Digital Microfluidics for Nucleic Acid Amplification
Abstract
:1. Introduction
Digital Microfluidics Configurations
2. Digital Microfluidics for Nucleic Acid Amplification
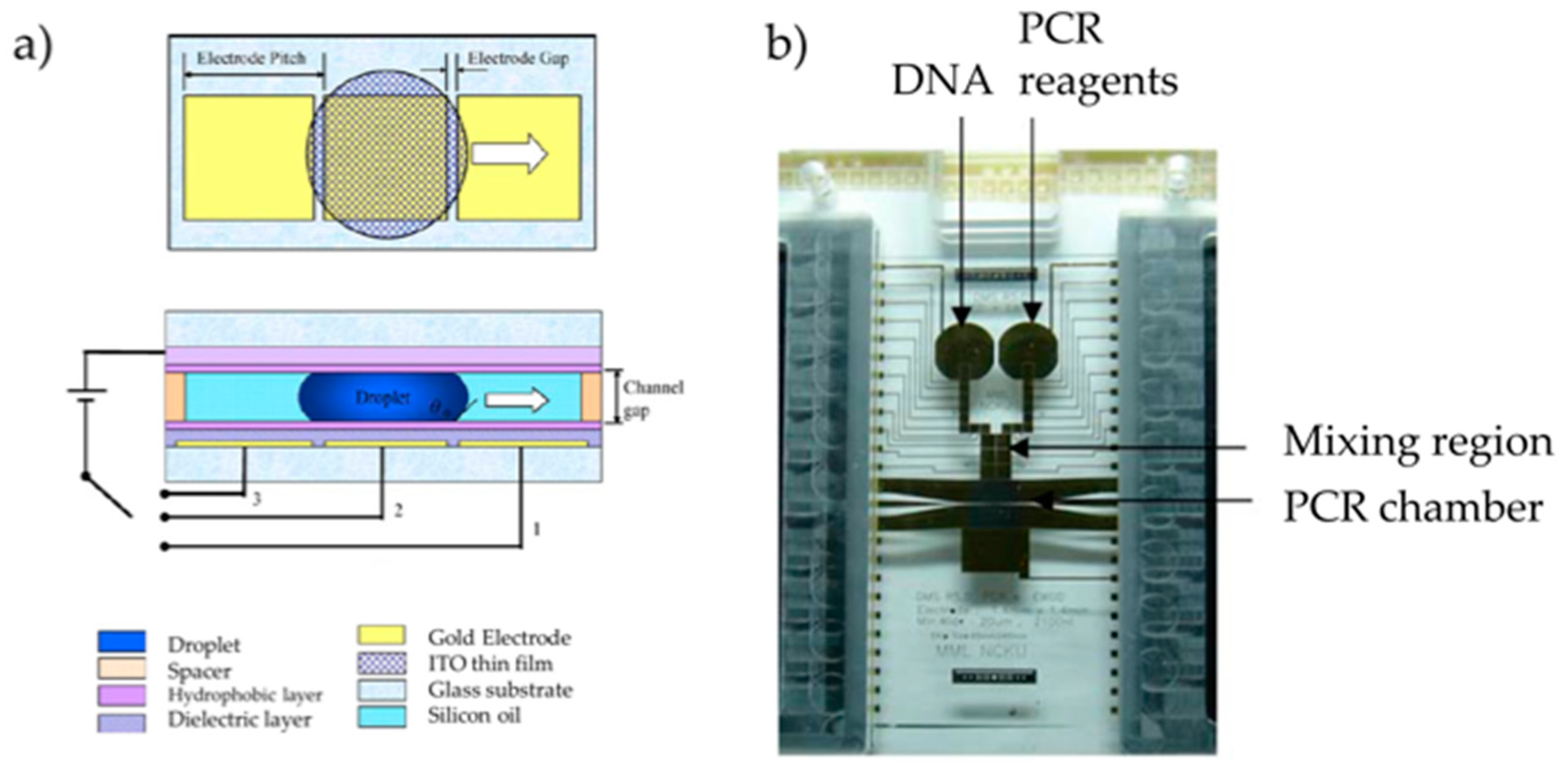
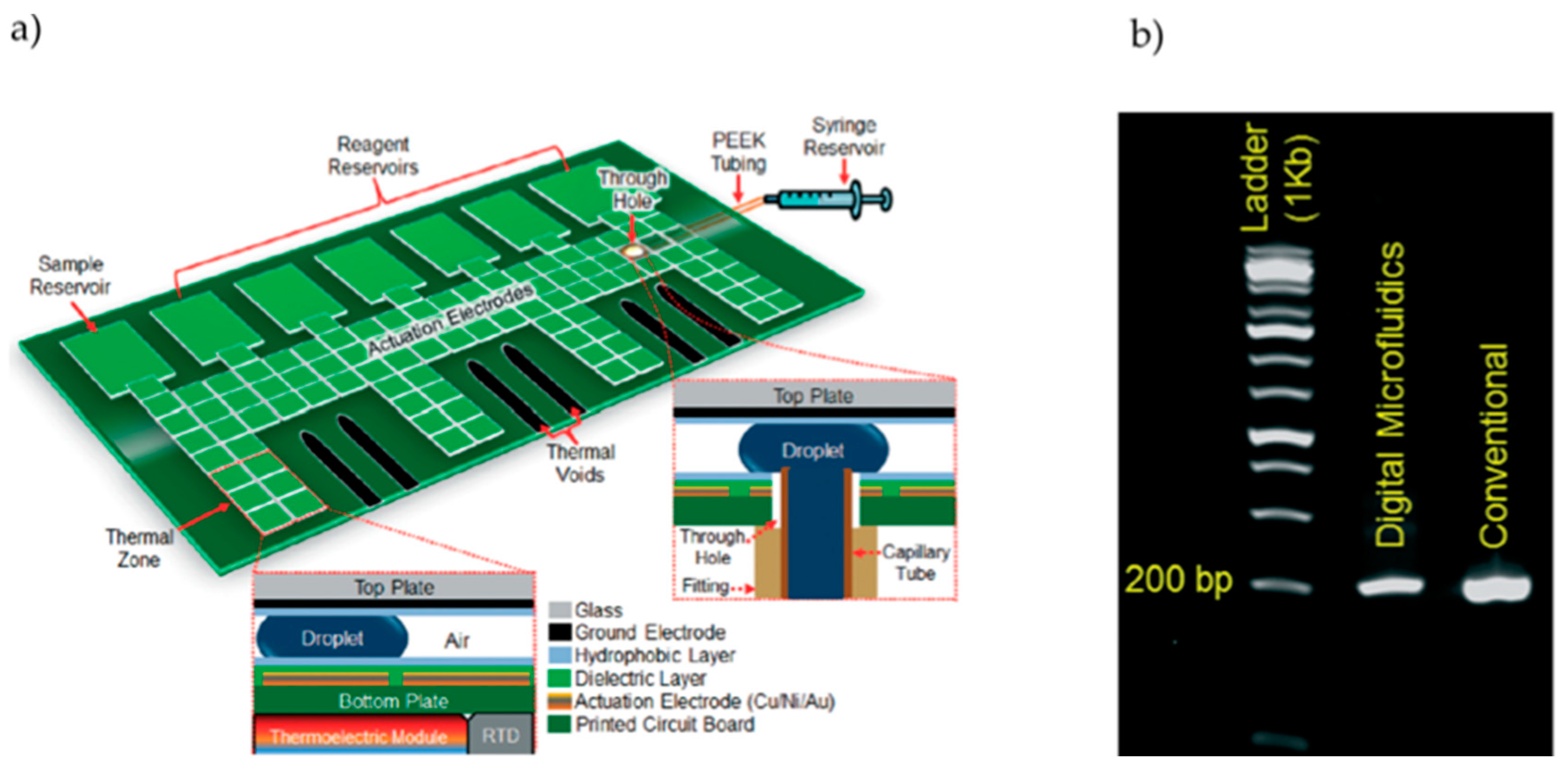
2.1. Development of DMF–PCR Platforms
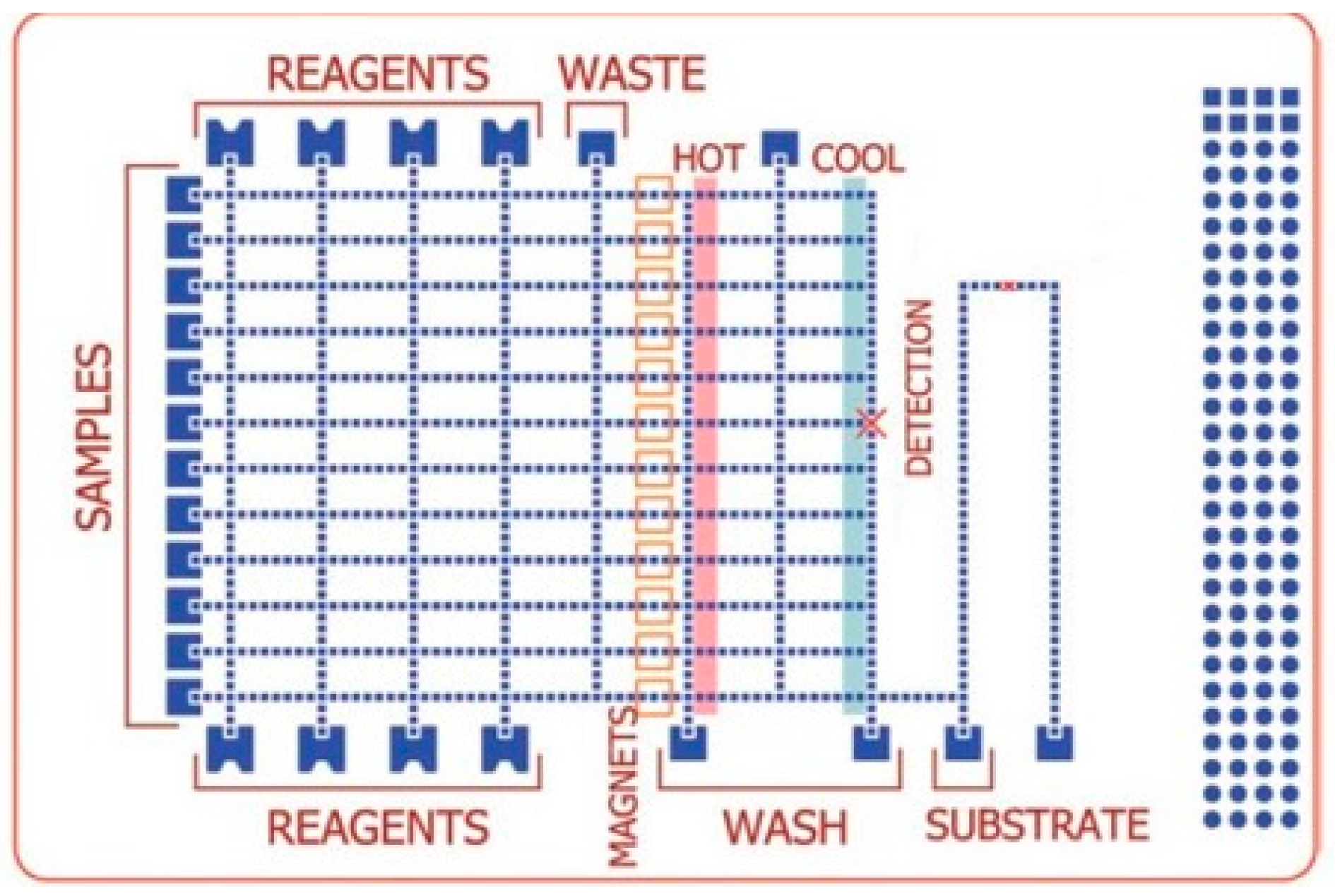
DMF–PCR Platform Validation
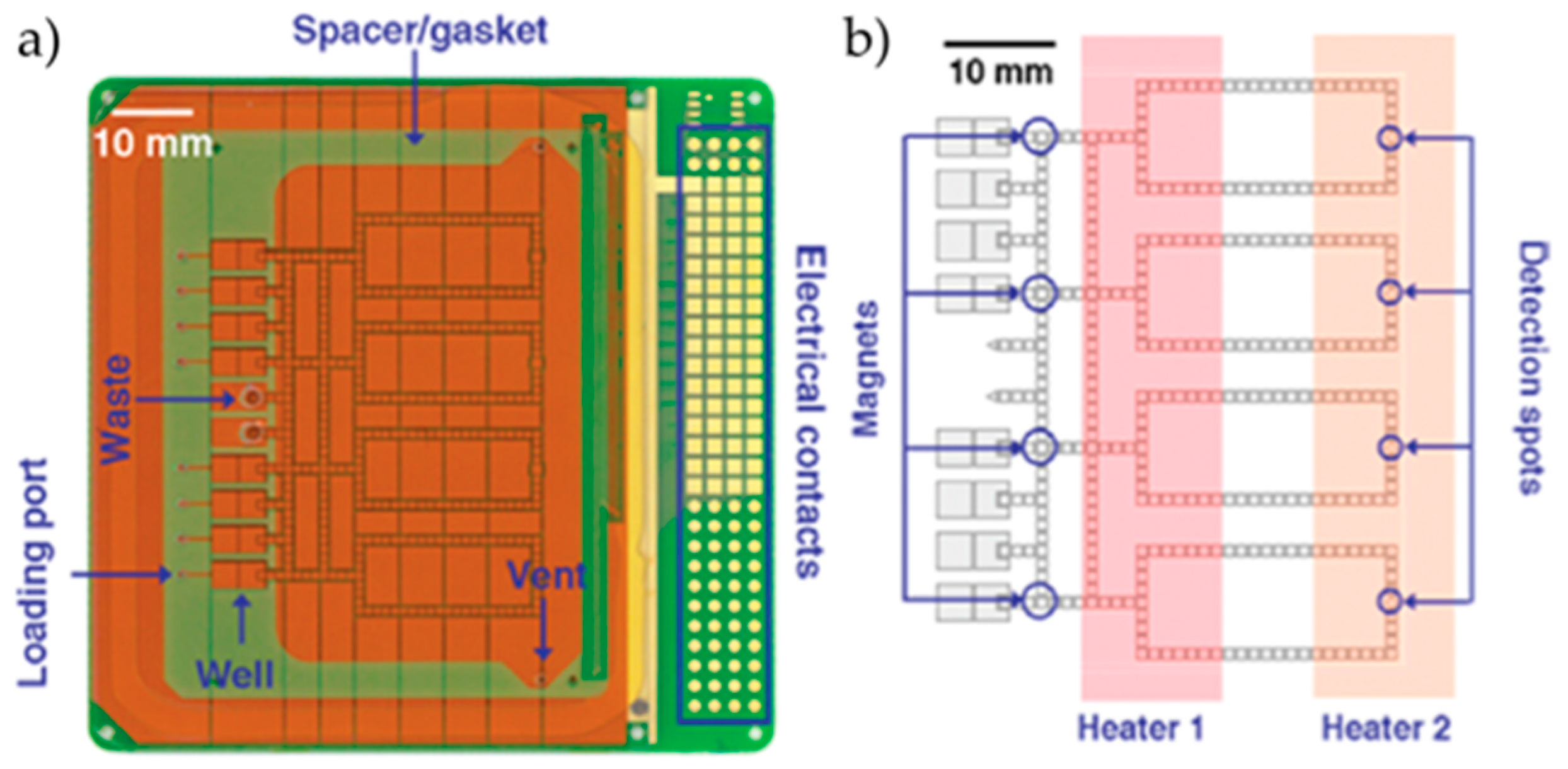
2.2. DMF—Conventional Microfluidics Hybrid Devices
2.3. DMF for Isothermal Nucleic Acid Amplification
3. Alternatives to EWOD for DMF-Assisted Nucleic Acid Amplification
4. Future Prospects
Acknowledgments
Conflicts of Interest
References
- Jebrail, M.J.; Wheeler, A.R. Let’s get digital: Digitizing chemical biology with microfluidics. Curr. Opin. Chem. Biol. 2010, 14, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Ng, A.H.C.; Fobel, R.; Wheeler, A.R. Digital Microfluidics. Annu. Rev. Anal. Chem. 2012, 5, 413–440. [Google Scholar] [CrossRef] [PubMed]
- Pesant, J.; Hareng, M.; Mourey, B.; Perbet, J. Electrodes for a Device Operating by Electrically Controlled Fluid Displacement. U.S. Patent US4569575 A, 11 February 1986. [Google Scholar]
- Pollack, M.G.; Fair, R.B.; Shenderov, A.D. Electrowetting-based actuation of liquid droplets for microfluidic applications. Appl. Phys. Lett. 2000, 77, 1725–1726. [Google Scholar] [CrossRef]
- Lee, J.; Moon, H.; Fowler, J.; Schoellhammer, T.; Kim, C.-J. Electrowetting and electrowetting-on-dielectric for microscale liquid handling. Sens. Actuators A Phys. 2002, 95, 259–268. [Google Scholar] [CrossRef]
- Berthier, J. Micro-Drops and Digital Microfluidics, 2nd ed.; Elsevier Inc.: Waltham, MA, USA, 2013. [Google Scholar]
