Bacteria Detection and Differentiation Using Impedance Flow Cytometry
Abstract
1. Introduction
2. Materials and Methods
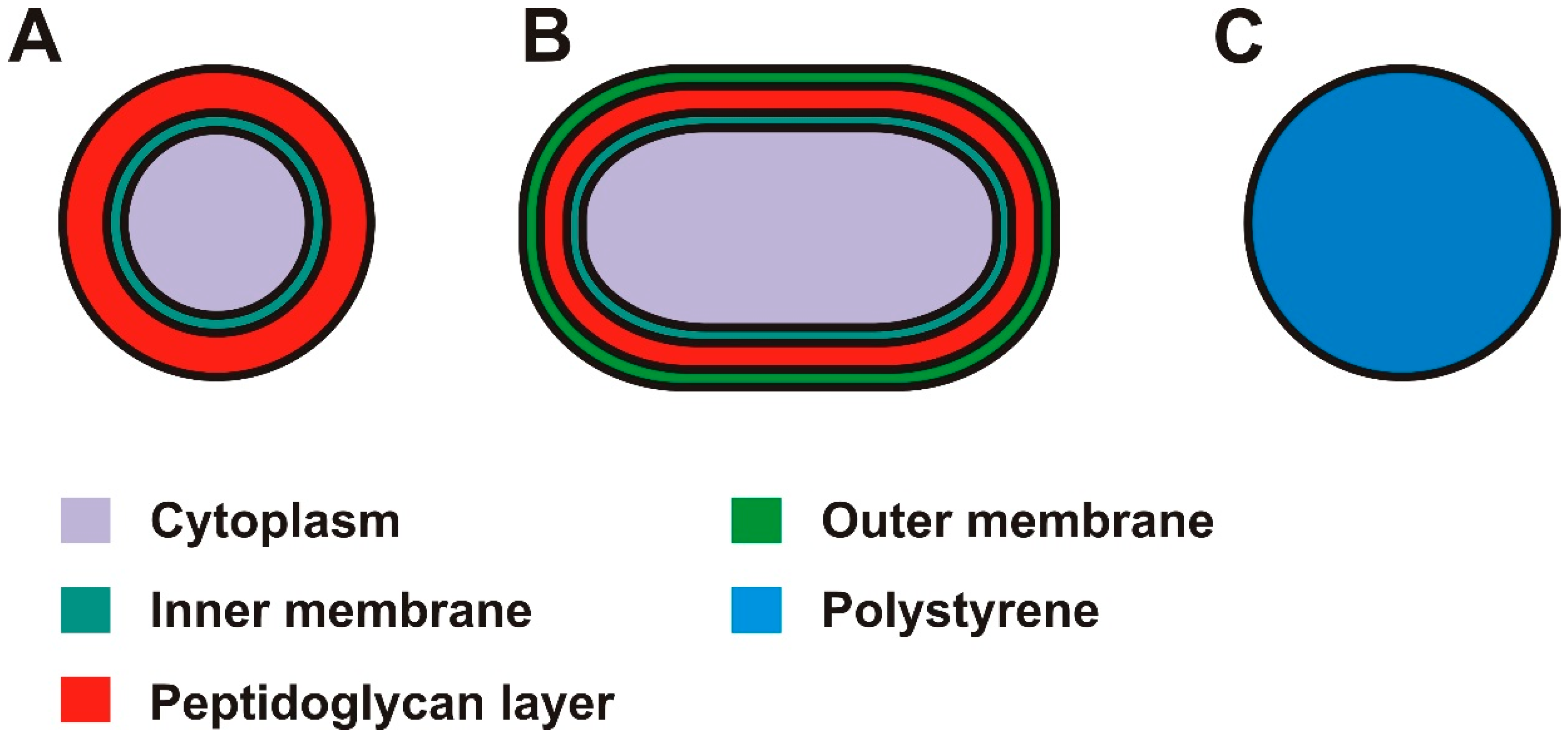
2.1. Sample Composition
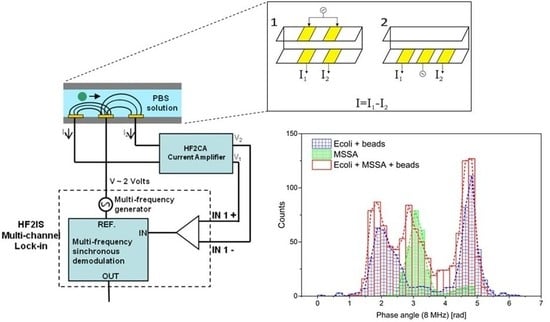
2.2. Detection Principle
2.3. Chip Fabrication
2.4. Measurement Setup
2.5. Samples
2.6. Data Acquisition and Analysis
3. Results and Discussion
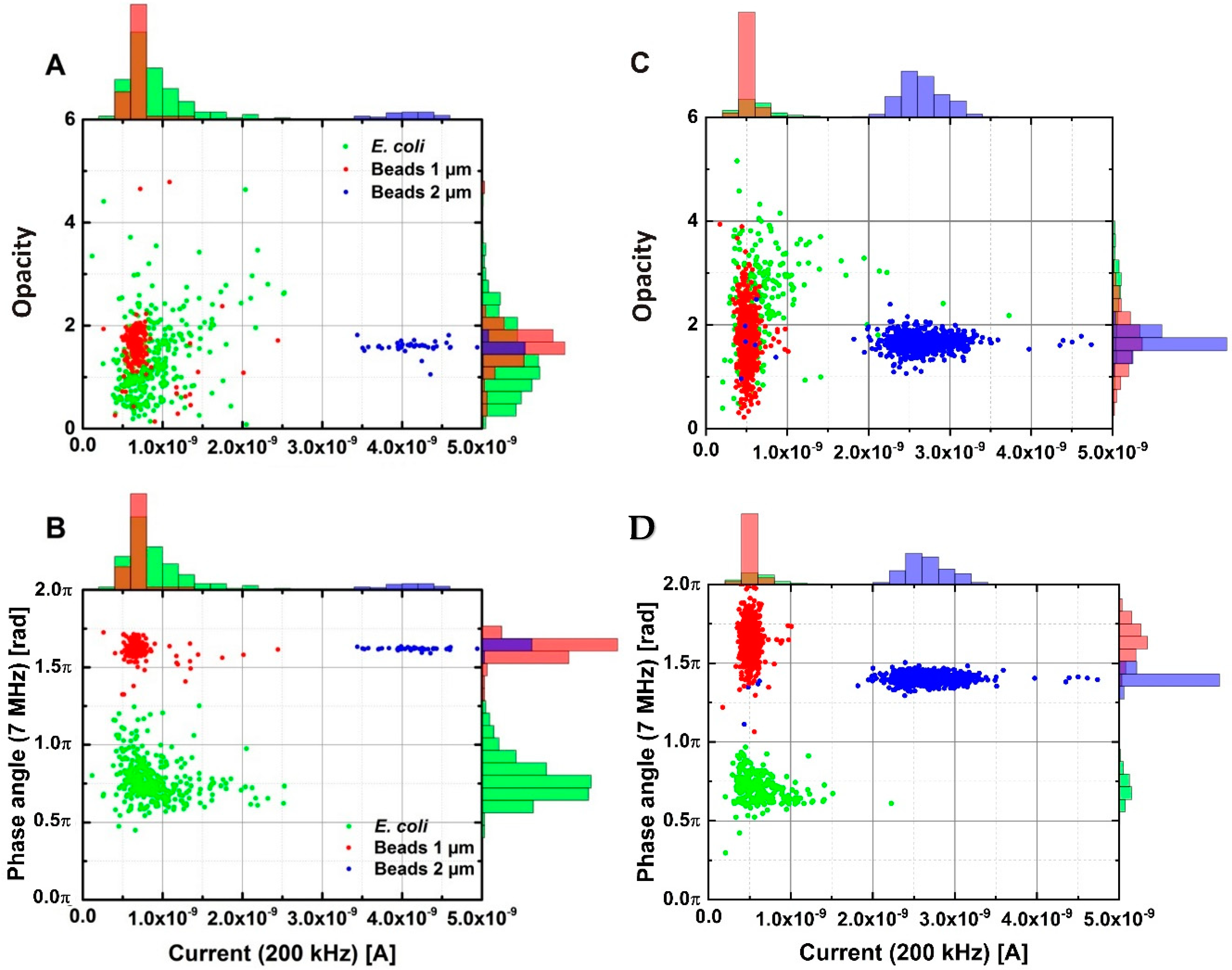
3.1. Separation of Biological and Non-Biological Samples in Diluted PBS and Tap Water
3.2. Determination of Bacteria Concentration
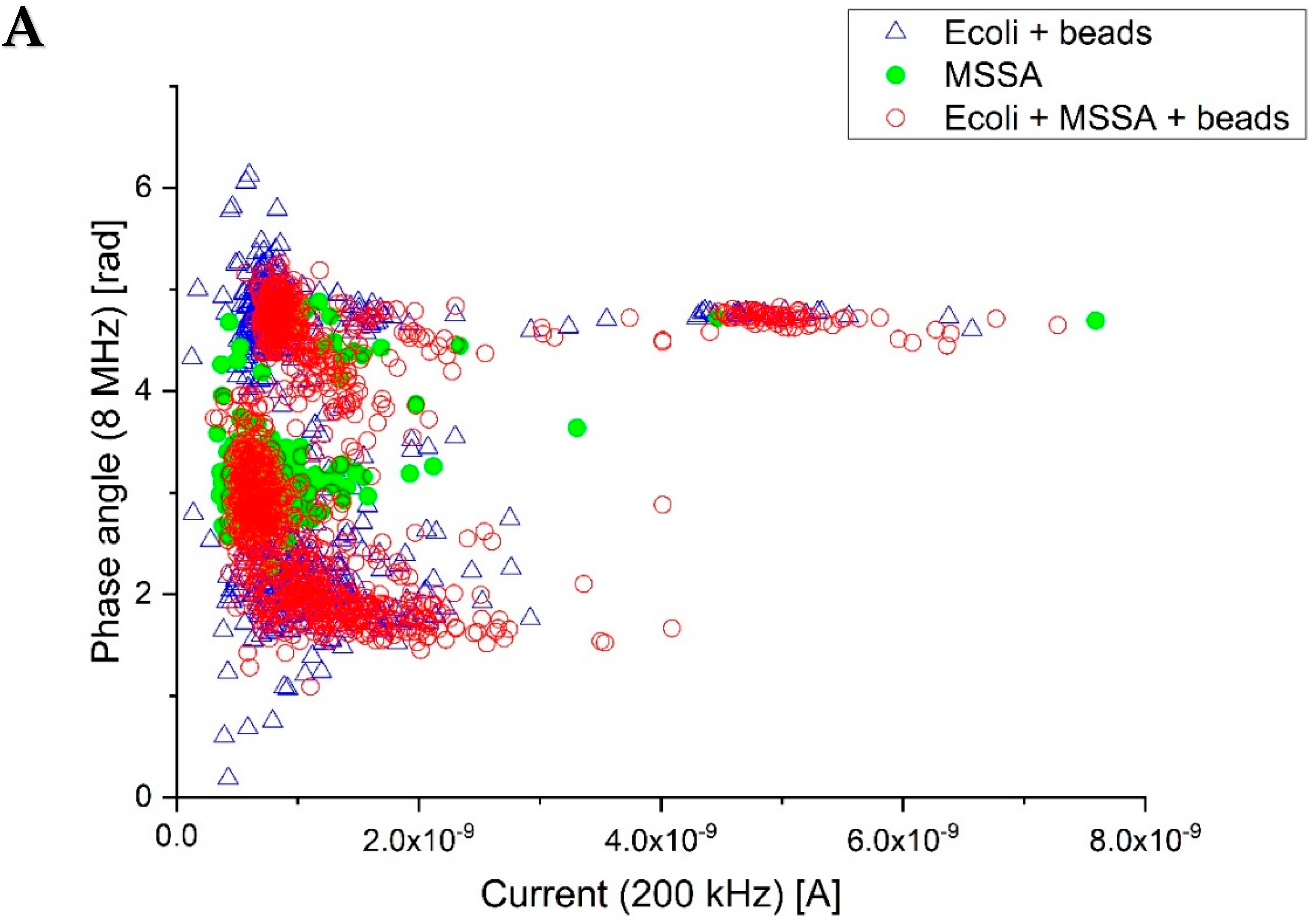
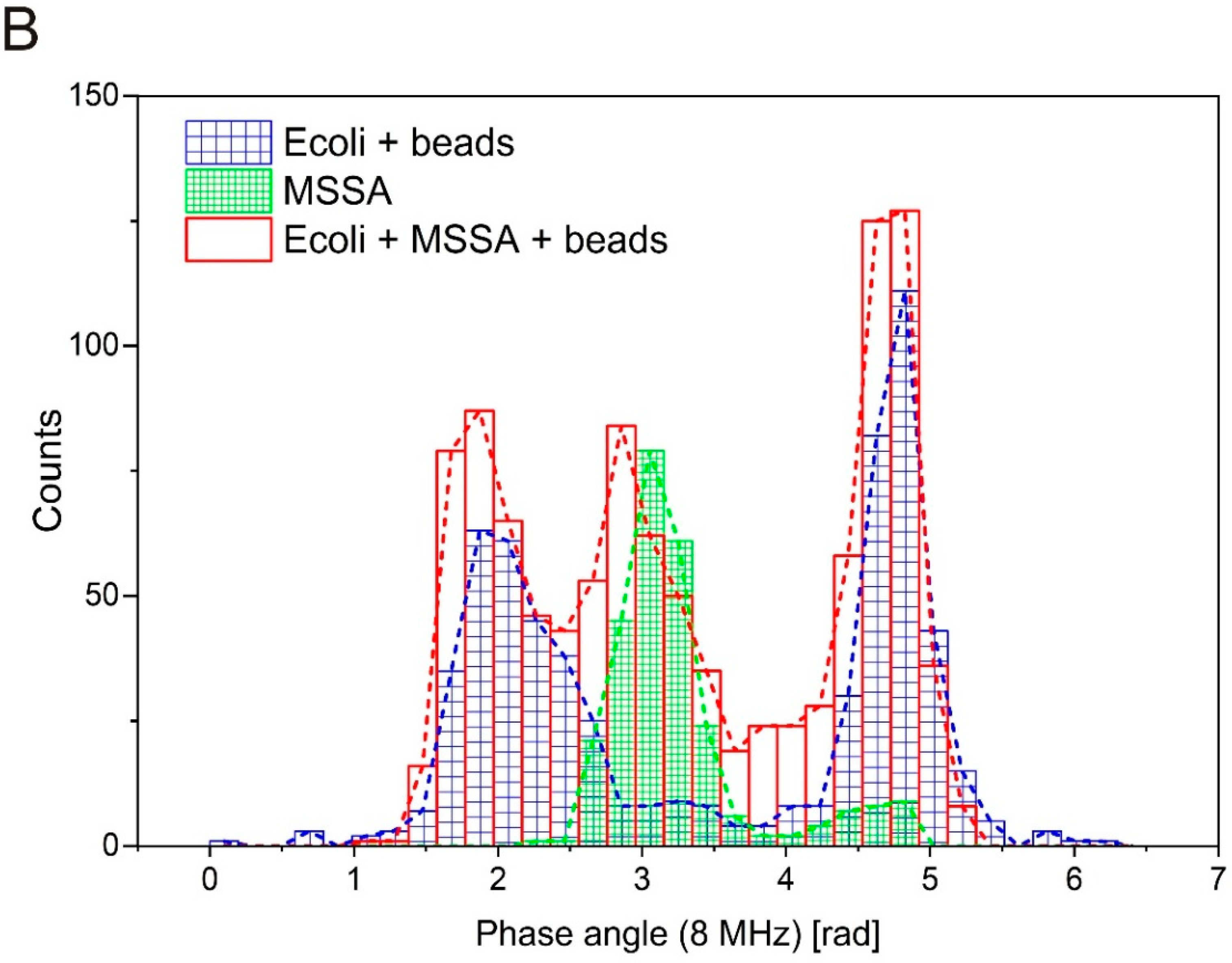
3.3. Differentiation between Gram-Positive and Gram-Negative Bacteria
3.4. General Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Højris, B.; Christensen, S.C.; Albrechtsen, H.J.; Smith, C.; Dahlqvist, M. A novel, optical, on-line bacteria sensor for monitoring drinking water quality. Sci. Rep. 2016, 6, 23935. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Roldan, R.; Tusell, P.; Cortina, J.L.; Courtois, S. On-line bacteriological detection in water. TrAC Trends Anal. Chem. 2013, 44, 46–57. [Google Scholar] [CrossRef]
- Hargesheimer, E.E.; Conio, O.; Popovicova, J.; Proaqua, C. Online Monitoring for Drinking Water Utilities; American Water Works Association: Denver, CO, USA, 2002. [Google Scholar]
