrasensitive detection of protein biomarkers. Biosens. Bioelectron. 2014, 61, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Li, Q.; Wang, J.; Yao, W.; Jin, Z.; Sui, Q.; Shi, J.; Zhang, F.; Jia, L.; Dong, W. Optical response of fiber-optic Fabry-Perot refractive-index tip sensor coated with polyelectrolyte multilayer ultra-thin films. J. Lightw. Technol. 2013, 31, 2321–2326. [Google Scholar] [CrossRef]
- Rosenberger, M.; Belle, S.; Hellmann, R. Detection of biochemical reaction and DNA hybridization using a planar Bragg grating sensor. Proc. SPIE 2011, 8073. [Google Scholar] [CrossRef]
- Wang, G.; Zheng, X.; Shum, P.; Li, C.; Ho, H.; Tong, L. Live cell index sensing based on the reflection mode of tilted fiber tip with gold nanoparticles. In Proceedings of the 9th International Conference on Optical Communications and Networks (ICOCN 2010), Nanjing, China, 24–27 October 2010; pp. 116–119. [Google Scholar]
- Consales, M.; Ricciardi, A.; Crescitelli, A.; Esposito, E.; Cutolo, A.; Cusano, A. Lab-on-fiber technology: Toward multifunctional optical nanoprobes. ACS Nano 2012, 6, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Chester, A.; Martellucci, S.; Scheggi, A. Optical Fiber Sensors; Springer Science & Business Media: Berlin, Germany, 2012; Volume 132. [Google Scholar]
- Mei, Z.; Wu, T.; Pion-Tonachini, L.; Qiao, W.; Zhao, C.; Liu, Z.; Lo, Y. Applying an optical space-time coding method to enhance light scattering signals in microfluidic devices. Biomicrofluidics 2011, 5, 034116. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Cho, S.; Chiu, Y.; Lo, Y. Lab-on-a-Chip Device and System for Point-of-Care Applications. Handb. Photonics Biomed. Eng. 2017, 87–121. [Google Scholar] [CrossRef]
- Welsh, J.; Holloway, J.; Wilkinson, J.; Englyst, N. Extracellular Vesicle Flow Cytometry Analysis and Standardization. Front. Cell Dev. Biol. 2017, 5, 78. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Diez-Silva, M.; Popescu, G.; Lykotrafitis, G.; Choi, W.; Feld, M.; Suresh, S. Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2008, 105, 13730–13735. [Google Scholar] [CrossRef] [PubMed]
