Revealing Nucleic Acid Mutations Using Förster Resonance Energy Transfer-Based Probes
Abstract
:1. Introduction
2. Förster Resonance Energy Transfer-Based Probes in Nucleic Acid Research and Molecular Diagnostics
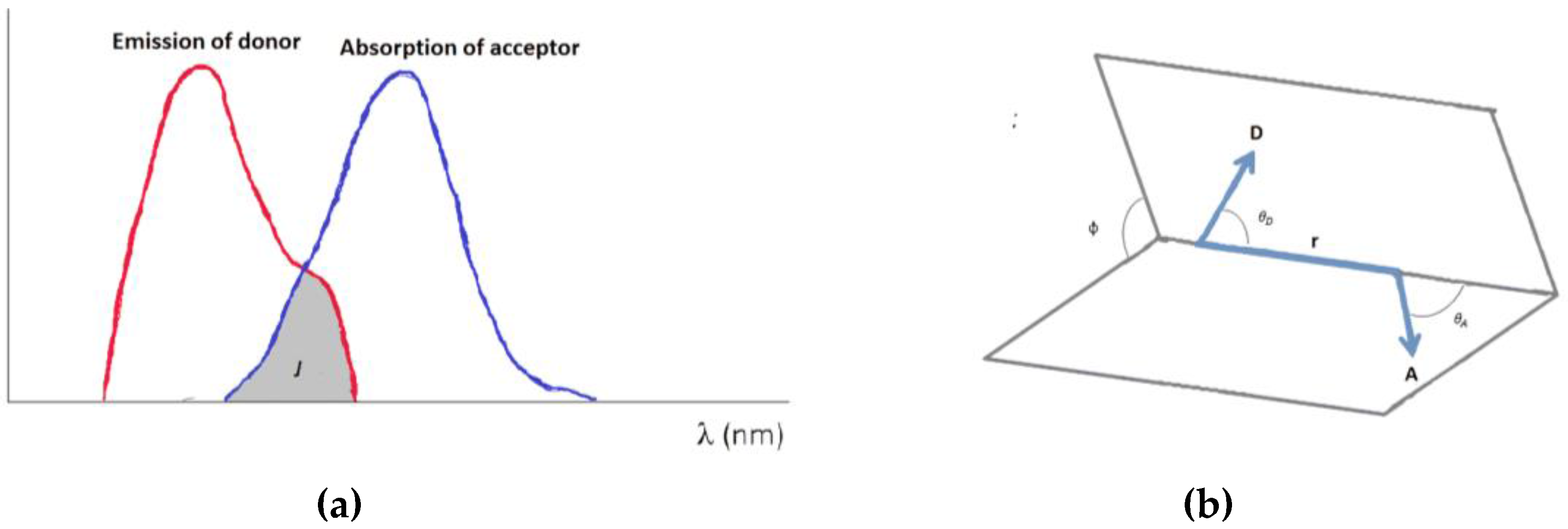
2.1. Theory of FRET
2.2. Challenges and Appeal of FRET Probes in Modern Nucleic Acid Research
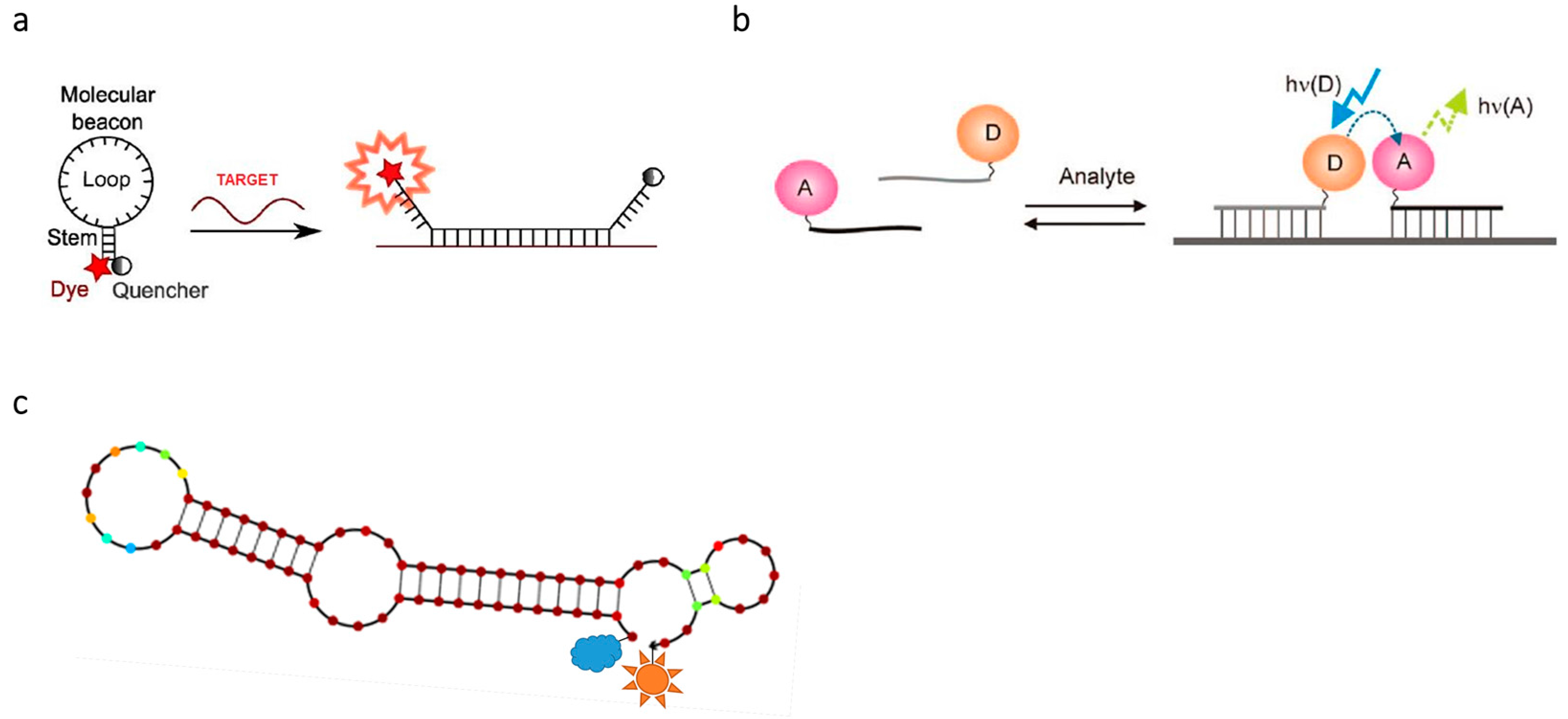
2.3. Design and Application of FRET Probes for Nucleic Acid Research
2.3.1. Design
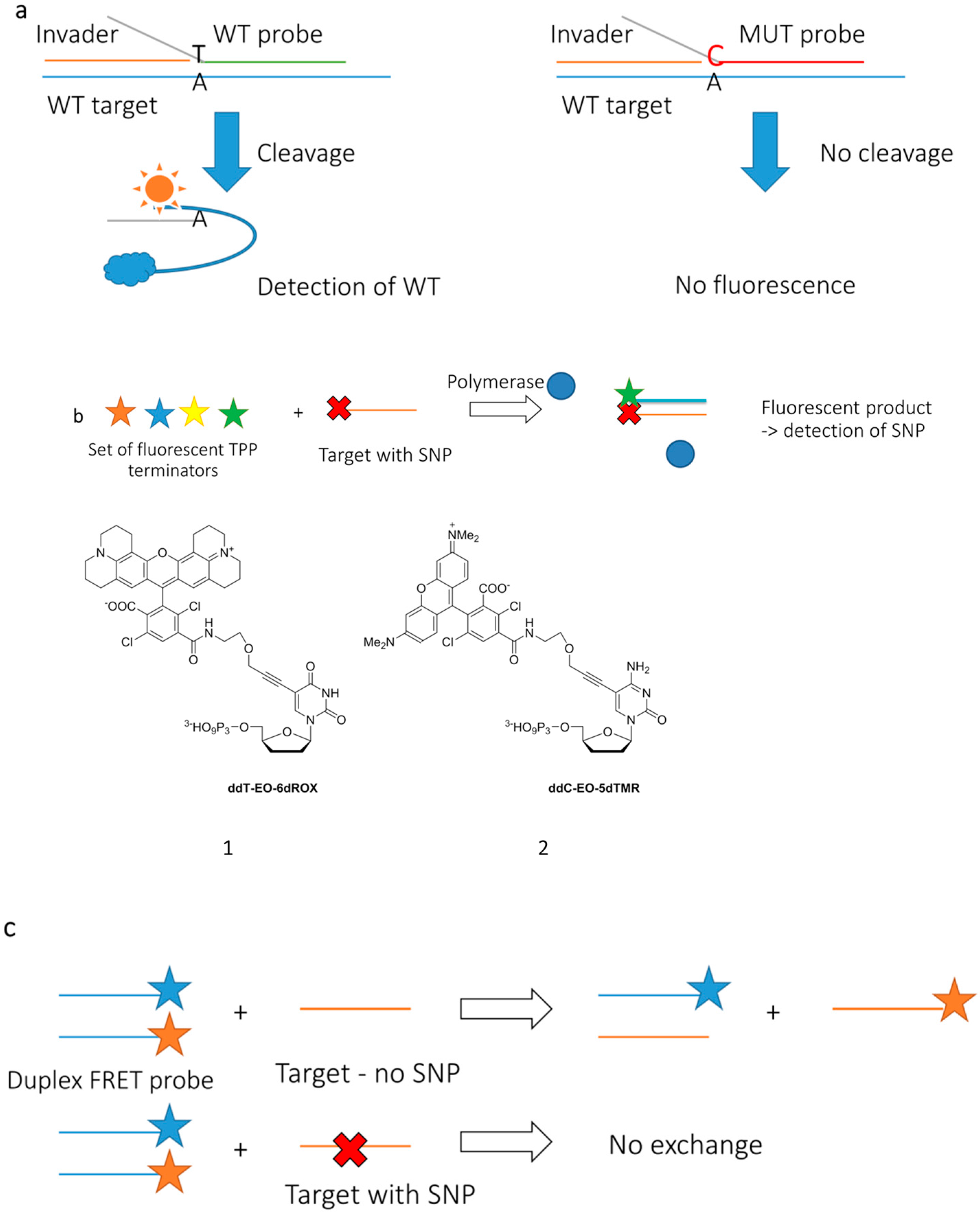
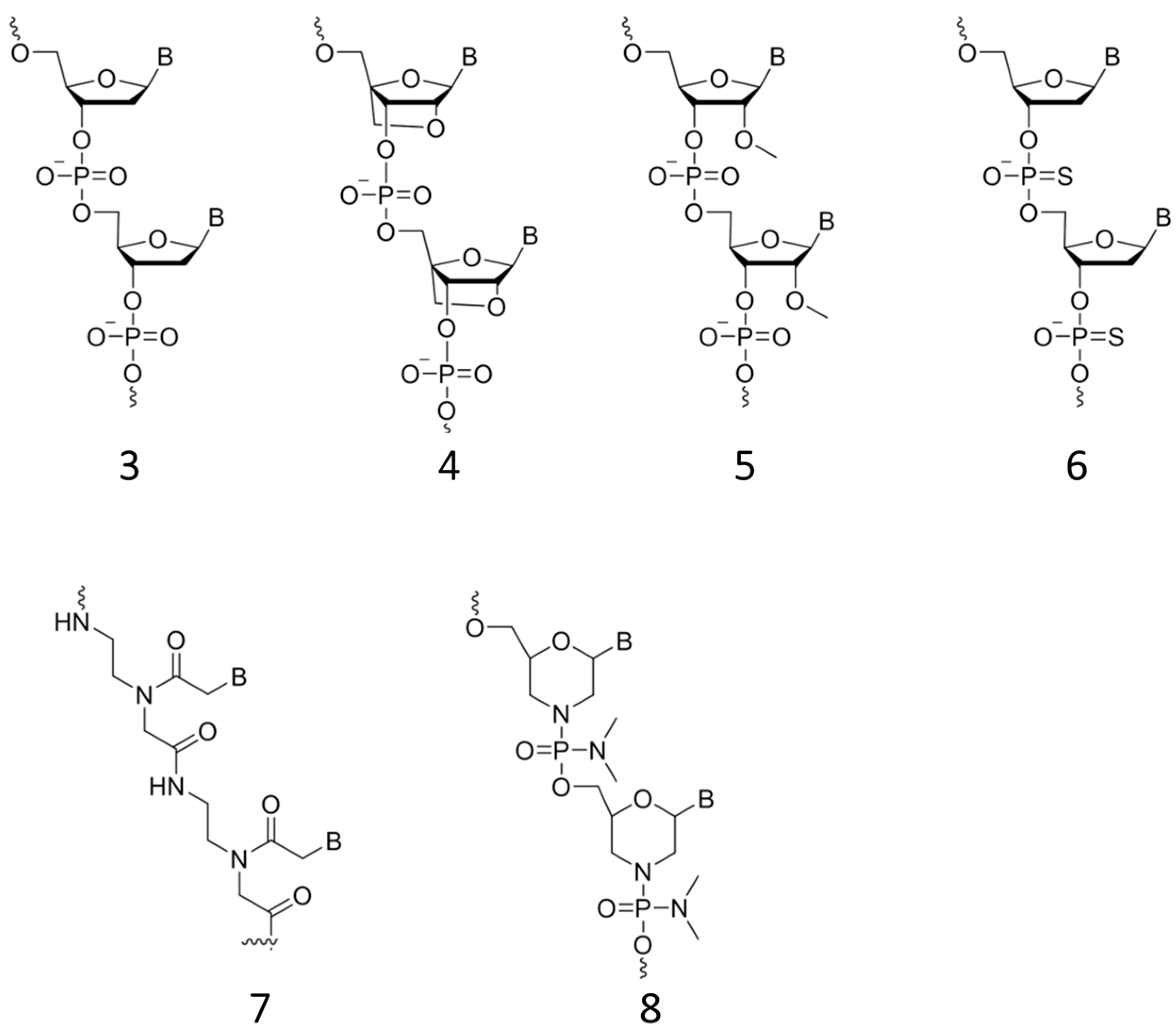
2.3.2. Approaching Challenges of SNP Detection by Modern FRET Probes: Chemistry of Backbone Modifications and Advanced Fluorescent Dyes
2.3.3. Up-to-Date Applications of FRET Probes: Summary and Specific Cases
2.4. Computational Strategies Help Designing Efficient FRET Probes
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Blackburn, G.M.; Gait, M.J.; Loakes, D.; Williams, D.M. Nucleic Acids in Chemistry and Biology, 3rd ed.; RCS: Great Britain, UK, 2006; pp. 20–67. [Google Scholar]
- Miotke, L.; Barducci, M.C.; Astakhova, K. Novel Signal-enhancing approaches for optical detection of nucleic acids—Going beyond target amplification. Chemosensors 2015, 3, 224–240. [Google Scholar] [CrossRef]
- Kraeva, R.I.; Krastev, D.B.; Roguev, A.; Ivanova, A.; Nedelcheva-Veleva, M.N.; Stoynov, S.S. Stability of mRNA/DNA and DNA/DNA duplexes affects mRNA transcription. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.X.; Zhang, D.Y.; Seelig, G. Conditionally fluorescent molecular probes for detecting single base changes in double-stranded DNA. Nat. Chem. 2013, 5, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.R.; Znosko, B.M. Positional and neighboring base pair effects on the thermodynamic stability of RNA single mismatches. Biochemistry 2010, 49, 8669–8679. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.E.; Kennelly, W.J.; Tsay, M.J.; Tuin, A.; Swenson, L.; Lee, H.R.; Morosyuk, S.; Hicks, D.A.; SantaLucia, J.J. Thermodynamic contributions of single internal rA·dA, rC·dC, rG·dG and rU·dT mismatches in RNA/DNA duplexes. Nucleic Acids Res. 2011, 39, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, K. Sensing of SNP in HIV-1 genome using fluorescent oligonucleotide probes. Front. Clin. Drug Res. HIV 2014. [Google Scholar] [CrossRef]
- Schlatterer, J.C.; Martin, J.S.; Laederach, A.; Brenowitz, M. Mapping the Kinetic Barriers of a Large RNA Molecule’s Folding Landscape. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, G.; Lee, R.A.; Jayaram, B.; Beveridge, D.L. A statistical thermodynamic model for investigating the stability of DNA sequences from oligonucleotides to genomes. J. Biophys. 2014, 106, 2465–2473. [Google Scholar] [CrossRef] [PubMed]
- Fontenete, S.; Leite, M.; Guimarães, N.; Madureira, P.; Ferreira, R.M.; Figueiredo, C.; Wengel, J.; Azevedo, N.F. Towards fluorescence in vivo hybridization (FIVH) detection of H. pylori in gastric mucosa using advanced LNA probes. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Brand, L. Resonance Energy Transfer: Methods and Application. Anal. Biochem. 1994, 218, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stryer, L. Fluorescence Energy Transfer as a Spectroscopic Ruler. Ann. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef] [PubMed]