- Hammes, F.; Egli, T. Cytometric methods for measuring bacteria in water: Advantages, pitfalls and applications. Anal. Bioanal. Chem. 2010, 397, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Gawad, S.; Cheung, K.; Seger, U.; Bertsch, A.; Renaud, P. Dielectric spectroscopy in a micromachined flow cytometer: Theoretical and practical considerations. Lab Chip 2004, 4, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Olesen, T.; Valvik, M.C.; Larsen, N.A.; Sandberg, R.H.; Unisensor, A.S. Optical Sectioning of a Sample and Detection of Particles in a Sample. U.S. Patent 8,780,181, 15 July 2014. [Google Scholar]
- Díaz, M.; Herrero, M.; García, L.A.; Quirós, C. Application of flow cytometry to industrial microbial bioprocesses. Biochem. Eng. J. 2010, 48, 385–407. [Google Scholar] [CrossRef]
- Gawad, S.; Schild, L.; Renaud, P.H. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip 2001, 1, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.; Pettigrew, D.; Reccius, C.H.; Gwyer, J.D.; van Berkel, C.; Holloway, J.; Davies, D.E.; Morgan, H. Leukocyte analysis and differentiation using high speed microfluidic single cell impedance cytometry. Lab Chip 2009, 9, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Morgan, H. Single-cell microfluidic impedance cytometry: A review. Microfluid. Nanofluid. 2010, 8, 423–443. [Google Scholar] [CrossRef]
- David, F.; Hebeisen, M.; Schade, G.; Franco-Lara, E.; Di Berardino, M. Viability and membrane potential analysis of Bacillus megaterium cells by impedance flow cytometry. Biotechnol. Bioeng. 2012, 109, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Ayliffe, H.E.; Frazier, A.B.; Rabbitt, R.D. Electric impedance spectroscopy using microchannels with integrated metal electrodes. J. Microelectromec. Syst. 1999, 8, 50–57. [Google Scholar] [CrossRef]