- Millington, D.S.; Sista, R.; Eckhardt, A.; Rouse, J.; Bali, D.; Goldberg, R.; Cotten, M.; Buckley, R.; Pamula, V. Digital Microfluidics: A Future Technology in the Newborn Screening Laboratory? Semin. Perinatol. 2010, 34, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.B. Digital microfluidics: Is a true lab-on-a-chip possible? Microfluid. Nanofluid. 2007, 3, 245–281. [Google Scholar] [CrossRef]
- Sista, R.; Hua, Z.; Thwar, P.; Sudarsan, A.; Srinivasan, V.; Eckhardt, A.; Pollack, M.; Pamula, V. Development of a digital microfluidic platform for point of care testing. Lab Chip 2008, 8, 2091–2104. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, M.; Freire, S.L.S.; Yang, H.; Wheeler, A.R. All-terrain droplet actuation. Lab Chip 2008, 8, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.C.; Kim, C.-J. “Cj” Droplet Actuation by Electrowetting-on-Dielectric (EWOD): A Review. J. Adhes. Sci. Technol. 2012, 26, 1747–1771. [Google Scholar]
- Nolan, T.; Bustin, S. PCR Technology: Current Innovations, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Ahrberg, C.D.; Manz, A.; Chung, B.G. Polymerase chain reaction in microfluidic devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Pinto, J.; Vinhas, R.; Calmeiro, T.; Martins, R.; Fortunato, E.; Baptista, P.V. Quantitative real-time monitoring of RCA amplification of cancer biomarkers mediated by a flexible ion sensitive platform. Biosens. Bioelectron. 2017, 91, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Branquinho, R.; Pinto, J.V.; Wojcik, P.J.; Martins, R.; Fortunato, E.; Baptista, P.V. Ion sensing (EIS) real-time quantitative monitorization of isothermal DNA amplification. Biosens. Bioelectron. 2014, 52, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Bernacka-Wojcik, I.; Águas, H.; Carlos, F.F.; Lopes, P.; Wojcik, P.J.; Costa, M.N.; Veigas, B.; Igreja, R.; Fortunato, E.; Baptista, P.V.; et al. Single nucleotide polymorphism detection using gold nanoprobes and bio-microfluidic platform with embedded microlenses. Biotechnol. Bioeng. 