- Clegg, R.M. Fluorescence resonance energy transfer. Anal. Biotechnol. 1995, 6, 103–110. [Google Scholar] [CrossRef]
- Morrison, L.E.; Stols, L.M. Sensitive Fluorescence-Based Thermodynamic and Kinetic Measurements of DNA Hybridization in Solution. Biochemistry 1993, 32, 3095–3104. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P. The concept of lambda-ratiometry in Fluorescence Sensing and Imaging. J. Fluoresc. 2010, 1099–1128. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Vogel, S.S.; van der Meer, B.W.; Blank, P.S. Estimating the distance separating fluorescent protein FRET pairs. Methods 2014, 66, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Losa, A.; Curutchet, C.; Krueger, B.P.; Hartsell, L.R.; Mennucci, B. Fretting about FRET: Failure of the Ideal Dipole Approximation. Biophys. J. 2009, 96, 4779–4788. [Google Scholar] [CrossRef] [PubMed]
- Didenko, V.V. DNA Probes Using Fluorescence Resonance Energy Transfer (FRET): Designs and Applications. Biotechniques 2001, 31, 1106–1121. [Google Scholar] [PubMed]
- Lee, S.P.; Porter, D.; Chirikjian, J.G.; Knutson, J.R.; Han, M.K. A fluorometric assay for DNA cleavage reactions characterized with BamHI restriction endonuclease. Anal. Biochem. 1994, 220, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Censullo, M.L.; Kim, H.G.; Knutson, J.R.; Han, M.K. Characterization of endonucleolytic activity of HIV-1 in-tegrase using a fluorogenic substrate. Anal. Biochem. 1995, 227, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.G.; Nordlund, T.M. Sequence dependence of energy transfer in DNA oligonucleotides. Biophys. J. 2000, 78, 1042–1058. [Google Scholar] [CrossRef]
- Kawahara, S.; Uchimaru, T.; Murata, S. Efficiency enhancement of long-range energy transfer by sequential multistep FRET using fluorescence labeled DNA. Nucleic Acids Symp. Ser. 1999, 42, 241–242. [Google Scholar] [CrossRef]
- Eftink, M.R. Fluorescence quenching: Theory and applications. In Topics in Fluorescence Spectroscopy; Lakowicz, J.R., Ed.; Plenum Press: New York, NY, USA, 1991; pp. 53–126. [Google Scholar]
- Rudi, K.; Zimonja, M.; Hannevik, S.E.; Drømtorp, S.M. Multiplex real-time single nucleotide polymorphism detection and quantification by quencher extension. Biotechniques 2006, 40, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Socher, E.; Knoll, A.; Seitz, O. Dual fluorophore PNA FIT-probes—Extremely responsive and bright hybridization probes for the sensitive detection of DNA and RNA. Org. Biomol. Chem. 2012, 10, 7363–7371. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J.; Hohng, S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Pinard, R.; de Winter, A.; Saekis, G.J.; Gerstein, M.B.; Tartaro, K.R.; Plant, R.N.; Egholm, M.; Rothberg, J.M.; Leamon, J.H. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genom. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Daudé, F.; Welsh, J.; Vogt, T.; McClelland, M. DNA rehybridization during PCR: The “Cot effect” and its consequences. Nucleic Acids Res. 1996, 24, 2080–2086. [Google Scholar] [CrossRef]
