A Label-Free Fluorescent AND Logic Gate Aptasensor for Sensitive ATP Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. The Process Used for Detection
2.3. The Selectivity Assays
2.4. ATP Detection in Real Urine Samples
2.5. Fluorescence Spectroscopy
3. Results and Discussion
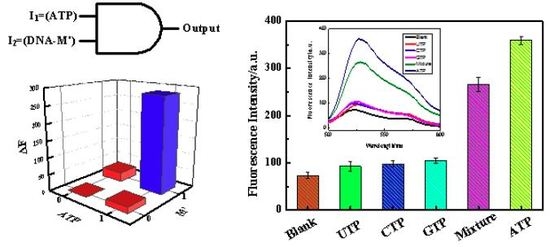
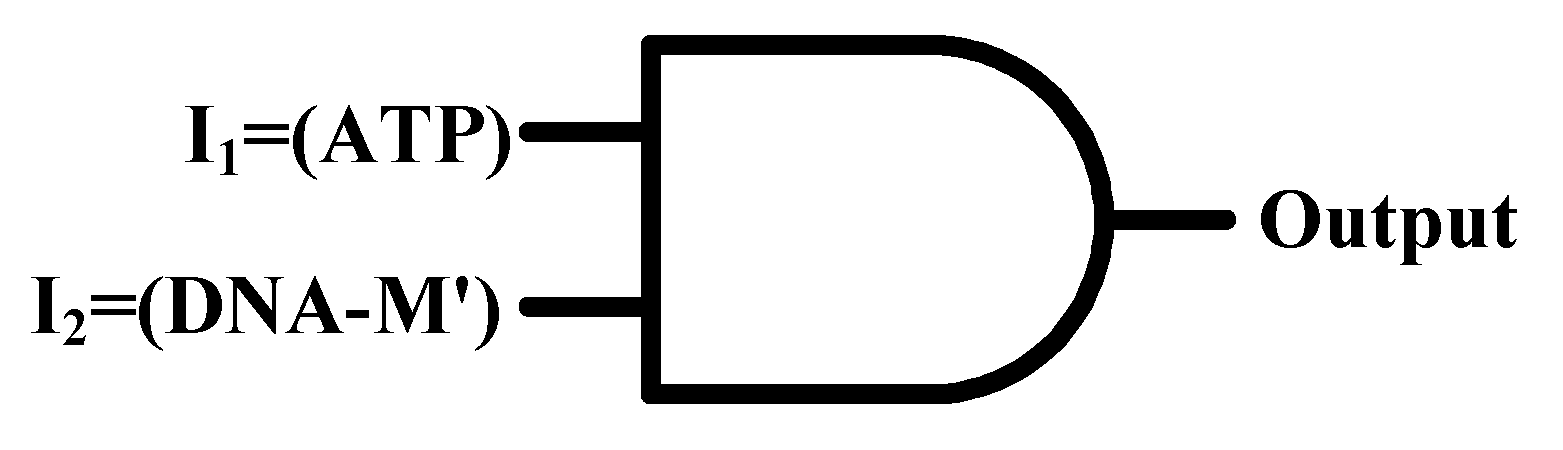
3.1. Construction of an AND Logic Gate
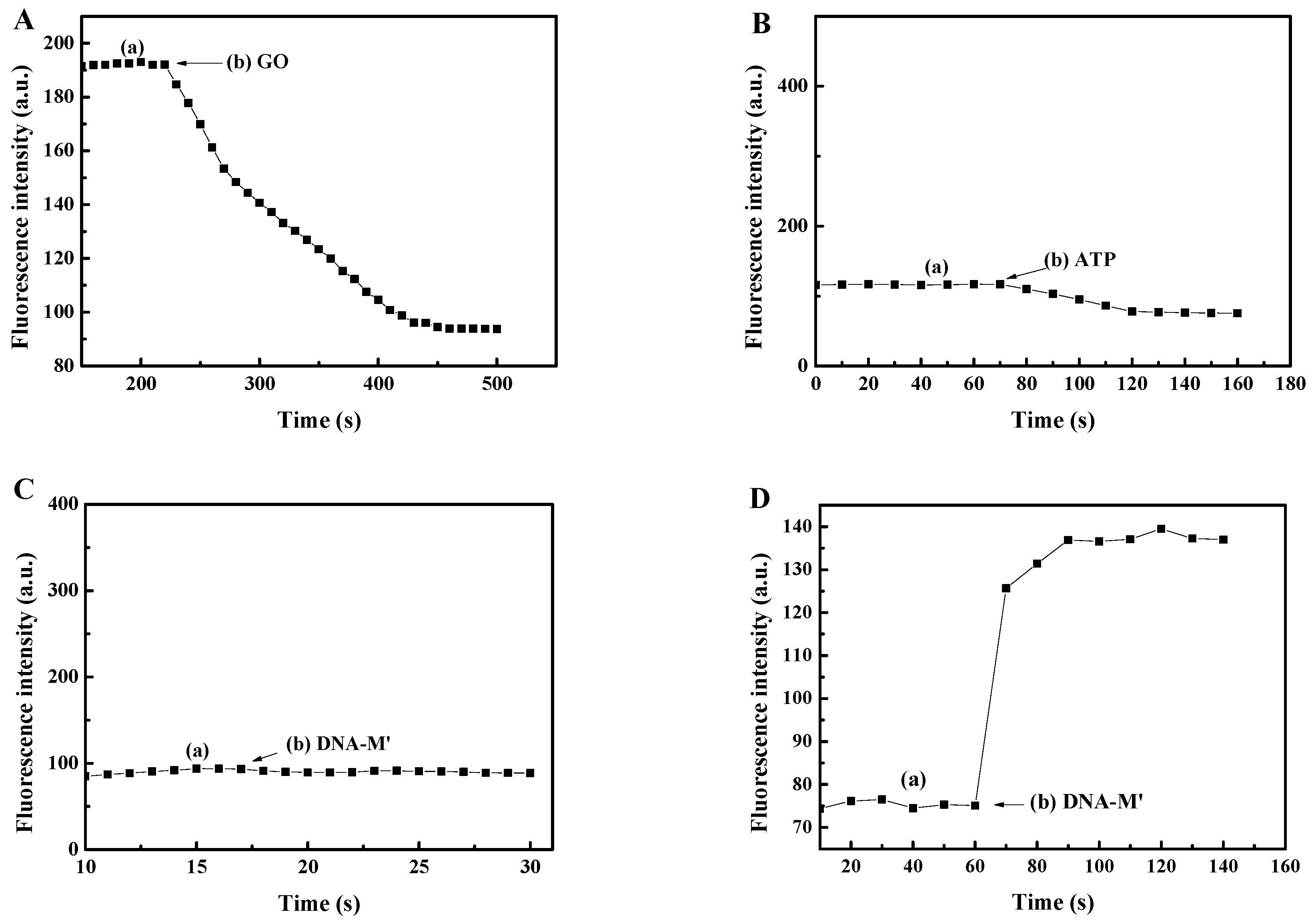
3.2. An Investigation into the Reaction Time
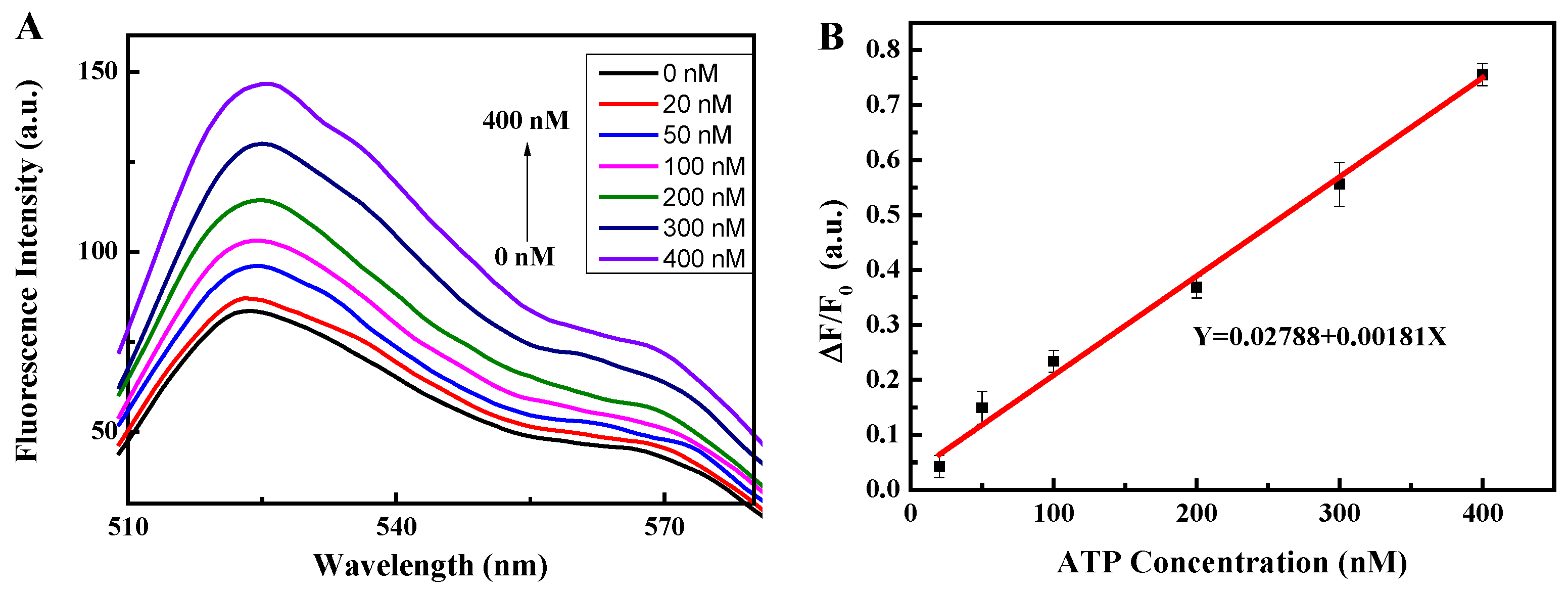
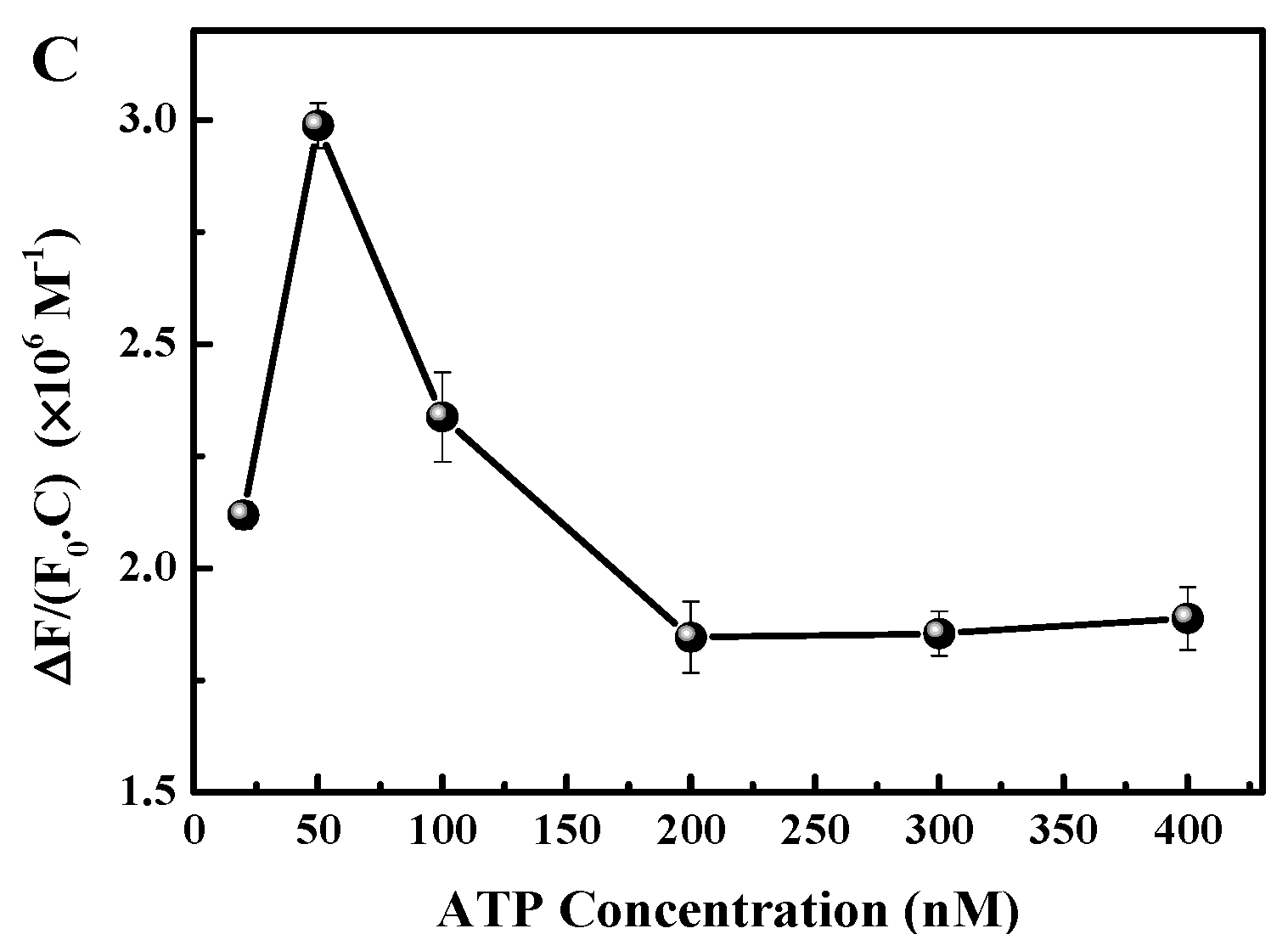
3.3. Analysis of the Linearity and Sensitivity
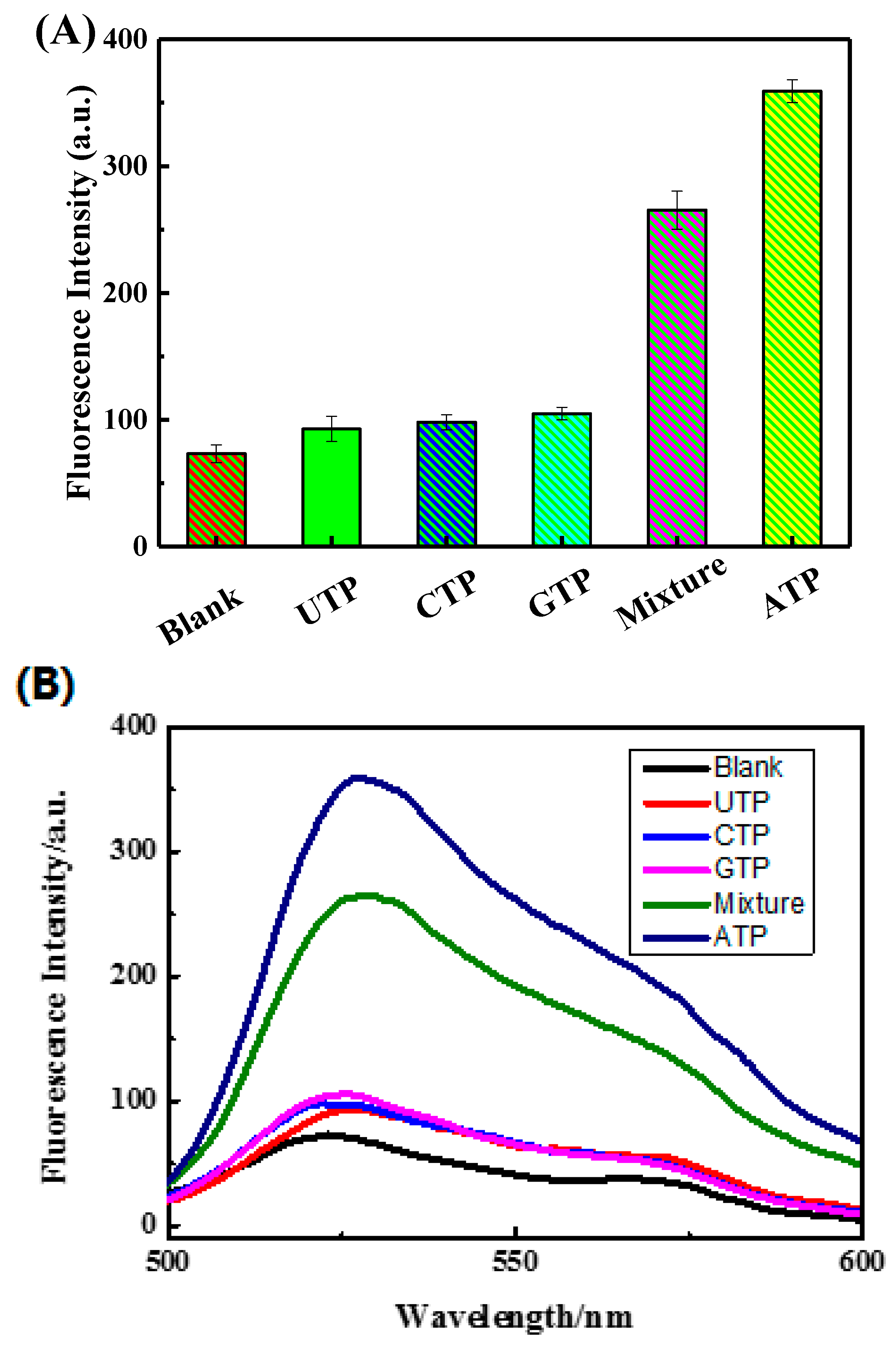
3.4. Analysis of the Selectivity
3.5. Detection of ATP Concentration in Real Urine Samples
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Imamura, H.; Nhat, K.P.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Yang, S.; Peng, Z.; Liu, C.; Li, J.; Zhong, W.; Yang, R.; Tan, W. Two-photon graphene oxide/aptamer nanosensing conjugate for in vitro or in vivo molecular probing. Anal. Chem. 2014, 86, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Mo, R.; Jiang, T.; Gu, Z. Enhanced anticancer efficacy by ATP-mediated liposomal drug delivery. Angew. Chem. Int. Ed. Engl. 2014, 53, 5815–5820. [Google Scholar] [CrossRef] [PubMed]
- Naito, M.; Ishii, T.; Matsumoto, A.; Miyata, K.; Miyahara, Y.; Kataoka, K. A phenylboronate-functionalized polyion complex micelle for ATP-triggered release of siRNA. Angew. Chem. Int. Ed. Engl. 2012, 51, 10751–10755. [Google Scholar] [CrossRef] [PubMed]
