Accurate Needle Localization Using Two-Dimensional Power Doppler and B-Mode Ultrasound Image Analyses: A Feasibility Study
Abstract
1. Introduction
2. Methods
2.1. Overview of the Proposed Needle Localization Method
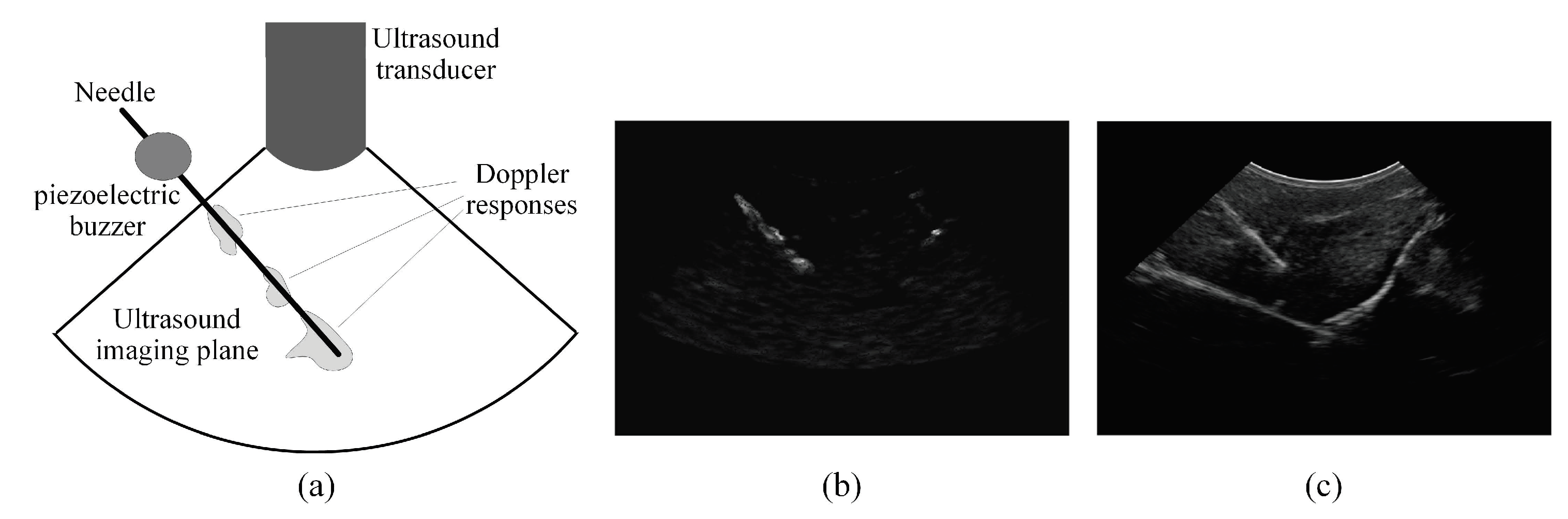
2.2. Analyzing the Power Doppler Ultrasound Image to Obtain an Initial Estimation of the Needle Axis and Identify the Candidate Regions of the Needle
2.3. Analyzing the B-Mode Ultrasound Image to Obtain Accurate Localization of the Needle Axis
2.4. Analyzing the Power Doppler and B-Mode Ultrasound Images to Obtain Accurate Localization of the Needle Tip
3. Experiments
3.1. Experimental Setup
3.2. Tuning the Parameters of the DBSCAN Clustering Algorithm
3.3. Performance Evaluations and Comparisons
3.4. Analyzing the Effect of the Needle Insertion Depth on the Accuracy of Localizing the Needle
3.5. Analyzing the Effect of the Needle Size on the Accuracy of Localizing the Needle
4. Results
4.1. Results of the Performance Evaluations and Comparisons
4.2. Results of Analyzing the Effect of the Needle Insertion Depth on the Accuracy of Localizing the Needle
4.3. Results of Analyzing the Effect of the Needle Size on the Accuracy of Localizing the Needle
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hadjerci, O.; Hafiane, A.; Conte, D.; Makris, P.; Vieyres, P.; Delbos, A. Computer-aided detection system for nerve identification using ultrasound images: A comparative study. Inform. Med. Unlocked 2016, 3, 29–43. [Google Scholar] [CrossRef]
- Jose, J.; Smith, M.K.; Kaplan, L.D.; Lesniak, B.P.; Levi, A.D. Ultrasound-Guided Needle Localization of the Saphenous Nerve for Removal of Neuroma in the Infrapatellar Branches: Technical Report. Neurosurgery 2014, 75, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Rowbotham, E.L.; Grainger, A.J. Ultrasound-Guided Intervention Around the Hip Joint. Musculoskelet. Imaging 2011, 197, W122–W127. [Google Scholar] [CrossRef] [PubMed]
- Sibbitt, W.L. Ultrasound-Guided Injections. In Musculoskeletal Ultrasound in Rheumatology Review; Kohler, M.J., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 265–318. [Google Scholar]
- Holm, H.H.; Skjoldbye, B. Interventional ultrasound. Ultrasound Med. Biol. 1996, 22, 773–789. [Google Scholar] [CrossRef]