- Gupta, G.D.; Kumar, V. Identification of nucleic acid binding sites on translin-associated factor X (TRAX) protein. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Ronander, E.; Bengtsson, D.C.; Joergensen, L.; Jensen, A.T.; Arnot, D.E. Analysis of single-cell gene transcription by RNA fluorescent in situ hybridization (FISH). J. Vis. Exp. 2012, 68. [Google Scholar] [CrossRef] [PubMed]
- Sabina, J.; Leamon, J.H. Bias in whole genome amplification: causes and considerations. Methods Mol. Biol. 2015, 1347, 15–41. [Google Scholar] [PubMed]
- Johnson, K.A. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochem. Biophys. Acta 2010, 1804, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, C.J. Imaging genome abnormalities in cancer research. Cell Chromosome 2004, 3. [Google Scholar] [CrossRef] [PubMed]
- Raulf, A.; Spahn, C.; Zessin, P.J.M.; Finan, K.; Bernhardt, S.; Heckel, A.; Heilemann, M. Click chemistry facilitates direct labeling and super-resolution imaging of nucleic acids and proteins. RSC Adv. 2014, 4, 30462–30466. [Google Scholar] [CrossRef] [PubMed]
- Kinjo, M.; Rigler, R. Ultrasensitive hybridization analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 1995, 23, 1795–1799. [Google Scholar] [CrossRef]
- Bolze, F.; Lux, J.; Peña, E.; Heinlein, M.; Wong, M.S.; Nicoud, J.F. Two-photon dyes and nucleic acid detection. Nonlinear Opt. Quantum Opt. 2012, 45, 29–39. [Google Scholar]
- Huang, B.; Babcock, H.; Zhuang, X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell 2010, 143, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Persson, F.; Bingen, P.; Staudt, T.; Engelhardt, J.; Tegenfeldt, J.O.; Hell, S.W. Fluorescence nanoscopy of single DNA molecules by using Stimulated Emission Depletion (STED). Angew. Chem. Int. Ed. Engl. 2011, 50, 5581–5583. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.D.; Kenyon, J.C.; Symmons, M.F.; Lever, A.M. Characterizing 3D RNA structure by single molecule FRET. Methods 2016. [Google Scholar] [CrossRef] [PubMed]
- Hardin, J.W.; Warnasooriya, C.; Kondo, Y.; Nagai, K.; Rueda, D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 2015, 43, 10963–10974. [Google Scholar] [CrossRef] [PubMed]
- Krainer, G.; Hartmann, A.; Schlierf, M. farFRET: Extending the Range in Single-Molecule FRET Experiments beyond 10 nm. Nano Lett. 2015, 15, 5826–5829. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, D.; Jenei, A.; Nagy, P.; Vereb, G.; Szöllősi, J. Understanding FRET as a research tool for cellular studies. Int. J. Mol. Sci. 2015, 16, 6718–6756. [Google Scholar] [CrossRef] [PubMed]
- Sustarsic, M.; Kapanidis, A.N. Taking the ruler to the jungle: Single-Molecule FRET for understanding biomolecular structure and dynamics in live cells. Curr. Opin. Struct. Biol. 2015, 34, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Obika, S.; Morio, K.; Hari, Y.; Imanishi, T. Facile synthesis and conformation of 3′-O,4′-C-methyleneribonucleosides. Chem. Commun. 1999. [Google Scholar] [CrossRef]