2015, 112, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Lee, G.-B.; Huang, F.-C.; Chen, Y.-Y.; Lin, J.-L. Integrated polymerase chain reaction chips utilizing digital microfluidics. Biomed. Microdevices 2006, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Tsaloglou, M.; Parry-jones, L.; Jacobs, A.; Watson, R.; Turner, C.; Amos, R.; Hadwen, B.; Buse, J.; et al. Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab Chip 2015, 15, 3065–3075. [Google Scholar] [CrossRef] [PubMed]
- Kühnemund, M.; Witters, D.; Nilsson, M.; Lammertyn, J. Circle-to-circle amplification on a digital microfluidic chip for amplified single molecule detection. Lab Chip 2014, 14, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Rouse, J.L.; Eckhardt, A.E.; Srinivasan, V.; Pamula, V.K.; Schell, W.A.; Benton, J.L.; Mitchell, T.G.; Pollack, M.G. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 2010, 82, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Jebrail, M.J.; Renzi, R.F.; Sinha, A.; Van De Vreugde, J.; Gondhalekar, C.; Ambriz, C.; Meagher, R.J.; Branda, S.S. A solvent replenishment solution for managing evaporation of biochemical reactions in air-matrix digital microfluidics devices. Lab Chip 2015, 15, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Norian, H.S.; Shepard, K.; Kymissis, J.; Field, R. An integrated CMOS quantitative-polymerase-chain-reaction lab-on-chip for point-of-care diagnostics. Lab Chip 2014, 14, 4076–4084. [Google Scholar] [CrossRef] [PubMed]
- Rival, A.; Jary, D.; Delattre, C.; Fouillet, Y.; Castellan, G.; Bellemin-Comte, A.; Gidrol, X. An EWOD-based microfluidic chip for single-cell isolation, mRNA purification and subsequent multiplex qPCR. Lab Chip 2014, 14, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Fouillet, Y.; Jary, D.; Brachet, A.; Berthier, J.; Blervaque, R.; Davous, L.; Roux, J.; Achard, J.; Peponnet, C. EWOD digital microfluidics for Lab on a Chip. In Proceedings of the ASME 4th International Conference on Nanochannels, Microchannels and Minichannels, Limerick, Ireland, 19–21 June 2006; ASME Press: Limerick, Ireland, 2006; pp. 1255–1264. [Google Scholar]