- Gourine, A.V.; Llaudet, E.; Dale, N.; Spyer, K.M. Atp is a mediator of chemosensory transduction in the central nervous system. Nature 2005, 436, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, S.; Chen, X.; Bian, L. A g-quadruplex based fluorescent oligonucleotide turn-on probe towards iodides detection in real samples. Food Chem. 2017, 230, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Zhang, Y.S.; Lin, Y.B.; Lu, Y.J.; Lin, Z.Y.; Wong, K.Y.; Chen, G.N. A novel fluorescent biosensor for detection of target DNA fragment from the transgene cauliflower mosaic virus 35s promoter. Biosens. Bioelectron. 2013, 41, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, J.; Yang, X.; Zhao, M.; Chen, M.; Yao, W.; Tan, W.; Lan, X. A label-free aptasensor for highly sensitive detection of ATP and thrombin based on metal-enhanced picogreen fluorescence. Biosens. Bioelectron. 2015, 63, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.G.; Ren, W.; Jia, J.; Feng, J.; Gao, Z.F.; Li, N.B.; Luo, H.Q. Fluorometric detection of mutant DNA oligonucleotide based on toehold strand displacement-driving target recycling strategy and exonuclease iii-assisted suppression. Biosens. Bioelectron. 2016, 77, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Yin, B.C.; Wang, P.; Ye, B.C. Highly sensitive detection of micrornas based on isothermal exponential amplification-assisted generation of catalytic g-quadruplex dnazyme. Biosens. Bioelectron. 2013, 42, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhou, W.; Li, X.; Chai, Y.; Yuan, R.; Xiang, Y. Coupling hybridization chain reaction with catalytic hairpin assembly enables non-enzymatic and sensitive fluorescent detection of microRNA cancer biomarkers. Biosens. Bioelectron. 2016, 77, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Liu, J.-W.; Duan, L.-Y.; Liu, S.-J.; Jiang, J.-H. Aptamer-based fluorometric determination of ATP by using target-cycling strand displacement amplification and copper nanoclusters. Microchim. Acta 2017, 184, 4183–4188. [Google Scholar] [CrossRef]