- Soppera, O.; Jradi, S.; Lougnot, D. Photopolymerization with microscale resolution: Influence of the physico-chemical and photonic parameters. J. Polym. Sci. Part A Polym. Chem. 2008, 46, 3783–3794. [Google Scholar] [CrossRef]
- Ribeiro, R.; Guerreiro, A.; Ecoffet, C.; Soppera, O.; Jorge, P. New theoretical and experimental methods for the design of fiber optic tweezers. In Proceedings of the Fifth European Workshop on Optical Fibre Sensors. International Society for Optics and Photonics, Krakow, Poland, 20 May 2013. [Google Scholar]
- Ribeiro, R.; Queirós, R.; Guerreiro, A.; Ecoffet, C.; Soppera, O.; Jorge, P. Fiber optical beam shaping using polymeric structures. In Proceedings of the OFS2014 23rd International Conference on Optical Fiber Sensors, International Society for Optics and Photonics, Santander, Spain, 2 June 2014. [Google Scholar]
- Ribeiro, R.; Queirós, R.; Ecoffet, C.; Soppera, O.; Oliva, A.; Guerreiro, A.; Jorge, P. Rapid fabrication of polymeric micro lenses for optical fiber trapping and beam shaping. In Proceedings of the SPIE NanoScience+ Engineering. International Society for Optics and Photonics, San Diego, CA, USA, 16 September 2014. [Google Scholar]
- Ribeiro, R. Optical Fiber Tools for Single Cell Trapping and Manipulation. Ph.D. Thesis, University of Porto, Porto, Portugal, 2017. [Google Scholar]
- Sultanova, N.; Kasarova, S.; Nikolov, I. Dispersion Properties of Optical Polymers. Acta Phys. Pol. Ser. A Gen. Phys. 2009, 116, 585–587. [Google Scholar] [CrossRef]
- Sigrist-Photometer, A. Refractive Index; Sigrist-Photometer: Ennetbürgen, Switzerland, 2017. [Google Scholar]
- De Sa, J. Pattern Recognition: Concepts, Methods and Applications; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Paiva, J.S.; Dias, D.; Cunha, J.P.S. Beat-ID: Towards a computationally low-cost single heartbeat biometric identity check system based on electrocardiogram wave morphology. PLoS ONE 2017, 12, e0180942. [Google Scholar] [CrossRef] [PubMed]