- Lindegaard, D.; Madsen, A.S.; Astakhova, I.V.; Malakhov, A.D.; Babu, B.R.; Korshun, V.A.; Wengel, J. Pyrene-perylene as a FRET pair coupled to the N2′-functionality of 2′-amino-LNA. Bioorg. Med. Chem. 2008, 16, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Myznikova, A.; Samokhina, E.; Astakhova, I.K. Rapid genotyping using pyrene-perylene locked nucleic acid complexes. Artif. DNA PNA XNA 2013, 4, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Dias, N.; Stein, C.A. Antisense Oligonucleotides: Basic Concepts and Mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluresce up hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Chen, H.; Draney, D.R.; Volcheck, W.; Schutz-Geschwender, A.; Olive, D.M. A nonfluorescent, broad-range quencher dye for Förster resonance energy transfer assays. Anal. Biochem. 2009, 388, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. An Elegant Biosensor Molecular Beacon Probe: Challenges and Recent Solutions. Scientifica 2012. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ju, J.; Turro, N.J. Fluorescent hybridization probes for nucleic acid detection. Anal. Bioanal. Chem. 2012, 402, 3115–3125. [Google Scholar] [CrossRef] [PubMed]
- Taygi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 2009, 6, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Martí, A.A.; Li, X.; Jockusch, S.; Stevens, N.; Li, Z.; Raveendra, B.; Kalachikov, S.; Morozova, I.; Russo, J.J.; Akins, D.L.; et al. Design and characterization of two-dye and three-dye binary fluorescent probes for mRNA detection. Tetrahedron 2007, 63, 3591–3600. [Google Scholar] [CrossRef] [PubMed]
- Dahan, L.; Huang, L.; Kedmi, R.; Behlke, M.A.; Peer, D. SNP Detection in mRNA in Living Cells Using Allele Specific FRET probe. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Savinov, A.; Perez, C.F.; Block, S.M. Single-molecule Studies of Riboswitch Folding. Biochem. Biophys. Acta 2014, 1839, 1030–1045. [Google Scholar] [CrossRef] [PubMed]
- Grimm, J.B.; English, B.P.; Chen, J.; Slaughter, J.P.; Zhang, Z.; Revyakin, A.; Patel, R.; Macklin, J.J.; Normanno, D.; Singer, R.H.; et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Rockey, W.M.; Hernandez, F.J.; Huang, S.Y.; Cao, S.; Howell, C.A.; Thomas, G.S.; Liu, X.Y.; Lapteva, N.; Spencer, D.M.; McNamara, J.O., II; et al. Truncation of an RNA Aptamer to Prostate-Specific Membrane Antigen Using Computational Structural Modeling. Nucleic Acid Ther. 2011, 21, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Boutorine, A.S.; Novopashina, D.S.; Krasheninina, O.A.; Nozeret, K.; Venyaminova, A.G. Fluorescent Probes for Nucleic Acid Visualization in Fixed and Live Cells. Molecules 2013, 18, 15357–15397. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Zhang, Y.; Cai, J.; Cai, W.; Gao, T. Aptamer-Based Fluorescent Biosensors. Curr. Med. Chem. 2011, 18, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M. The Invader assay for SNP genotyping. Mutat. Res. 2005, 573, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, B.B.; Lee, L.G.; Spurgeon, S.L.; Khan, S.H.; Menchen, S.M.; Heiner, C.R.; Chen, S.M. New dye-labeled terminators for improved DNA sequencing patterns. Nucl. Acid Res. 1997, 25, 4500–4504. [Google Scholar] [CrossRef]