- Zhou, F.; Li, B.; Ma, J. A linear DNA probe as an alternative to a molecular beacon for improving the sensitivity of a homogenous fluorescence biosensing platform for DNA detection using target-primed rolling circle amplification. RSC Adv. 2015, 5, 4019–4025. [Google Scholar] [CrossRef]
- Zeng, S.; Huang, H.; Huang, Y.; Liu, X.; Qin, J.; Zhao, S.; Chen, Z.-F.; Liang, H. Label-free and amplified colorimetric assay of ribonuclease h activity and inhibition based on a novel enzyme-responsive dnazyme cascade. RSC Adv. 2015, 5, 43105–43109. [Google Scholar] [CrossRef]
- Lu, C.H.; Li, J.; Lin, M.H.; Wang, Y.W.; Yang, H.H.; Chen, X.; Chen, G.N. Amplified aptamer-based assay through catalytic recycling of the analyte. Angew. Chem. Int. Ed. Engl. 2010, 49, 8454–8457. [Google Scholar] [CrossRef] [PubMed]
- He, S.N.; Qu, L.; Tan, Y.; Liu, F.; Wang, Y.; Zhang, W.; Cai, Z.M.; Mou, L.S.; Jiang, Y.Y. A fluorescent aptasensor with product-triggered amplification by exonuclease iii digestion for highly sensitive ATP detection. Anal. Methods 2017, 9, 4837–4842. [Google Scholar] [CrossRef]
- Ning, Y.; Wei, K.; Cheng, L.; Hu, J.; Xiang, Q. Fluorometric aptamer based determination of adenosine triphosphate based on deoxyribonuclease i-aided target recycling and signal amplification using graphene oxide as a quencher. Microchim. Acta 2017, 184, 1847–1854. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Hua, X.; Feng, X.; Su, S.; Huang, Y.; Fan, Q.; Wang, L.; Huang, W. An improved turn-on aptasensor for thrombin detection using split aptamer fragments and graphene oxide. Chin. J. Chem. 2015, 33, 981–986. [Google Scholar] [CrossRef]