- Hatt, C.R.; Ng, G.; Parthasarathy, V. Enhanced needle localization in ultrasound using beam steering and learning-based segmentation. Comput. Med. Imaging Gr. 2015, 41, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.J.; Perlas, A.; Chan, V.W.; Brull, R. Needle Visualization in Ultrasound-Guided Regional Anesthesia: Challenges and Solutions. Reg. Anesth. Pain Med. 2008, 33, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.; Rohling, R. Enhancement of needle visibility in ultrasound-guided percutaneous procedures. Ultrasound Med. Biol. 2004, 30, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Fenster, A. A real-time biopsy needle segmentation technique using Hough Transform. Med. Phys. 2003, 30, 2222–2233. [Google Scholar] [CrossRef] [PubMed]
- Kaya, M.; Bebek, O. Needle localization using Gabor filtering in 2D ultrasound images. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Hong Kong, China, 31 May–7 June 2014; pp. 4881–4886. [Google Scholar]
- Fogel, I.; Sagi, D. Gabor filters as texture discriminator. Biol. Cybern. 1989, 61, 103–113. [Google Scholar] [CrossRef]
- Hacihaliloglu, I.; Beigi, P.; Ng, G.; Rohling, R.N.; Salcudean, S.; Abolmaesumi, P. Projection-based phase features for localization of a needle tip in 2D curvilinear ultrasound. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 347–354. [Google Scholar]
- Daoud, M.I.; Rohling, R.N.; Salcudean, S.E.; Abolmaesumi, P. Needle detection in curvilinear ultrasound images based on the reflection pattern of circular ultrasound waves. Med. Phys. 2015, 42, 6221–6233. [Google Scholar] [CrossRef] [PubMed]
- Mwikirize, C.; Nosher, J.L.; Hacihaliloglu, I. Signal attenuation maps for needle enhancement and localization in 2D ultrasound. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Tsui, B.C.; Doyle, K.; Chu, K.; Pillay, J.; Dillane, D. Case series: Ultrasound-guided supraclavicular block using a curvilinear probe in 104 day-case hand surgery patients. Can. J. Anesth. 2009, 56, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Levi, D.S. Intra-articular hip injections using ultrasound guidance: Accuracy using a linear array transducer. PM R 2013, 5, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Daoud, M.I.; Abolmaesumi, P.; You, W.; Salcudean, S.E.; Rohling, R.N. Signature-based algorithm for improved needle localization in ultrasound images: A feasibility study. In Proceedings of the 2011 IEEE International Ultrasonics Symposium, Orlando, FL, USA, 18–21 October 2011; pp. 1575–1578. [Google Scholar]
- Klein, S.M.; Fronheiser, M.P.; Reach, J.; Nielsen, K.C.; Smith, S.W. Piezoelectric vibrating needle and catheter for enhancing ultrasound-guided peripheral nerve blocks. Anesth. Analg. 2007, 105, 1858–1860. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fronheiser, M.P.; Idriss, S.F.; Wolf, P.D.; Smith, S.W. Vibrating interventional device detection using real-time 3-D color Doppler. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2008, 55, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Adebar, T.K.; Fletcher, A.E.; Okamura, A. 3-D ultrasound-guided robotic needle steering in biological tissue. IEEE Trans. Biomed. Eng. 2014, 61, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Greer, J.D.; Adebar, T.K.; Hwang, G.L.; Okamura, A.M. Real-Time 3D Curved Needle Segmentation Using Combined B-Mode and Power Doppler Ultrasound. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014, Boston, MA, USA, 14–18 September 2014; Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 381–388. [Google Scholar]
- Glasbey, C.A. An analysis of histogram-based thresholding algorithms. CVGIP Gr. Models Image Process. 1993, 55, 532–537. [Google Scholar] [CrossRef]
- Tran, T.N.; Drab, K.; Daszykowski, M. Revised DBSCAN algorithm to cluster data with dense adjacent clusters. Chemometr. Intell. Lab. Syst. 2013, 120, 92–96. [Google Scholar] [CrossRef]
- Toft, P. The Radon Transform. Theory and Implementation. Ph.D. Thesis, Institut for Matematisk Modellering, Danmarks Tekniske Universitet, Lyngby, Denmark, 1996. [Google Scholar]
- Ayres, F.J.; Rangayyan, R.M. Design and performance analysis of oriented feature detectors. J. Electron. Imaging 2007, 16, 023007. [Google Scholar] [CrossRef]
- Kamarainen, J.K.; Kyrki, V.; Kalviainen, H. Invariance properties of Gabor filter-based features—Overview and applications. IEEE Trans. Image Process. 2006, 15, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Oloumi, F.; Rangayyan, R.M.; Oloumi, F.; Eshghzadeh-Zanjani, P.; Ayres, F.J. Detection of Blood Vessels in Fundus Images of the Retina using Gabor Wavelets. In Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE EMBS), Lyon, France, 22–26 August 2007; pp. 6451–6454. [Google Scholar]
- Adebar, T.K.; Okamura, A.M. 3D segmentation of curved needles using Doppler ultrasound and vibration. In Proceedings of the International Conference on Information Processing in Computer-Assisted Interventions–IPCAI 2013, Heidelberg, Germany, 26 June 2013; Barratt, D., Cotin, S., Fichtinger, G., Jannin, P., Navab, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 61–70. [Google Scholar]
- Beigi, P.; Rohling, R.; Salcudean, S.E.; Ng, G.C. Spectral analysis of the tremor motion for needle detection in curvilinear ultrasound via spatiotemporal linear sampling. Int. J. Comput. Assist. Radiol. Surg. 2016, 11, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Schafhalter-Zoppoth, I.; McCulloch, C.E.; Gray, A.T. Ultrasound visibility of needles used for regional nerve block: An in vitro study. Reg. Anesth. Pain Med. 2004, 29, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Pourtaherian, A.; Scholten, H.; Kusters, L.; Zinger, S.; Mihajlovic, N.; Kolen, A.; Zuo, F.; Ng, G.; Korsten, H.; de With, P. Medical instrument detection in 3-dimensional ultrasound data volumes. IEEE Trans. Med. Imaging 2017, 36, 1664–1675. [Google Scholar] [CrossRef] [PubMed]