- Stavroulakis, P.; Stamp, M. Handbook of Information and Communication Security; Springer Science & Business Media: Berlin, Germany, 2010. [Google Scholar]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Sands, T.; Tayal, D.; Morris, M.; Monteiro, S. Robust stock value prediction using support vector machines with particle swarm optimization. In Proceedings of the 2015 IEEE Congress on Evolutionary Computation (CEC), Sendai, Japan, 25–28 May 2015; pp. 3327–3331. [Google Scholar]
- Ahmed, N.; Natarajan, T.; Rao, K. Discrete cosine transform. IEEE Trans. Comput. 1974, 100, 90–93. [Google Scholar] [CrossRef]
- Chui, C. Wavelets: A tutorial in theory and applications. In Wavelet Analysis and Its Applications; Chui, C.K., Ed.; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- Dasgupta, N.; Runkle, P.; Carin, L.; Couchman, L.; Yoder, T.; Bucaro, J.; Dobeck, G.J. Class-based target identification with multiaspect scattering data. IEEE J. Ocean. Eng. 2003, 28, 271–282. [Google Scholar] [CrossRef]
- Roberts, P.; Jaffe, J. Multiple angle acoustic classification of zooplankton. J. Acoust. Soc. Am. 2007, 121, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Cabreira, A.; Tripode, M.; Madirolas, A. Artificial neural networks for fish-species identification. ICES J. Mar. Sci. 2009, 66, 1119–1129. [Google Scholar] [CrossRef]
- Malkin, R.; Alexandrou, D. Acoustic classification of abyssopelagic animals. IEEE J. Ocean. Eng. 1993, 18, 63–72. [Google Scholar] [CrossRef]
- Azimi-Sadjadi, M.; Yao, D.; Huang, Q.; Dobeck, G. Underwater target classification using wavelet packets and neural networks. IEEE Trans. Neural Netw. 2000, 11, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Yovel, Y.; Au, W. How can dolphins recognize fish according to their echoes? A statistical analysis of fish echoes. PLoS ONE 2010, 5, e14054. [Google Scholar] [CrossRef] [PubMed]