- Lin, R.; Simon, M.G.; Lee, A.P.; Lopez-Prieto, J. Label-free detection of DNA amplification in droplets using electrical impedance. In Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Seattle, WA, USA, 2–6 October 2011; Volume 3, pp. 1683–1685. [Google Scholar]
- Haandbæk, N.; Bürgel, S.C.; Heer, F.; Hierlemann, A. Characterization of subcellular morphology of single yeast cells using high frequency microfluidic impedance cytometer. Lab Chip 2014, 14, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.; Morgan, H. Single Cell Impedance Cytometry for Identification and Counting of CD4 T-Cells in Human Blood Using Impedance Labels. Anal. Chem. 2010, 82, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Evander, M.; Ricco, A.J.; Morser, J.; Kovacs, G.T.; Leung, L.L.; Giovangrandi, L. Microfluidic impedance cytometer for platelet analysis. Lab Chip 2013, 13, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, J.; Clausen, C.H.; Rodriguez-Trujillo, R.; Svendsen, W.E. Study of Paclitaxel-Treated HeLa Cells by Differential Electrical Impedance Flow Cytometry. Biosensors 2014, 4, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Mernier, G.; Hasenkamp, W.; Piacentini, N.; Renaud, P. Multiple-frequency impedance measurements in continuous flow for automated evaluation of yeast cell lysis. Sens. Actuators B Chem. 2012, 170, 2–6. [Google Scholar] [CrossRef]