- Didenko, V.V. Fluorescent energy transfer nucleic acid probes: Designs and Protocols. In Methods Molecular Biology; Humana Press: Totowa, NJ, USA, 2006. [Google Scholar]
- Morrison, L.E.; Halder, T.C.; Stols, L.M. Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal. Biochem. 1989, 183, 231–244. [Google Scholar] [CrossRef]
- Sixou, S.; Szoka, F.C.J.; Green, G.A.; Giusti, B.; Zon, G.; Chin, D.J. Intra-cellular oligonucleotide hybridization detected by fluorescence resonance energy transfer (FRET). Nucleic Acids Res. 1994, 22, 662–668. [Google Scholar] [CrossRef]
- Majlessi, M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Inoue, H.; Hayase, A.; Iwai, S.; Miura, K.; Ohtsuka, E. Synthesis and hybrdiation studies on two complementary nona(2′-O-methyl) ribonucleotides. Nucleic Acids Res. 1987, 15, 6131–6148. [Google Scholar] [CrossRef]
- Okabe, K.; Harada, Y.; Zhang, J.; Tadakuma, H.; Tani, T.; Funatsu, T. Real time monitoring of endogenous cytoplasmic mRNA using linear antisense 2′-O-methyl RNA probes in living cells. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Thomas, G.; Koller, E.; Southwell, A.L.; Hayden, M.R.; Seth, P.P. Biophysical and biological characterization of hairpin and molecular beacon RNase H Active Antisense Oligonucleotide. ACS Chem. Biol. 2015, 10, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Iannitti, T.; Morales-Medina, J.C.; Palmieri, B. Phosphorothioate oligonucleotides: Effectiveness and toxicity. Curr. Drug Targets 2014, 15, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.T.; Cheng, S.H.; Liu, C.P.; Souris, J.S.; Chen, C.T.; Mou, C.Y.; Lo, L.W. Recent Advances in Nanoparticle-Based Förster Resonance Energy Transfer for Biosensing, Molecular Imaging and Drug Release Profiling. Int. J. Mol. Sci. 2012, 13, 16598–16623. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, J.; Hong, Y. The morpholino molecular beacon for specific RNA visualization in vivo. Chem. Commun. 2016, 52, 3191–3194. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, H.; Wang, Z.; Feng, M.; Jin, H.; Wu, Y.; Zhang, L.; Zhang, L.; Tang, X. Sensitive Detection of Single-Nucleotide Mutation in the BRAF Mutation Site (V600E) of Human Melanoma Using Phosphate- Pyrene-Labeled DNA Probes. Anal. Chem. 2016, 88, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.N.; Seo, Y.J. A multiplex fluorophore molecular beacon: Detection of the target sequence using large Stokes shift and multiple emission signal properties. Chem. Commun. 2015, 51, 2939–2941. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, J.; Schorr, S.; Iqbal, A.; Wilson, T.J.; Lilley, D.M. Orientation of cyanine fluorophores terminally attached to DNA via long, flexible tethers. Biophys. J. 2011, 101, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- Ranjit, S.; Gurunathan, K.; Levitus, M. Photophysics of backbone fluorescent DNA modifications: Reducing uncertainties in FRET. J. Phys. Chem. B 2009, 113, 7861–7866. [Google Scholar] [CrossRef] [PubMed]