- Berthier, J.; Mourier, V.; Sarrut, N.; Jary, D.; Fouillet, Y.; Pouteau, P.; Caillat, P.; Peponnet, C. Some examples of micro-devices for biotechnology developed at the Department of Technologies for Life Sciences and Healthcare of the LETI. Int. J. Nanotechnol. 2010, 7, 802–818. [Google Scholar] [CrossRef]
- Wulff-Burchfield, E.; Schell, W.A.; Eckhardt, A.E.; Pollack, M.G.; Hua, Z.; Rouse, J.L.; Pamula, V.K.; Srinivasan, V.; Benton, J.L.; Alexander, B.D.; et al. Microfluidic platform versus conventional real-time polymerase chain reaction for the detection of Mycoplasma pneumoniae in respiratory specimens. Diagn. Microbiol. Infect. Dis. 2010, 67, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Schell, W.A.; Johnson, M.D.; Alexander, B.D.; Perfect, J.R.; Smith, P.B.; Benjamin, D.K.; Mitchell, T.G.; Benton, J.L.; Poore, M.; Rouse, J.L.; et al. Evaluation of a digital microfluidic real-time PCR platform to detect DNA of Candida albicans in blood. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2237–2245. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, M.; Watson, M.W.L.; Wheeler, A.R. Hybrid microfluidics: A digital-to-channel interface for in-line sample processing and chemical separations. Lab Chip 2009, 9, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.W.L.; Jebrail, M.J.; Wheeler, A.R. Multilayer hybrid microfluidics: A digital-to-channel interface for sample processing and separations. Anal. Chem. 2010, 82, 6680–6686. [Google Scholar] [CrossRef] [PubMed]
- Ugsornrat, K.; Afzulpurkar, N.V.; Wisitsoraat, A.; Tuantranont, A. Design, Simulation, and Experimental Study of a Droplet-Based PCR by EWOD. Sens. Mater. 2010, 22, 271–284. [Google Scholar]