- Morgan, H.; Sun, T.; Holmes, D.; Gawad, S.; Green, N.G. Single cell dielectric spectroscopy. J. Phys. D Appl. Phys. 2007, 40, 61–70. [Google Scholar] [CrossRef]
- Haandbæk, N.; Bürgel, S.C.; Heer, F.; Hierlemann, A. Resonance-enhanced microfluidic impedance cytometer for detection of single bacteria. Lab Chip 2014, 14, 3313–3324. [Google Scholar] [CrossRef] [PubMed]
- Grossman, N.; Ron, E.Z.; Woldringh, C.L. Changes in cell dimensions during amino acid starvation of Escherichia coli. J. Bacteriol. 1982, 152, 35–41. [Google Scholar] [PubMed]
- Freeman, J.T.; Anderson, D.J.; Sexton, D.J. Seasonal peaks in Escherichia coli infections: Possible explanations and implications. Clin. Microbiol. Infect. 2009, 15, 951–953. [Google Scholar] [CrossRef] [PubMed]
- Mitra, K.; Ubarretxena-Belandia, I.; Taguchi, T.; Warren, G.; Engelman, D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA 2004, 101, 4083–4088. [Google Scholar] [CrossRef] [PubMed]
- Sanchis, A.; Brown, A.P.; Sancho, M.; Martinez, G.; Sebastian, J.L.; Munoz, S.; Miranda, J.M. Dielectric characterization of bacterial cells using dielectrophoresis. Bioelectromagnetics 2007, 28, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, P.J. Cell Wall Thickness, Size Distribution, Refractive Index Ratio and Dry Weight Content of Living Bacteria (Stapylococcus aureus). Nature 1970, 226, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Zhang, Y.; Wang, T.H.; Yang, S. Continuous dielectrophoretic bacterial separation and concentration from physiological media of high conductivity. Lab Chip 2011, 11, 2893–2900. [Google Scholar] [CrossRef] [PubMed]