- Lv, H.; Li, S.; Liu, Y.; Wang, G.; Li, X.; Lu, Y.; Wang, J. A reversible fluorescent inhibit logic gate for determination of silver and iodide based on the use of graphene oxide and a silver–selective probe DNA. Microchim. Acta 2015, 182, 2513–2520. [Google Scholar] [CrossRef]
- Luo, J.; Shen, X.; Li, B.; Li, X.; Zhou, X. Signal amplification by strand displacement in a carbon dot based fluorometric assay for ATP. Mikrochim. Acta 2018, 185, 392. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, H.; Bai, J. Homogeneous selecting of a quadruplex-binding ligand-based gold nanoparticle fluorescence resonance energy transfer assay. Anal. Chem. 2009, 81, 5709–5715. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.C.; Zuo, P.; Huo, H.; Zhong, X.; Ye, B.C. Dnazyme self-assembled gold nanoparticles for determination of metal ions using fluorescence anisotropy assay. Anal. Biochem 2010, 401, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Saberi, Z.; Rezaei, B.; Khayamian, T. A fluorescent aptasensor for analysis of adenosine triphosphate based on aptamer-magnetic nanoparticles and its single-stranded complementary DNA labeled carbon dots. Luminescence 2018, 33, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Hai, X.M.; Li, N.; Wang, K.; Zhang, Z.Q.; Zhang, J.; Dang, F.Q. A fluorescence aptasensor based on two-dimensional sheet metal-organic frameworks for monitoring adenosine triphosphate. Anal. Chim. Acta 2018, 998, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Dragan, A.I.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. Characterization of picogreen interaction with dsdna and the origin of its fluorescence enhancement upon binding. Biophys. J. 2010, 99, 3010–3019. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Liu, J.; Zhou, Y.; Guan, Z.; Yang, S.; Li, C.; Chen, A. Highly sensitive fluorescent detection of small molecules, ions, and proteins using a universal label-free aptasensor. Chem. Commun. 2013, 49, 5465–5467. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.H.; Gao, Z.F.; Li, N.; Shi, Y.; Li, N.B.; Luo, H.Q. Thiazole orange as a fluorescent probe: Label-free and selective detection of silver ions based on the structural change of i-motif DNA at neutral ph. Talanta 2016, 156–157, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Pu, W.; Zhao, H.; Huang, C.; Wu, L.; Xua, D. Fluorescent detection of silver(i) and cysteine using sybr green i and a silver(i)-specific oligonucleotide. Microchim. Acta 2012, 177, 137–144. [Google Scholar] [CrossRef]
- Liu, H.; Ma, C.; Ning, F.; Chen, H.; He, H.; Wang, K.; Wang, J. A facile label-free g-quadruplex based fluorescent aptasensor method for rapid detection of ATP. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 175, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Wang, R.; Sun, F.; Chen, H.; Wang, Z.; Na, N.; Ouyang, J. A nuclease-assisted label-free aptasensor for fluorescence turn-on detection of ATP based on the in situ formation of copper nanoparticles. Biosens. Bioelectron. 2017, 87, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Voelcker, N.H.; Guckian, K.M.; Saghatelian, A.; Ghadiri, M.R. Sequence-addressable DNA logic. Small 2008, 4, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.-Q.; Qin, W.-S.; Liu, X.-F.; Huang, W. An optical-logic system based on cationic conjugated polymer/DNA/intercalating dyes assembly for label-free detection of conformational conversion of DNA i-motif structure. Polym. Chem. 2011, 2, 1341. [Google Scholar] [CrossRef]