- Kim, H.; Jebrail, M.J.; Sinha, A.; Bent, Z.W.; Solberg, O.D.; Williams, K.P.; Langevin, S.A.; Renzi, R.F.; Van De Vreugde, J.L.; Meagher, R.J.; et al. A Microfluidic DNA Library Preparation Platform for Next-Generation Sequencing. PLoS ONE 2013, 8, e68988. [Google Scholar]
- Welch, E.R.F.; Lin, Y.Y.; Madison, A.; Fair, R.B. Picoliter DNA sequencing chemistry on an electrowetting-based digital microfluidic platform. Biotechnol. J. 2011, 6, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Boles, D.J.; Benton, J.L.; Siew, G.J.; Levy, M.H.; Thwar, P.K.; Sandahl, M.A.; Rouse, J.L.; Perkins, L.C.; Sudarsan, A.P.; Pamula, V.; et al. Droplet-Based Pyrosequencing Using Digital Microfluidics. Anal. Chem. 2011, 83, 8439–8447. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.H.; Jin Shin, D.; Zhang, Y.; Wang, T.H. Topography-assisted electromagnetic platform for blood-to-PCR in a droplet. Biosens. Bioelectron. 2013, 50, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, T.H. Full-range magnetic manipulation of droplets via surface energy traps enables complex bioassays. Adv. Mater. 2013, 25, 2903–2908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Park, S.; Liu, K.; Tsuan, J.; Yang, S.; Wang, T.-H. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip 2011, 11, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Pipper, J.; Inoue, M.; Ng, L.F.-P.; Neuzil, P.; Zhang, Y.; Novak, L. Catching bird flu in a droplet. Nat. Med. 2007, 13, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Reboud, J.; Bourquin, Y.; Wilson, R.; Pall, G.S.; Jiwaji, M.; Pitt, A.R.; Graham, A.; Waters, A.P.; Cooper, J.M. Shaping acoustic fields as a toolset for microfluidic manipulations in diagnostic technologies. Proc. Natl. Acad. Sci. USA 2012, 109, 15162–15167. [Google Scholar] [CrossRef] [PubMed]