| Range of Needle Insertion Angles | Tissue Type | Failure Rate | Angel Error () | Axis Error (mm) | Tip Error (mm) |
|---|---|---|---|---|---|
| Shallow angles (–) | Bovine muscle | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 | |
| Bovine liver | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.3 ± 0.1 | ||
| Porcine muscle | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.3 ± 0.1 | ||
| Moderate angles (–) | Bovine muscle | 0.4 ± 0.2 | 0.4 ± 0.1 | 0.5 ± 0.1 | |
| Bovine liver | 0.3 ± 0.2 | 0.3 ± 0.1 | 0.4 ± 0.1 | ||
| Porcine muscle | 0.3 ± 0.2 | 0.4 ± 0.1 | 0.4 ± 0.1 | ||
| Steep angles (–) | Bovine muscle | 0.8 ± 0.2 | 0.5 ± 0.1 | 0.6 ± 0.1 | |
| Bovine liver | 0.7 ± 0.2 | 0.5 ± 0.1 | 0.5 ± 0.1 | ||
| Porcine muscle | 0.8 ± 0.2 | 0.6 ± 0.1 | 0.5 ± 0.1 | ||
| All angles (–) | Bovine muscle | 0.5 ± 0.3 | 0.4 ± 0.1 | 0.5 ± 0.1 | |
| Bovine liver | 0.4 ± 0.3 | 0.3 ± 0.2 | 0.4 ± 0.1 | ||
| Porcine muscle | 0.5 ± 0.3 | 0.4 ± 0.2 | 0.4 ± 0.1 |
| Range of Needle Insertion Angles | Tissue Type | Failure Rate | Angel Error () | Axis Error (mm) | Tip Error (mm) |
|---|---|---|---|---|---|
| Shallow angles (–) | Bovine muscle | 0.9 ± 0.6 | 1.1 ± 0.3 | 1.1 ± 0.4 | |
| Bovine liver | 1.3 ± 0.5 | 1.2 ± 0.5 | 1.3 ± 0.5 | ||
| Porcine muscle | 1.1 ± 0.4 | 1.1 ± 0.3 | 1.2 ± 0.3 | ||
| Moderate angles (–) | Bovine muscle | 1.9 ± 0.4 | 1.7 ± 0.2 | 1.6 ± 0.2 | |
| Bovine liver | 2.5 ± 1.2 | 1.8 ± 0.5 | 1.8 ± 0.7 | ||
| Porcine muscle | 2.2 ± 1.2 | 1.5 ± 0.4 | 1.6 ± 0.6 | ||
| Steep angles (–) | Bovine muscle | 2.9 ± 0.6 | 2.3 ± 0.3 | 2.2 ± 0.4 | |
| Bovine liver | 2.9 ± 1.2 | 2.4 ± 0.4 | 2.5 ± 0.5 | ||
| Porcine muscle | 2.7 ± 1.1 | 2.0 ± 0.5 | 2.1 ± 0.5 | ||
| All angles (–) | Bovine muscle | 1.8 ± 1.0 | 1.7 ± 0.5 | 1.6 ± 0.5 | |
| Bovine liver | 2.2 ± 1.2 | 1.7 ± 0.7 | 1.8 ± 0.7 | ||
| Porcine muscle | 1.9 ± 1.2 | 1.5 ± 0.5 | 1.6 ± 0.6 |
| Range of Needle Insertion Angles | Tissue Type | Failure Rate | Angel Error () | Axis Error (mm) | Tip Error (mm) |
|---|---|---|---|---|---|
| Shallow angles (–) | Bovine muscle | 1.7 ± 0.9 | 1.2 ± 0.6 | 1.2 ± 0.7 | |
| Bovine liver | 1.8 ± 0.8 | 1.0 ± 0.4 | 1.1 ± 0.4 | ||
| Porcine muscle | 1.8 ± 0.3 | 1.1 ± 0.5 | 1.1 ± 0.5 | ||
| Moderate angles (–) | Bovine muscle | 2.0 ± 1.0 | 1.6 ± 0.7 | 1.6 ± 0.8 | |
| Bovine liver | 2.2 ± 1.1 | 1.5 ± 0.7 | 1.6 ± 0.9 | ||
| Porcine muscle | 2.3 ± 0.7 | 1.7 ± 0.6 | 1.8 ± 0.8 | ||