- Demierre, N.; Braschler, T.; Linderholm, P.; Seger, U.; van Lintel, H.; Renaud, P. Characterization and optimization of liquid electrodes for lateral dielectrophoresis. Lab Chip 2007, 7, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Moresco, J.; Clausen, C.H.; Svendsen, W. Improved anti-stiction coating of SU-8 molds. Sens. Actuators B Chem. 2010, 145, 698–701. [Google Scholar] [CrossRef]
- Serra, S.; Schneider, A.; Malecki, K. A simple bonding process of SU-8 to glass to seal a microfuidic device. In Proceedings of the Multi-Material Micro Manufacture, Borovets, Bulgaria, 3–5 October 2007; pp. 43–46. [Google Scholar]
- Clausen, C.H.; Skands, G.E.; Bertelsen, C.V.; Svendsen, W.E. Coplanar Electrode Layout Optimized for Increased Sensitivity for Electrical Impedance Spectroscopy. Micromachines 2015, 6, 110–120. [Google Scholar] [CrossRef]
- Verhille, S. Understanding Microbial Indicators for Drinking Water Assessment: Interpretation of Test Results and Public Health Significance. National Collaborating Center for Environmental Health. Available online: http://www.ncceh.ca/sites/default/files/Microbial_Indicators_Jan_2013_0.pdf (accessed on 16 October 2018).
- World Health Organisation (WHO). Guidelines for Drinking-Water Quality Incorporating First Addendum, 3rd ed.; WHO: Geneva, Switzerland, 2006; Volume 1, p. 285. [Google Scholar]
- Staley, J.T. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Ann. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]