- Petrut, T.; Ioana, C.; Mauuary, D.; Mallet, J.; Phillipe, O. Distributed data classification in underwater acoustic sensors based on local time-frequency coherence analysis. In Proceedings of the OCEANS 2014-TAIPEI, Taipei, Taiwan, 7–10 April 2014; pp. 1–6. [Google Scholar]
- Chen, P.; Liu, L.; Wang, X.; Chong, J.; Zhang, X.; Yu, X. Modulation Model of High Frequency Band Radar Backscatter by the Internal Wave Based on the Third-Order Statistics. Remote Sens. 2017, 9, 501. [Google Scholar] [CrossRef]
- Shaw, D.; Stone, K.; Ho, K.; Keller, J.; Luke, R.; Burns, B. Sequential feature selection for detecting buried objects using forward looking ground penetrating radar. In Proceedings of the SPIE Defense + Security. International Society for Optics and Photonics, Baltimore, MD, USA, 3 May 2016. [Google Scholar]
- Roberts, P.; Jaffe, J.; Trivedi, M. Multiview, broadband acoustic classification of marine fish: A machine learning framework and comparative analysis. IEEE J. Ocean. Eng. 2011, 36, 90–104. [Google Scholar] [CrossRef]
- Paiva, J.; Cardoso, J.; Pereira, T. Supervised Learning Methods for Pathological Arterial Pulse Wave Differentiation: A SVM and Neural Networks Approach. Int. J. Med. Inform. 2017, 109, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Pereira, T.; Paiva, J.; Correia, C.; Cardoso, J. An automatic method for arterial pulse waveform recognition using KNN and SVM classifiers. Med. Biol. Eng. Comput. 2016, 54, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.; Polmear, M.; Mineva, N.; Romagnoli, M.; Sonenshein, G.; Georgakoudi, I. Endogenous light scattering as an optical signature of circulating tumor cell clusters. Biomed. Opt. Express 2016, 7, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Diggle, P. Statistical Analysis of Spatial and Spatio-Temporal Point Patterns; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Zhai, S.; Jiang, T. Target detection and classification by measuring and processing bistatic UWB radar signal. Measurement 2014, 47, 547–557. [Google Scholar] [CrossRef]