| Steep angles (–) | Bovine muscle | 2.6 ± 1.2 | 1.9 ± 0.5 | 2.1 ± 0.3 | |
| Bovine liver | 2.7 ± 1.0 | 1.9 ± 0.7 | 2.0 ± 0.5 | ||
| Porcine muscle | 2.9 ± 1.5 | 2.0 ± 0.4 | 2.0 ± 0.5 | ||
| All angles (–) | Bovine muscle | 2.0 ± 1.1 | 1.5 ± 0.7 | 1.6 ± 0.7 | |
| Bovine liver | 2.2 ± 1.0 | 1.4 ± 0.7 | 1.5 ± 0.7 | ||
| Porcine muscle | 2.3 ± 1.0 | 1.6 ± 0.6 | 1.6 ± 0.7 |
| Range of Needle Insertion Angles | Tissue Type | Angel Error () | Axis Error (mm) | Tip Error (mm) |
|---|---|---|---|---|
| – | Bovine muscle | 0.4 ± 0.2 | 0.3 ± 0.2 | 0.3 ± 0.1 |
| Bovine liver | 0.3 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.1 | |
| Porcine muscle | 0.3 ± 0.2 | 0.3 ± 0.2 | 0.4 ± 0.1 | |
| – | Bovine muscle | 0.5 ± 0.3 | 0.4 ± 0.1 | 0.5 ± 0.2 |
| Bovine liver | 0.3 ± 0.2 | 0.3 ± 0.1 | 0.4 ± 0.1 | |
| Porcine muscle | 0.4 ± 0.2 | 0.4 ± 0.1 | 0.4 ± 0.1 | |
| – | Bovine muscle | 0.5 ± 0.3 | 0.4 ± 0.2 | 0.5 ± 0.2 |
| Bovine liver | 0.5 ± 0.2 | 0.4 ± 0.2 | 0.4 ± 0.1 | |
| Porcine muscle | 0.5 ± 0.2 | 0.4 ± 0.1 | 0.4 ± 0.1 | |
| – | Bovine muscle | 0.6 ± 0.3 | 0.5 ± 0.2 | 0.6 ± 0.2 |
| Bovine liver | 0.5 ± 0.3 | 0.4 ± 0.2 | 0.5 ± 0.2 | |
| Porcine muscle | 0.6 ± 0.2 | 0.5 ± 0.1 | 0.5 ± 0.1 |
| Range of Needle Insertion Angles | Failure Rate | Angel Error () | Axis Error (mm) | Tip Error (mm) |
|---|---|---|---|---|
| Shallow angles (–) | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.3 ± 0.1 | |
| Moderate angles (–) | 0.3 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.1 | |
| Steep angles (–) | 0.7 ± 0.2 | 0.5 ± 0.1 | 0.6 ± 0.1 | |
| All angles (–) | 0.4 ± 0.3 | 0.4 ± 0.2 | 0.4 ± 0.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daoud, M.I.; Shtaiyat, A.; Zayadeen, A.R.; Alazrai, R. Accurate Needle Localization Using Two-Dimensional Power Doppler and B-Mode Ultrasound Image Analyses: A Feasibility Study. Sensors 2018, 18, 3475. https://doi.org/10.3390/s18103475
Daoud MI, Shtaiyat A, Zayadeen AR, Alazrai R. Accurate Needle Localization Using Two-Dimensional Power Doppler and B-Mode Ultrasound Image Analyses: A Feasibility Study. Sensors. 2018; 18(10):3475. https://doi.org/10.3390/s18103475
Chicago/Turabian StyleDaoud, Mohammad I., Ahmad Shtaiyat, Adnan R. Zayadeen, and Rami Alazrai. 2018. "Accurate Needle Localization Using Two-Dimensional Power Doppler and B-Mode Ultrasound Image Analyses: A Feasibility Study" Sensors 18, no. 10: 3475. https://doi.org/10.3390/s18103475
APA StyleDaoud, M. I., Shtaiyat, A., Zayadeen, A. R., & Alazrai, R. (2018). Accurate Needle Localization Using Two-Dimensional Power Doppler and B-Mode Ultrasound Image Analyses: A Feasibility Study. Sensors, 18(10), 3475. https://doi.org/10.3390/s18103475