- Barroso, M.M. Quantum Dots in Cell Biology. J. Histochem. Cytochem. 2011, 59, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Algar, W.R. A long-wavelength quantum dot-concentric FRET configuration: Characterization and application in a multiplexed hybridization assay. Analyst 2016, 141, 3636–3647. [Google Scholar] [CrossRef] [PubMed]
- Hemmilä, I.; Laitala, V. Progress in Lanthanides as Luminescent Probes. J. Fluoresc. 2005, 15, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Qui, X.; Hildebrandt, N. Rapid and Multiplexed MicroRNA Diagnostic Assay Using Quantum Dot-Based Förster Resonance Energy Transfer. ACS Nano 2015, 9, 8449–8457. [Google Scholar]
- Jin, Z.; Geiβler, D.; Qiu, X.; Wegner, K.; Hildebrandt, N. A Rapid, Amplification-Free, and Sensitive Diagnostic Assay for Single-Step Multiplexed Fluorescence Detection of MicroRNA. Angew. Chem. Int. Ed. 2015, 54, 10024–10029. [Google Scholar] [CrossRef] [PubMed]
- Kashida, H.; Osawa, T.; Morimoto, K.; Kamiya, Y.; Asanuma, H. Molecular design of Cy3 derivative for highly sensitive in-stem molecular beacon and its application to the wash-free FISH. Bioorg. Med. Chem. 2015, 23, 1758–1762. [Google Scholar] [CrossRef] [PubMed]
- Häner, R.; Biner, S.M.; Langenegger, S.M.; Meng, T.; Malinovskii, V.L. A highly sensitive, excimer-controlled molecular beacon. Angew. Chem. Int. Ed. 2010, 49, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Beta, M.; Krishnakumar, S.; Elchuri, S.V.; Salim, B.; Narayanan, J. A comparative fluorescent beacon based method for serum microRNA quantification. Anal. Sci. 2015, 31, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Oquare, B.Y.; Taylor, J.S. Synthesis of Peptide Nucleic Acid FRET Probes via an orthogonally protected building block for post synthetic labeling of Peptide Nucleic Acid at the 5-Position of Uracil. Bioconjugate Chem. 2008, 19, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Khologar, S.A.; Novopashina, D.S.; Meschaninova, M.I.; Venyaminova, A.G. Multipyrene tandem probes for point mutation detection in DNA. J. Nucleic Acids 2013, 2013. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, K.; Shen, Y.; Smith, J.; Bloch, S.; Achilefu, S.; Wooley, K.; Taylor, J.S. Imaging mRNA expression levels in living cells with PNA*DNA binary FRET probes delivered by cationic shell-crosslinked nanoparticles. Org. Biomol. Chem. 2013, 11, 3159–3167. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.K.; Samokhina, E.; Babu, B.R.; Wengel, J. Novel (phenylethynyl)pyrene-LNA constructs for fluorescence SNP sensing in polymorphic nucleic acid targets. ChemBioChem 2012, 13, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Durán, C.F.; Esnal, I.; Valois-Escamilla, I.; Urías-Benavides, A.; Bañuelos, J.; Arbeloa, I.L.; García-Moreno, I.; Peña-Cabrera, E. Near-IR BODIPY dyes á la Carte-Programmed Orthogonal Functionalization of Rationally Designed Building Blocks. Chem. Eur. J. 2016, 22, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, W.; Wu, J.; Zhou, B.; Niu, G.; Zhang, H.; Ge, J.; Wang, P. Deep-red to near-infrared fluorescent dyes: Synthesis, photophysical properties, and application in cell imaging. Spectrochim. Acta A 2016, 164, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Mhlanga, M.M.; Malmberg, L. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods 2001, 25, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, K.M.; Dai, Q.; Alman, B.A.; Chan, W.C. Are quantum dots toxic? Exploring the discrepancy between cell culture and animal studies. Acc. Chem. Res. 2013, 46, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, H.E.; Gahlaut, N.; Mohandessi, S.; Yu, D.; Turner, J.R.; Miller, L.W. Time-resolved luminescence resonance energy transfer imaging of protein-protein interactions in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 13582–13587. [Google Scholar] [CrossRef] [PubMed]
- Ansbacher, T.; Srivastava, H.K.; Stein, T.; Baer, R.; Merkx, M.; Shurki, A. Calculation of transtion dipole moment in fluorescent proteins—Towards efficient energy transfer. Phys. Chem. Chem. Phys. 2012, 14, 4109–4117. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.L.; Lukyanov, K.A.; Yampolsky, I.V.; Mishin, A.S.; Bommarius, A.S.; Duraj-Thatte, A.M.; Azizi, B.; Tolbert, L.M.; Solntsev, K.M. Fluorescence imaging using synthetic GFP chromophores. Curr. Opin. Chem. Biol. 2015, 27, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, R.J.; Musial, M. Couple-cluster theory in quantum chemistry. Rev. Mod. Phys. 2007, 79, 291–352. [Google Scholar] [CrossRef]
- Warshel, A.; Levitt, M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976, 103, 227–249. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed. 2009, 48, 1198–1229. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.C.; Kollman, P.A. A combined ab initio quantum mechanical and molecular mechanical method for carrying out simulations on complex molecular systems: Applications to the CH3Cl + Cl− exchange reaction and gas phase protonation of polyethers. J. Comput. Chem. 1986, 7, 718–730. [Google Scholar] [CrossRef]
- Field, M.J.; Bash, P.A.; Karplus, M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J. Comput. Chem. 1990, 11, 700–733. [Google Scholar] [CrossRef]
- Setinmann, C.; Kongsted, J. Electronic energy transfer in polarizable heterogeneous environments: A systematic investigation of different quantum chemical approaches. J. Chem. Theory Comput. 2015, 11, 4283–4293. [Google Scholar] [CrossRef] [PubMed]
- Curutchet, C.; Munoz-Loza, A.; Monti, S.; Kongsted, J.; Scholes, G.D.; Mennucci, B. Electronic energy transfer in condensed phase studied by a polarizable QM/MM model. J. Chem. Theory Comput. 2009, 5, 1838–1848. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Lin, W.; Zheng, K.; Zhu, S. FRET-based Small-Molecule Fluorescent Probes: Rational Design and Bioimaging applications. Acc. Chem. Res. 2013, 46, 1462–1473. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.F.; Dumat, B.; Wranne, M.S.; Lawson, C.P.; Preus, S.; Bood, M.; Graden, H.; Wilhemsson, L.M.; Grøtli, M. Development of bright fluorescent quadracyclic adenine analogues: TDDFT-calculation supported rational design. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- List, N.H.; Pimenta, F.M; Holmegaard, L.; Jensen, R.L.; Etzerodt, M; Schwabe, T.; Kongsted, J.; Ogilby, P.R; Christiansen, O. Effect of chromophore encapsulation on linear and nonlinear optical properties: The case of “miniSOG”, a protein-encased flavin. Phys. Chem. Chem. Phys. 2014, 16, 9950–9959. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.J.; Aidas, K.; Kongsted, J. Solvatochromic Shifts in Uracil: A Combined MD-QM/MM Study. J. Chem. Theory Comput. 2010, 6, 249–256. [Google Scholar] [CrossRef] [PubMed]