| Sample | Measurement Time (s) | Beads (#) | Beads (/mL) | E. coli (#) | E. coli (/mL) | Plate Count (/mL) |
|---|---|---|---|---|---|---|
| A | 1551.46 | 377 | 191,202 | 7 | 6221 | 3000 |
| B | 1554.65 | 426 | 201,612 | 92 | 48,181 | 154,000 |
| C | 1551.71 | 398 | 192,603 | 224 | 118,006 | 326,000 |
| D | 1551.9 | 366 | 185,596 | 483 | 274,769 | 643,000 |
| E | 1861.85 | 445 | 186,478 | 843 | 396,767 | 1.00 × 106 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clausen, C.H.; Dimaki, M.; Bertelsen, C.V.; Skands, G.E.; Rodriguez-Trujillo, R.; Thomsen, J.D.; Svendsen, W.E. Bacteria Detection and Differentiation Using Impedance Flow Cytometry. Sensors 2018, 18, 3496. https://doi.org/10.3390/s18103496
Clausen CH, Dimaki M, Bertelsen CV, Skands GE, Rodriguez-Trujillo R, Thomsen JD, Svendsen WE. Bacteria Detection and Differentiation Using Impedance Flow Cytometry. Sensors. 2018; 18(10):3496. https://doi.org/10.3390/s18103496
Chicago/Turabian StyleClausen, Casper Hyttel, Maria Dimaki, Christian Vinther Bertelsen, Gustav Erik Skands, Romen Rodriguez-Trujillo, Joachim Dahl Thomsen, and Winnie E. Svendsen. 2018. "Bacteria Detection and Differentiation Using Impedance Flow Cytometry" Sensors 18, no. 10: 3496. https://doi.org/10.3390/s18103496
APA StyleClausen, C. H., Dimaki, M., Bertelsen, C. V., Skands, G. E., Rodriguez-Trujillo, R., Thomsen, J. D., & Svendsen, W. E. (2018). Bacteria Detection and Differentiation Using Impedance Flow Cytometry. Sensors, 18(10), 3496. https://doi.org/10.3390/s18103496