- Kong, L.; Xu, J.; Xu, Y.; Xiang, Y.; Yuan, R.; Chai, Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on sybr green i dye. Biosens. Bioelectron. 2013, 42, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Cheng, Y.; Wang, L.; Li, Y. An ‘‘off-on’’ phosphorescent aptasensor switch for the detection of ATP. Talanta 2018, 190, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, X.; Ma, D.-L.; Lu, L.; Qu, B. A label-free triplex-to-G-qadruplex molecular switch for sensitive fluorescent detection of acetamiprid. Talanta 2018, 189, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Song, C.; Yang, K.; Hong, W.; Lu, Y.; Yu, P.; Mao, L. Photoinduced regeneration of an aptamer-based electrochemical sensor for sensitively detecting adenosine triphosphate. Anal. Chem. 2018, 90, 4968–4971. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Su, X.; Xu, Q.; Xu, G.; Lin, J.; Luo, X. Antifouling aptasensor for the detection of adenosine triphosphate in biological media based on mixed self-assembled aptamer and zwitterionic peptide. Biosens. Bioelectron. 2018, 101, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Mashhadizadeh, M.H.; Naseri, N.; Mehrgardi, M.A. A simple non-enzymatic strategy for adenosine triphosphate electrochemical aptasensor using silver nanoparticle-decorated graphene oxide. J. Iran. Chem. Soc. 2017, 14, 2007–2016. [Google Scholar] [CrossRef]
- Shahsavar, K.; Hosseini, M.; Shokri, E.; Ganjali, M.R.; Ju, H. A sensitive colorimetric aptasensor with a triplehelix molecular switch based on peroxidase-like activity of a dnazyme for ATP detection. Anal. Methods 2017, 9, 4726–4731. [Google Scholar] [CrossRef]