- Sanei, S.; Chambers, J. EEG Signal Processing; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Shankar, P. A general statistical model for ultrasonic backscattering from tissues. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2000, 47, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Caixinha, M.; Jesus, D.; Velte, E.; Santos, M.; Santos, J. Using ultrasound backscattering signals and Nakagami statistical distribution to assess regional cataract hardness. IEEE Trans. Biomed. Eng. 2014, 61, 2921–2929. [Google Scholar] [CrossRef] [PubMed]
- Trop, I.; Destrempes, F.; El Khoury, M.; Robidoux, A.; Gaboury, L.; Allard, L.; Chayer, B.; Cloutier, G. The added value of statistical modeling of backscatter properties in the management of breast lesions at US. Radiology 2014, 275, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Chin-Hsing, C.; Jiann-Der, L.; Ming-Chi, L. Classification of underwater signals using wavelet transforms and neural networks. Math. Comput. Model. 1998, 27, 47–60. [Google Scholar] [CrossRef]
- Rao, K.; Yip, P. Discrete Cosine Transform: Algorithms, Advantages, Applications; Academic press: Cambridge, MA, USA, 2014. [Google Scholar]
- Chui, C. An Introduction to Wavelets; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Ganapathi, S.; Kumar, S.; Deivasigamani, M. Noise reduction in underwater acoustic signals for tropical and subtropical coastal waters. In Proceedings of the 2016 IEEE/OES China Ocean Acoustics (COA), Harbin, China, 9–11 January 2016; pp. 1–6. [Google Scholar]
- Pallant, J. SPSS Survival Manual; McGraw-Hill Education: Buckingham, UK, 2013. [Google Scholar]
- Izenman, A. Modern Multivariate Statistical Techniques; Springer: Berlin, Germany, 2008; Volume 1. [Google Scholar]
- Xanthopoulos, P.; Pardalos, P.; Trafalis, T. Linear discriminant analysis. In Robust Data Mining; Springer: Berlin, Germany, 2013; pp. 27–33. [Google Scholar]
- Aktas, M.; Akgun, T.; Demircin, M.; Buyukaydin, D. Deep learning based multi-threat classification for phase-OTDR fiber optic distributed acoustic sensing applications. In Proceedings of the Fiber Optic Sensors and Applications XIV, International Society for Optics and Photonics, Anaheim, CA, USA, 27 April 2017. [Google Scholar]