- Guttenberg, Z.; Müller, H.; Habermüller, H.; Geisbauer, A.; Pipper, J.; Felbel, J.; Kielpinski, M.; Scriba, J.; Wixforth, A. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab Chip 2005, 5, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.N.; Valley, J.K.; Wang, Y.; Wu, M.C. Distributed Circuit Model for Multi-Color Microfluidic Device. J. Lightware Technol. 2015, 33, 3486–3493. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci. Rep. 2015, 5, 13174. [Google Scholar] [CrossRef] [PubMed]
- Coelho, B.J.; Veigas, B.; Águas, H.; Fortunato, E.; Martins, R.; Baptista, P.V.; Igreja, R. Digital Microfluidics Devices for Nucleic Acid Amplification: Redifining the Paradigm of Molecular Diagnostics. In Proceedings of the Materiais 2017: VIII International Symposium on Materials, Aveiro, Portugal, 9–12 April 2017. [Google Scholar]
- Marques, A.C.; Santos, L.; Costa, M.N.; Dantas, J.M.; Duarte, P.; Goncalves, A.; Martins, R.; Salgueiro, C.A.; Fortunato, E. Office Paper Platform for Bioelectrochromic Detection of Electrochemically Active Bacteria using Tungsten Trioxide Nanoprobes. Sci. Rep. 2015, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.N.; Veigas, B.; Jacob, J.M.; Santos, D.S.; Gomes, J.; Baptista, P.V.; Martins, R.; Inácio, J.; Fortunato, E. A low cost, safe, disposable, rapid and self-sustainable paper-based platform for diagnostic testing: Lab-on-paper. Nanotechnology 2014, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Jacob, J.M.; Costa, M.N.; Santos, D.S.; Viveiros, M.; Inácio, J.; Martins, R.; Barquinha, P.; Fortunato, E.; Baptista, P.V. Gold on paper-paper platform for Au-nanoprobe TB detection. Lab Chip 2012, 12, 4802–4808. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Ferreira, I.; Fortunato, E. Electronics with and on paper. Phys. Status Solidi Rapid Res. Lett. 2011, 5, 332–335. [Google Scholar] [CrossRef]