- Mao, Y.; Fan, T.; Gysbers, R.; Tan, Y.; Liu, F.; Lin, S.; Jiang, Y. A simple and sensitive aptasensor for colorimetric detection of adenosine triphosphate based on unmodified gold nanoparticles. Talanta 2017, 168, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Ge, J.; Xu, H.; Zhang, L.; Hu, Y.; Huang, Z.; Li, Z. A label-free aptasensor for highly sensitive ATP detection by using exonuclease i and oligonucleotide-templated fluorescent copper nanoparticles. Anal. Methods 2017, 9, 2710–2714. [Google Scholar] [CrossRef]
- Jia, J.; Feng, J.; Chen, H.G.; Luo, H.Q.; Li, N.B. A simple electrochemical method for the detection of ATP using target-induced conformational change of dual-hairpin DNA structure. Sens. Actuators B Chem. 2016, 222, 1090–1095. [Google Scholar] [CrossRef]
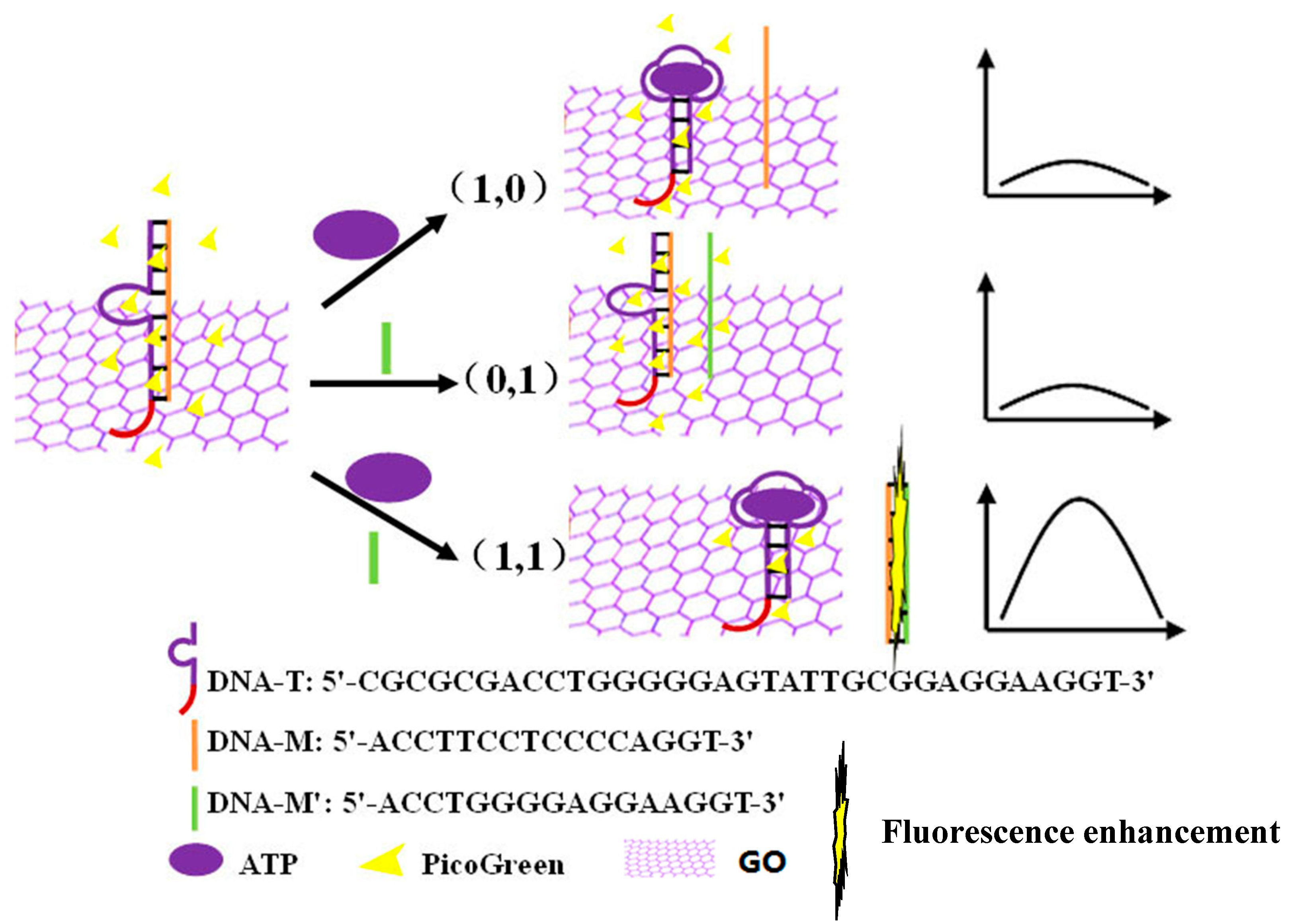

| Oligonucleotide | Sequence |
|---|---|
| DNA-T | 5′-CGC GCG ACC TGG GGG AGT ATT GCG GAG GAA GGT-3′ |
| DNA-M | 5′-ACC TTC CTC CCC AGG T-3′ |
| DNA-M′ | 5′-ACC TGG GGA GGA AGG T-3′ |
| I1 = (ATP) | I2 = (DNA-M′) | Output = (ΔF) |
|---|---|---|
| 0 | 0 | 0 |
| 0 | 1 | 0 |
| 1 | 0 | 0 |
| 1 | 1 | 1 |
| Methods | LOD | Linear Range | Correlation Coefficient (R2) | Reference |
|---|---|---|---|---|
| A Universal and Label-free Aptasensor | 23.4 nM | 0–10 mM | N/A | [33] |
| An “off-on” Phosphorescent Aptasensor Switch | 0.9 nM | 2 nM–9 µM | 0.995 | [34] |
| Hollow NiFe PBA Derived NiOxFeOy@mC | 1.62 fM | 8.26 fM–8.26 nM | 0.996 | [35] |
| Photoinduced Regeneration | 0.5 nM | 1 nM–100 μM | 0.996 | [36] |
| Mixed Self-assembled Aptamer and Zwitterionic Peptide | 0.1 pM | 0.1 pM–5 nM | 0.9984 | [37] |
| Silver Nanoparticle-decorated Graphene Oxide | 5.0 nM | 10–850 nM | 0.9901 | [38] |