- Du, W.; Chen, B.; Li, H.; Xu, C. A Novel Classification Method of Fish Based on Multi-Feature Fusion. Appl. Mech. Mater. Trans. Tech. Publ. 2015, 713–715, 1513–1519. [Google Scholar] [CrossRef]
- Choi, E.; Lee, C. Feature extraction based on the Bhattacharyya distance. Pattern Recognit. 2003, 36, 1703–1709. [Google Scholar] [CrossRef]
- Guorong, X.; Peiqi, C.; Minhui, W. Bhattacharyya distance feature selection. In Proceedings of the 13th International Conference on Pattern Recognition, Vienna, Austria, 25–29 August 1996; Volume 2, pp. 195–199. [Google Scholar]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D. The mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Jones, F.; Nichols, M.; Pappas, S. Organic Coatings: Science and Technology; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Burguera, A.; Bonin-Font, F.; Lisani, J.; Petro, A.; Oliver, G. Towards automatic visual sea grass detection in underwater areas of ecological interest. In Proceedings of the 2016 IEEE 21st International Conference on Emerging Technologies and Factory Automation (ETFA), Berlin, Germany, 6–9 September 2016; pp. 1–4. [Google Scholar]
- Farnoud, N.; Kolios, M.; Krishnan, S. Ultrasound backscatter signal characterization and classification using autoregressive modeling and machine learning algorithms. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Cancun, Mexico, 17–21 September 2003; Volume 3, pp. 2861–2864. [Google Scholar]

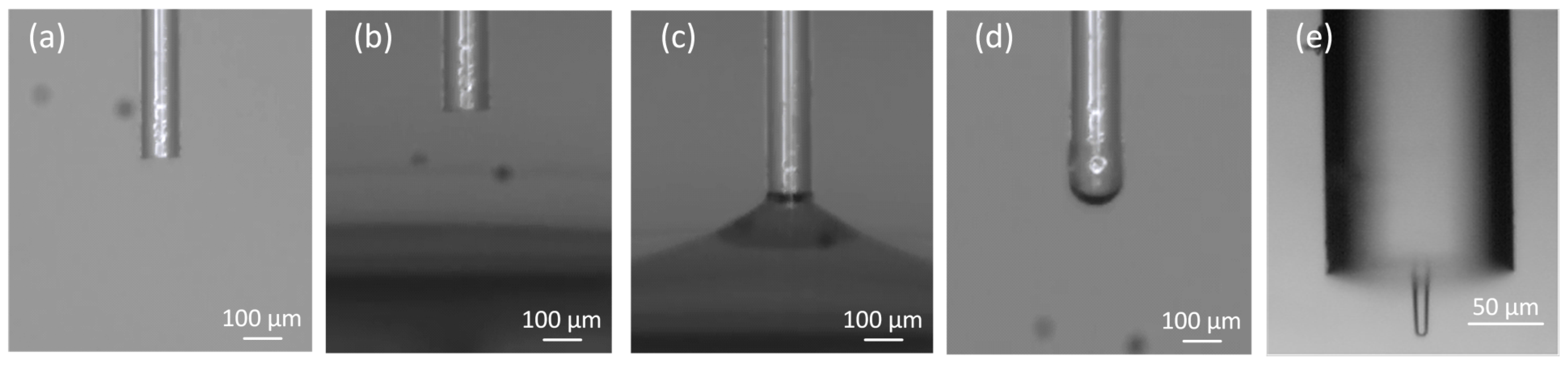
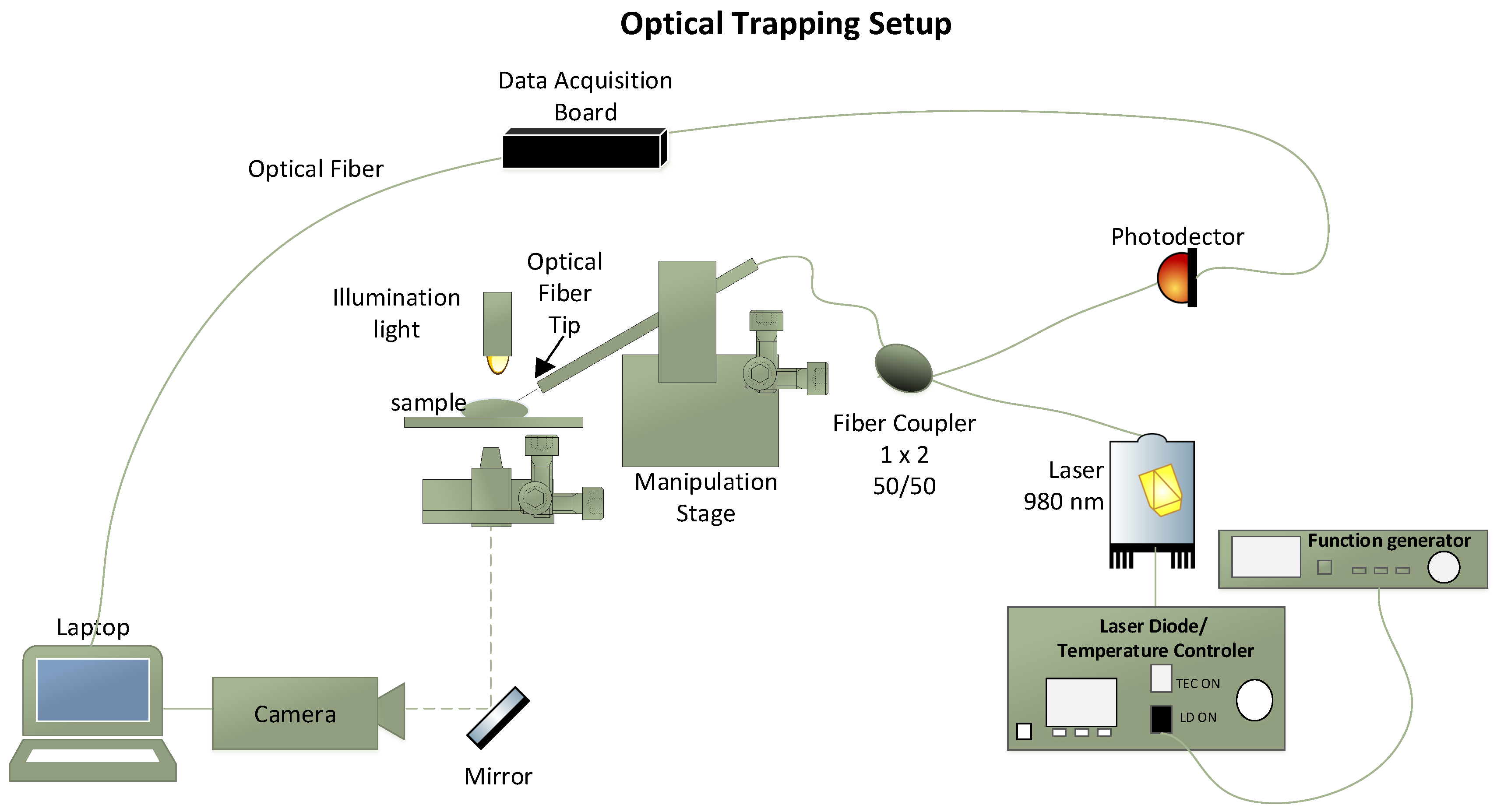
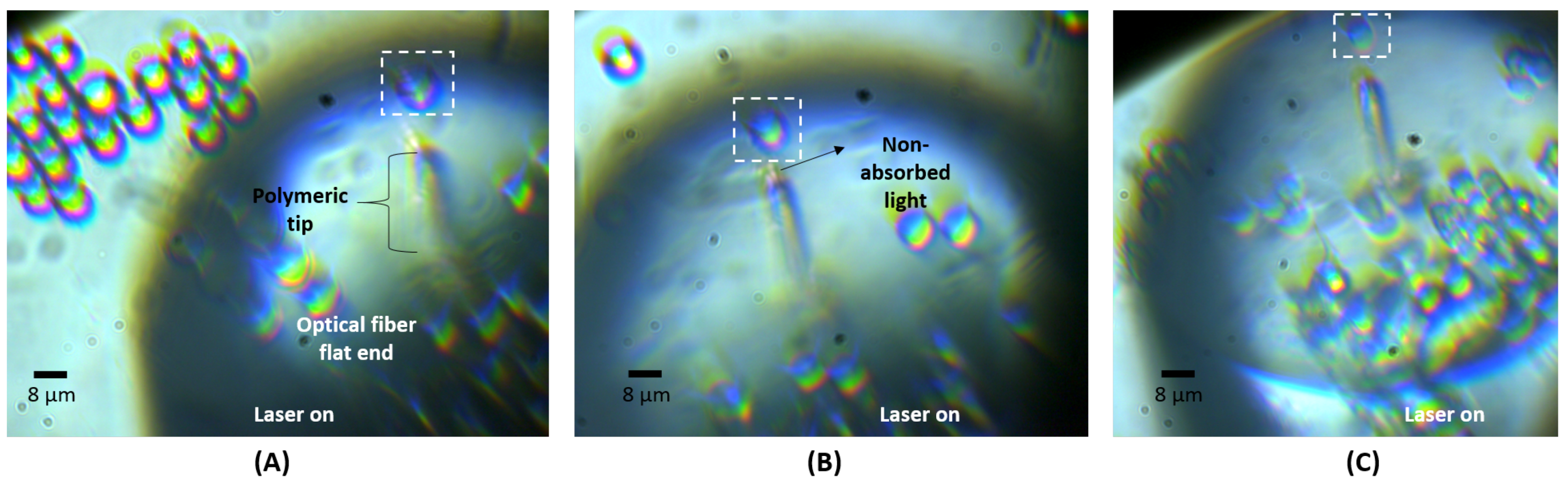

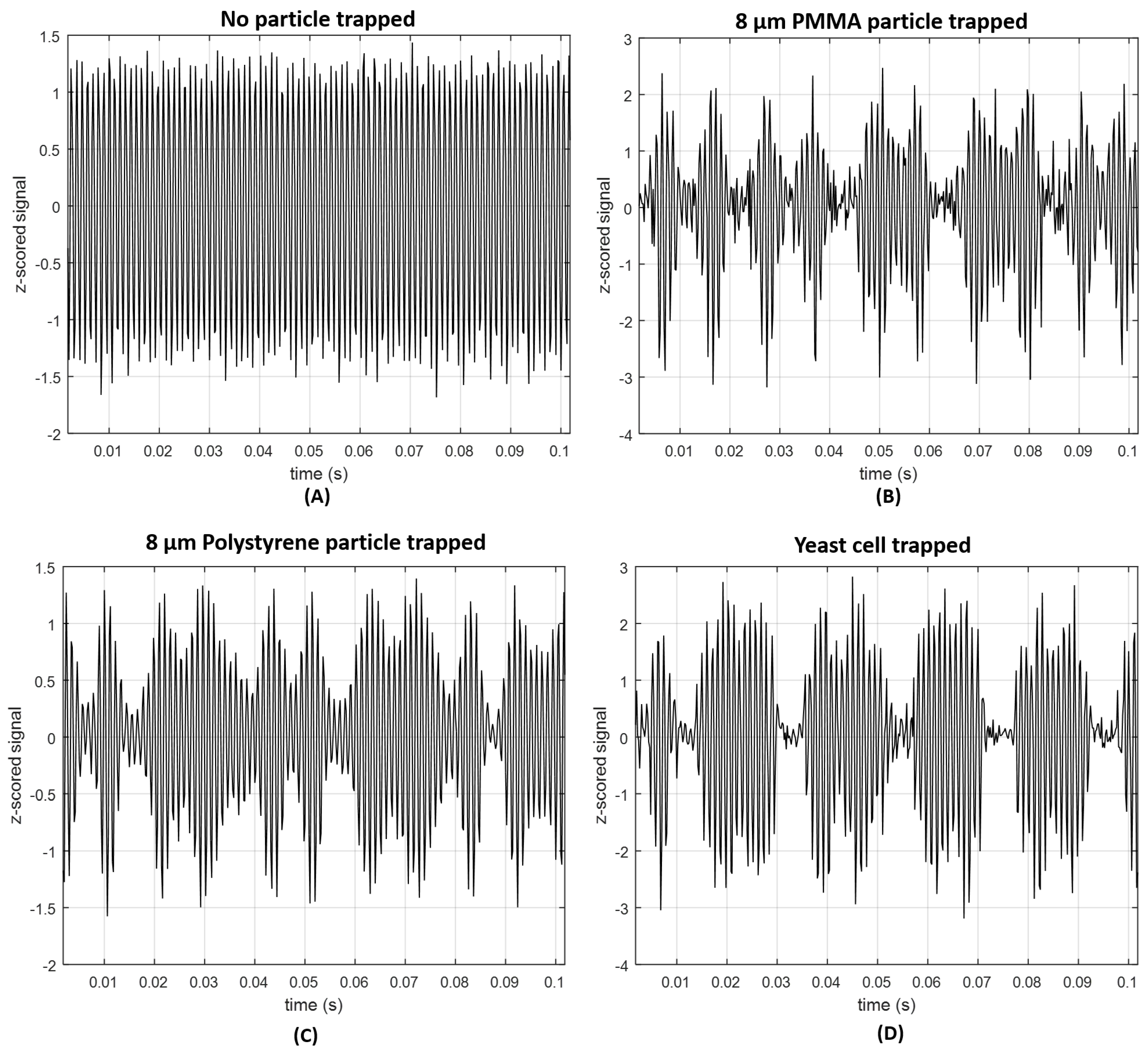

| Solution | Solvent | Particles Type | Particles Diameter (d) | Particles RI | Nr. of Particles Used |
|---|---|---|---|---|---|
| 1 | De-ionized water (n = 1.327) | Polystyrene microspheres | 8 m | 1.5731 [42] | 18 |
| 2 | PMMA microspheres | 8 m | 1.4843 [42] | 16 | |