- Coelho, B.J. A Digital Microfluidics Platform for Loop-Mediated Isothermal Amplification of DNA. Master’s Thesis, Faculdade de Ciências e Tecnologia—Universidade Nova de Lisboa, Caparica, Portugal, 2016. [Google Scholar]
- Bernacka-Wojcik, I.; Senadeera, R.; Wojcik, P.J.; Silva, L.B.; Doria, G.; Baptista, P.; Aguas, H.; Fortunato, E.; Martins, R. Inkjet printed and “doctor blade” TiO2 photodetectors for DNA biosensors. Biosens. Bioelectron. 2010, 25, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Fobel, R.; Kirby, A.E.; Ng, A.H.C.; Farnood, R.R.; Wheeler, A.R. Paper microfluidics goes digital. Adv. Mater. 2014, 26, 2838–2843. [Google Scholar] [CrossRef] [PubMed]
- Matos, D. Digital Microfluidics on Paper. Master’s Thesis, Faculdade de Ciências e Tecnologia—Universidade Nova de Lisboa, Caparica, Portugal, 2014. [Google Scholar]

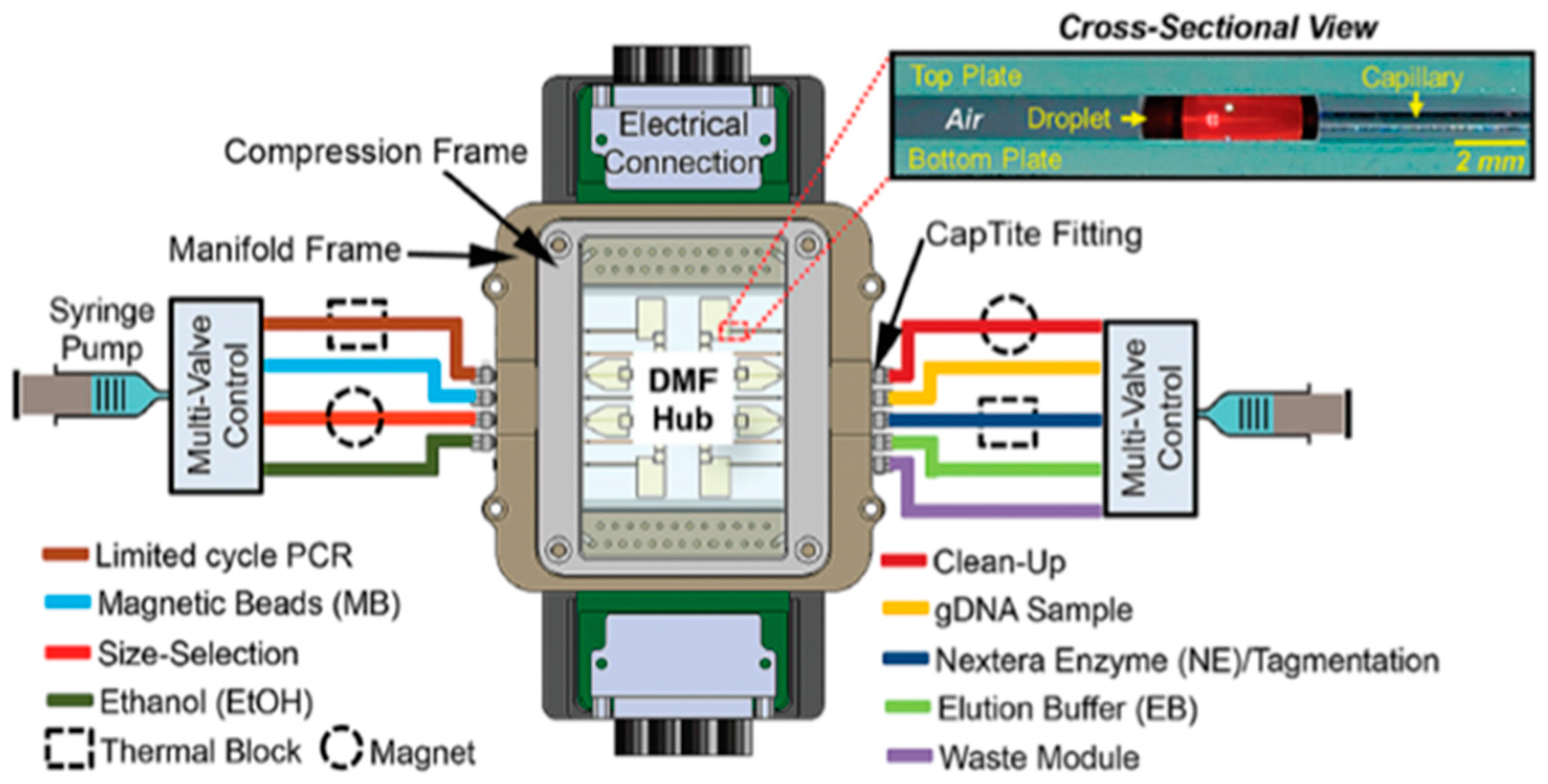
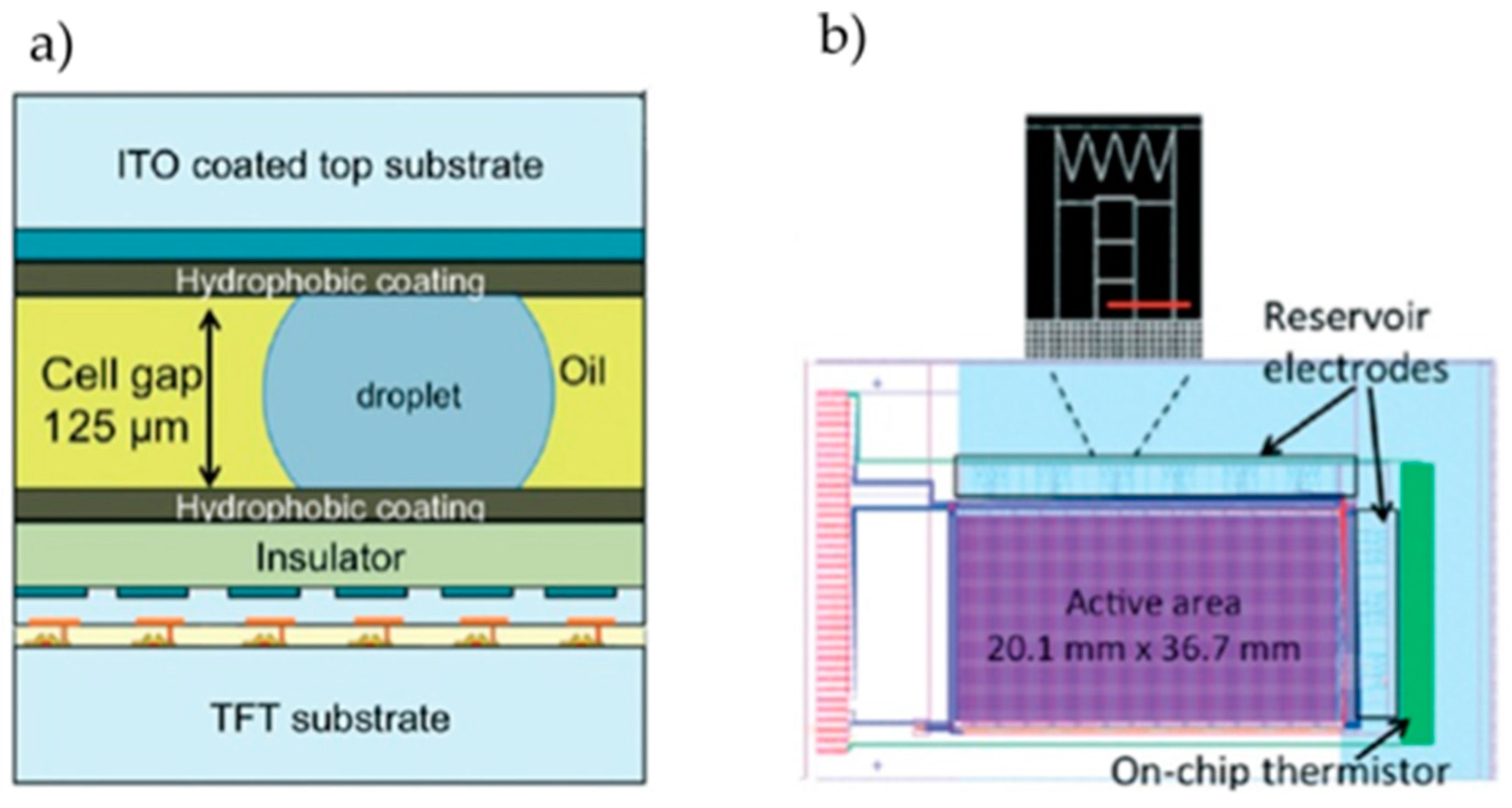

| Application | Reaction Volume | Actuation Voltage | Dielectric Material | Hydrophobic Material | Electrode Material | Substrate Material | Filler | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| DMF–PCR platforms | Dengue II virus detection | 1.46 µL | 12 VRMS (3 kHz) | Si3N4 | Teflon® AF | Au | Glass | Silicone oil | [17] |
| SNP genotyping | 64 nL | 60 VRMS (3 kHz) | Si3N4 | Teflon® AF | Au | Glass | Silicone oil | [25] | |
| POC testing, MRSA, S. aureus and C. albicans detection | 600 nL | - | Parylene C | Teflon® AF | Cr | PCB | Hexadecane | [9] | |
| MRSA, S. aureus, M. pneumoniae and C. albicans detection | Variable | - | - | - | - | PCB | Hexadecane/silicone oil | [21] | |
| S. aureus detection | 1.2 nL | 70 V–250 V | Parylene C | Teflon® AF | - | - | n-dodecane | [23] | |
| Cell genetic expression analysis | Variable | 48 V (3 kHz) | Si3N4 | SiOC | Ti/AlCu | Silicon wafer | Silicone oil | [24] | |