| Peroxidase-like Activity of DNAzyme | 2.4 nM | 5–230 nM | 0.9854 | [39] |
| Unmodified Gold Nanoparticles | 0.1 μM | 0–5 μM | 0.9989 | [40] |
| Oligonucleotide-templated Fluorescent Copper Nanoparticles | 500 nM | 1–80 mM | 0.9901 | [41] |
| Target-induced Conformational Change of Dual-hairpin DNA | 1.4 nM | 5 nM–1 μM | 0.9976 | [42] |
| GO-Based Aptamer Logic Gates | 142.6 pM | 20–400 nM | 0.9943 | This work |
| ATP | Proposed | Method | |||
|---|---|---|---|---|---|
| Samples | Added | Found | Recovery | Standard Deviation (n = 3) | Relative Standard Deviation (n = 3) |
| (nM) | (nM) | (%) | (nM) | (%) | |
| 1 | 100.00 | 114.368 | 104.2 | 7.75 | 7.4 |
| 102.745 | |||||
| 95.551 | |||||
| 2 | 200.00 | 211.597 | 102.5 | 4.95 | 2.4 |
| 203.427 | |||||
| 199.758 | |||||
| 3 | 300.00 | 308.788 | 102.0 | 1.95 | 0.6 |
| 305.366 | |||||
| 304.202 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Yang, C.; Niu, C.; Liu, C.; Cai, X.; Du, J.; Chen, Y. A Label-Free Fluorescent AND Logic Gate Aptasensor for Sensitive ATP Detection. Sensors 2018, 18, 3281. https://doi.org/10.3390/s18103281
Zhang J, Yang C, Niu C, Liu C, Cai X, Du J, Chen Y. A Label-Free Fluorescent AND Logic Gate Aptasensor for Sensitive ATP Detection. Sensors. 2018; 18(10):3281. https://doi.org/10.3390/s18103281
Chicago/Turabian StyleZhang, Jingjing, Chunzheng Yang, Chaoqun Niu, Chen Liu, Xuepin Cai, Jie Du, and Yong Chen. 2018. "A Label-Free Fluorescent AND Logic Gate Aptasensor for Sensitive ATP Detection" Sensors 18, no. 10: 3281. https://doi.org/10.3390/s18103281
APA StyleZhang, J., Yang, C., Niu, C., Liu, C., Cai, X., Du, J., & Chen, Y. (2018). A Label-Free Fluorescent AND Logic Gate Aptasensor for Sensitive ATP Detection. Sensors, 18(10), 3281. https://doi.org/10.3390/s18103281