| 3 | Living yeast cells | 6–7 m | 1.49–1.53 [43] | 16 |
| Class | 1: “No Particle” | 2: “PMMA Particle” | 3: “PS Particle” | 4: “Living Yeast Cell” |
|---|---|---|---|---|
| Nr. of Particles | 16 | 16 | 18 | 16 |
| Avg. nr. of signal portions per particle | 59 ± 0 | 57 ± 2 | 54 ± 5 | 59 ± 2 |
| Total nr. of signal portions (all particles) | 949 | 919 | 971 | 939 |
| Type | Group | Number | Feature/Parameter |
|---|---|---|---|
| Time Domain | Time Domain Statistics | 1 | Mean (M) |
| 2 | Standard Deviation (SD) | ||
| 3 | Root Mean Square (RMS) | ||
| 4 | Skewness (Skew) | ||
| 5 | Kurtosis (Kurt) | ||
| 6 | Interquartile Range (IQR) | ||
| 7 | Entropy ( E ) | ||
| Time Domain Histogram | 8 | ||
| 9 | |||
| Frequency Domain | Discrete Cosine Transform (DCT) | 10 | 1st Coefficient () |
| 11 | 2nd Coefficient () | ||
| 12 | 3rd Coefficient () | ||
| 13 | 4th Coefficient () | ||
| 14 | 5th Coefficient () | ||
| 15 | 6th Coefficient () | ||
| 16 | 7th Coefficient () | ||
| 17 | 8th Coefficient () | ||
| 18 | 9th Coefficient () | ||
| 19 | 10th Coefficient () | ||
| 20 | 11th Coefficient () | ||
| 21 | 12th Coefficient () | ||
| 22 | 13th Coefficient () | ||
| 23 | 14th Coefficient () | ||
| 24 | 15th Coefficient () | ||
| 25 | 16th Coefficient () | ||
| 26 | 17th Coefficient () | ||
| 27 | 18th Coefficient () | ||
| 28 | 19th Coefficient () | ||
| 29 | 20th Coefficient () | ||
| 30 | Number of coefficients that capture 98% of the original signal () | ||
| 31 | Total spectrum Area Under Curve (AUC) () | ||
| 32 | Maximum peak amplitude () | ||
| 33 | Total spectral power () | ||
| Wavelet Packet Decomposition | 34 | Haar Relative Power 1st level () | |
| 35 | Haar Relative Power 2nd level () | ||
| 36 | Haar Relative Power 3rd level () | ||
| 37 | Haar Relative Power 4th level () | ||
| 38 | Haar Relative Power 5th level () | ||
| 39 | Haar Relative Power 6th level () | ||
| 40 | Db10 Relative Power 1st level () | ||
| 41 | Db10 Relative Power 2nd level () | ||