| Bacteriophage M13mp18 detection | 1.5 µL | 80–100 VRMS (18 kHz) | Solder mask | Teflon® AF | Cu/Ni/Au | PCB | Air | [22] | |
| Validation of DMF–PCR platforms | C. albicans detection on human blood | Variable | - | - | - | - | PCB | Hexadecane/silicone oil | [27] |
| M. pneumoniae detection on human nasopharyngeal wash | Variable | - | - | - | - | PCB | Hexadecane/silicone oil | [28] | |
| Hybrid platforms | DNA amplification (only the primers are described) | 25 µL | - | SiO2 | Teflon® AF | Cr/Au | Glass | Silicone oil | [31] |
| Pre-DNA sequencing | 2.8 µL | 80–90 VRMS (15 kHz) | Parylene C | Teflon® AF | ITO | Glass | Air | [32] | |
| DMF platforms for isothermal amplification | CTX-M gene detection in E. coli bacteria | Minimum 270 nL | 20 V | Al2O3 | Cytop® | ITO | Glass | n-dodecane | [18] |
| P. aeruginosa detection | 5 µL | 120 VAC (1 kHz) | Parylene C | Teflon® AF | Cr | Glass | Vapor-Lock oil | [19] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, B.; Veigas, B.; Fortunato, E.; Martins, R.; Águas, H.; Igreja, R.; Baptista, P.V. Digital Microfluidics for Nucleic Acid Amplification. Sensors 2017, 17, 1495. https://doi.org/10.3390/s17071495
Coelho B, Veigas B, Fortunato E, Martins R, Águas H, Igreja R, Baptista PV. Digital Microfluidics for Nucleic Acid Amplification. Sensors. 2017; 17(7):1495. https://doi.org/10.3390/s17071495
Chicago/Turabian StyleCoelho, Beatriz, Bruno Veigas, Elvira Fortunato, Rodrigo Martins, Hugo Águas, Rui Igreja, and Pedro V. Baptista. 2017. "Digital Microfluidics for Nucleic Acid Amplification" Sensors 17, no. 7: 1495. https://doi.org/10.3390/s17071495
APA StyleCoelho, B., Veigas, B., Fortunato, E., Martins, R., Águas, H., Igreja, R., & Baptista, P. V. (2017). Digital Microfluidics for Nucleic Acid Amplification. Sensors, 17(7), 1495. https://doi.org/10.3390/s17071495