| 42 | Db10 Relative Power 3rd level () | ||
| 43 | Db10 Relative Power 4th level () | ||
| 44 | Db10 Relative Power 5th level () | ||
| 45 | Db10 Relative Power 6th level () |
| Type of Wavelet | Wavelet Energy Level | 4 Classes Comparisons | Class 1 vs. Class 2 | Class 1 vs. Class 3 | Class 1 vs. Class 4 | Class 2 vs. Class 3 | Class 2 vs. Class 4 | Class 3 vs. Class 4 |
|---|---|---|---|---|---|---|---|---|
| Haar | 1st | ** | ** | ** | * | |||
| 2nd | ** | ** | ** | * | ||||
| 3rd | ** | * | ** | * | ||||
| 4th | ** | * | ** | * | ||||
| 5th | ** | ** | ** | ** | * | ** | ** | |
| 6th | ** | ** | ** | * | * | ** | ** | |
| Db10 | 1st | ** | ** | ** | * | |||
| 2nd | ** | ** | ** | |||||
| 3rd | ** | ** | ** | ** | ||||
| 4th | ** | ** | ** | * | * | ** | ** | |
| 5th | ** | ** | ** | ** | * | ** | ** | |
| 6th | ** | * | ** | * | * | ** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paiva, J.S.; Ribeiro, R.S.R.; Cunha, J.P.S.; Rosa, C.C.; Jorge, P.A.S. Single Particle Differentiation through 2D Optical Fiber Trapping and Back-Scattered Signal Statistical Analysis: An Exploratory Approach. Sensors 2018, 18, 710. https://doi.org/10.3390/s18030710
Paiva JS, Ribeiro RSR, Cunha JPS, Rosa CC, Jorge PAS. Single Particle Differentiation through 2D Optical Fiber Trapping and Back-Scattered Signal Statistical Analysis: An Exploratory Approach. Sensors. 2018; 18(3):710. https://doi.org/10.3390/s18030710
Chicago/Turabian StylePaiva, Joana S., Rita S. R. Ribeiro, João P. S. Cunha, Carla C. Rosa, and Pedro A. S. Jorge. 2018. "Single Particle Differentiation through 2D Optical Fiber Trapping and Back-Scattered Signal Statistical Analysis: An Exploratory Approach" Sensors 18, no. 3: 710. https://doi.org/10.3390/s18030710
APA StylePaiva, J. S., Ribeiro, R. S. R., Cunha, J. P. S., Rosa, C. C., & Jorge, P. A. S. (2018). Single Particle Differentiation through 2D Optical Fiber Trapping and Back-Scattered Signal Statistical Analysis: An Exploratory Approach. Sensors, 18(3), 710. https://doi.org/10.3390/s18030710