- List, N.H.; Jensen, H.J.A.; Kongsted, J.; Hedegård, E.D. A unified framework for the polarizable embedding and continuum methods within multiconfigurational self-consistent-field theory. Adv. Quantum Chem. 2013, 66, 195–459. [Google Scholar]
- Schwabe, T.; Olsen, J.M.H.; Sneskov, K.; Kongsted, J.; Christiansen, O. Solvation effects on electronic transitions: Exploring the performance of advanced solvent potentials in polarizable embedding calculations. J. Chem. Theory Comput. 2011, 7, 2209–2217. [Google Scholar] [CrossRef] [PubMed]
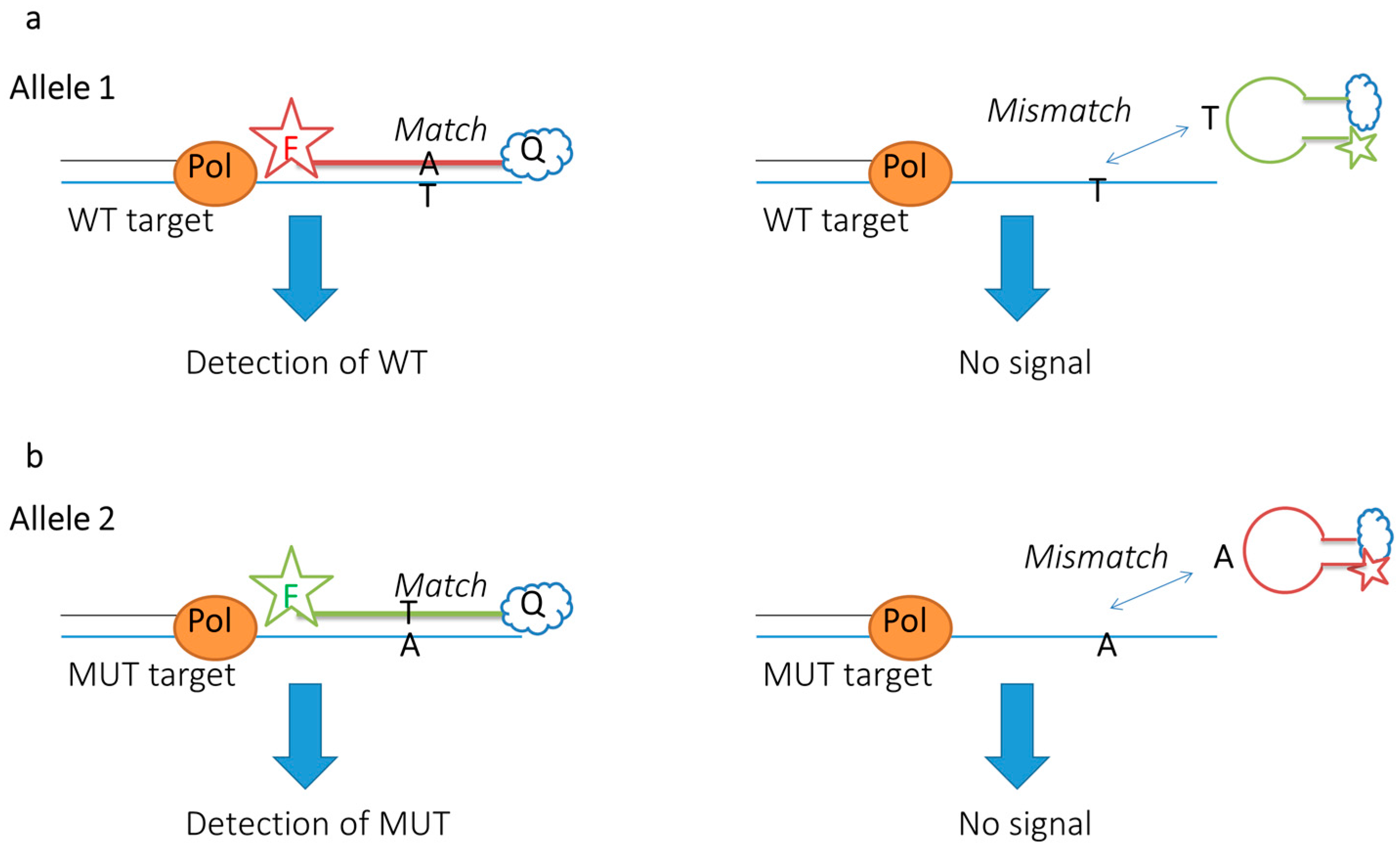
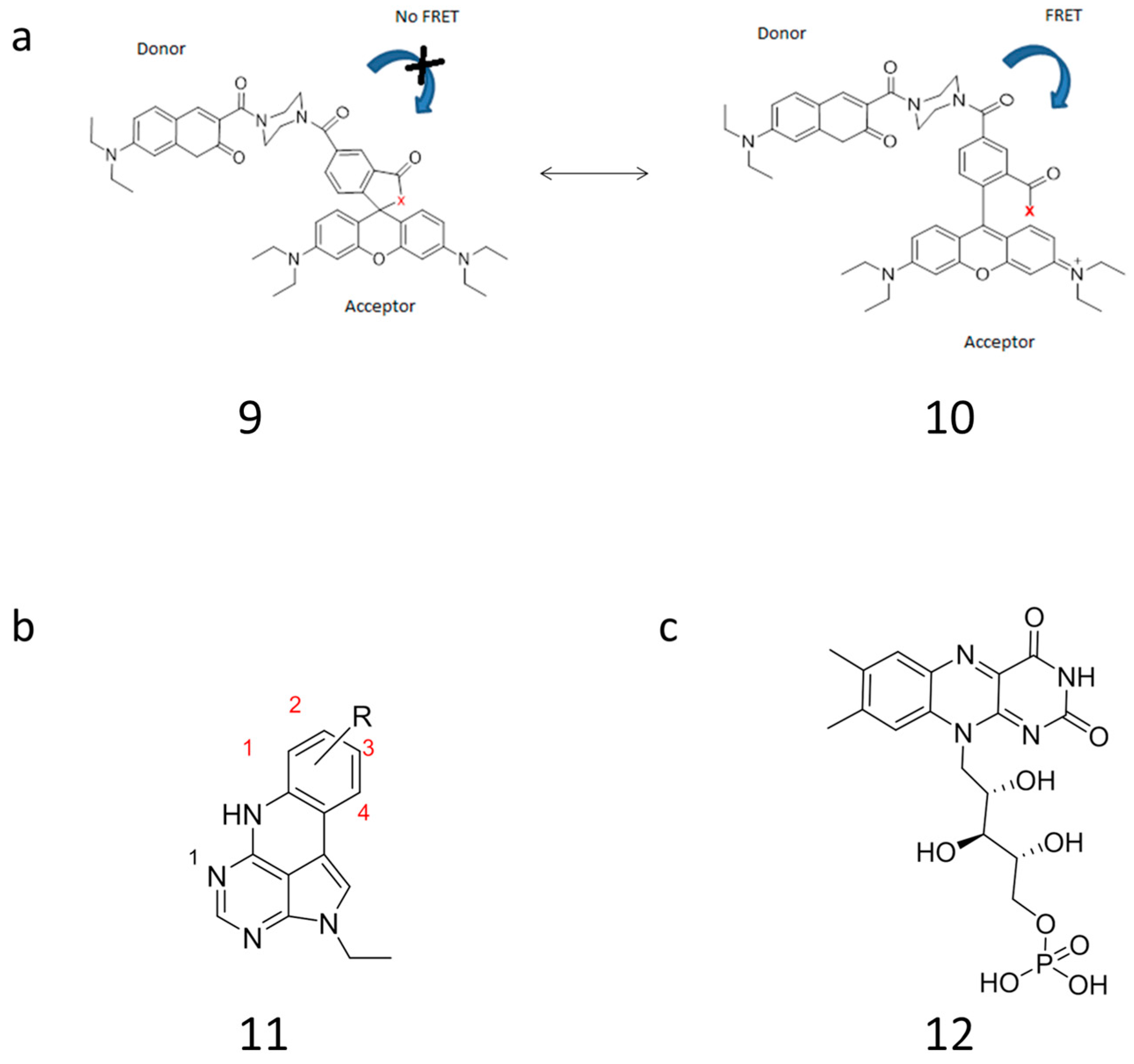
| Probe Design & Sequence (5′-3′) | Target/Assay | FRET Dyes | Backbone ModiFication | LOD | SNP Detection | Ref. |
|---|---|---|---|---|---|---|
| Unimolecular | ||||||
| MB with 3 dyes at 5′ end:d(GCU GAG AAG TTA GAA CCT ATG CTC AGC) | cDNA/in vitro hybridization | Terminal: Pyrene, FAM, TAMRA, Q: EB | None | 1 fM | + | [74] |
| In-stem MB: d(GXTG GXTG CCA GGG CAG TGA TCT CTC CAQQC CAQQC) | β-actin mRNA/FISH | X = Cy3; Q = Nitro methyl red | None | 0.2 μM | + | [83] |
| PNA-MB: H-Lys-(A)-GTCC GYA-Arg(TO)-ATAGCCG-Gly-NH2 | cDNA/in vitro hybridization | TO, ICC | PNA | 40 pM | − | [26] |
| In-stem LNA-MB: d(GGT CXX CTA GAG GGG TCA GAG GAT QQG ACC) | cDNA/in vitro hybridization | X = Pyrene, Q = PDI | None | 0.3 nM | + | [84] |
| MB with LNA in the loop: d(CCGACT ATCTGCACTAGATGCACCTTAC/Bio/CGG) | Serum miRNA/qRT-PCR | Terminal: FAM, Q: 3Dab | LNA, biotin | 0.5 μM | − | [85] |
| Bimolecular | ||||||
| BP with three dyes: Cy5-r(GUA UGU UUC ACU GGA UGA), r (AAG UGG AUC AAG dT(FAM)UG GU(TAMRA) | Sensorin mRNA from neurons/in vitro hybridization | FAM, Cy5, TAMRA | 2′-OMe-RNA | 26 nM | − | [55] |
| OP PNA-BP: d(CTCTTCTU(FAM)TTTT CCT)-K, K(Cy5)-TCC CTC TTC CG ATC | cDNA/in vitro hybridization | Cy5, FAM | Protected PNA | 0.2 μM | − | [86] |
| BP—Bispyrene: bis-Pyr-r(GAG CCG AUU UCA UCA)T, r(GGA GAA GGU GUC UGC GGA G) bis pyr | SNP C677T in MTHFR gene/in vitro hybridization | bis-Pyrene | 2′-OMe-RNA, 3′-inverted thymidine | Nd | + | [87] |
| PNA-DNA BP: d(TCT TCA CGT TGT TGT)-K-(ε)-Cy5, FAM-(ε)-K-d(ATG TCC TTT TCC TCT) | iNOS mRNA/cell line study | Cy5, FAM | PNA | 2 μM | − | [88] |
| PAH-DNA BP: X-CTL TCC ACLA, CALC CAA C-Y | HIV-1 RNA/cDNA/in vitro hybridization and RT-qPCR | Pyrene, Perylene | 2′-amino-LNA, LNA | 5 nM | + | [48] |
| Tb/fluorophore miRNA-complex: Tb-CGA TCA GTC-AGG-CAA-AGC-GG, TTA-CTG-TGC-ACA-GAG-GA-X | Colon-adeno-carcinoma-Hsa-miR-20a-5p, in vivo hybrdization and ligation | Tb; X = Cy3.5 | 5′ C6 thiol, 3′ C7 amine | 0.2 nM | + | [82] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Junager, N.P.L.; Kongsted, J.; Astakhova, K. Revealing Nucleic Acid Mutations Using Förster Resonance Energy Transfer-Based Probes. Sensors 2016, 16, 1173. https://doi.org/10.3390/s16081173
Junager NPL, Kongsted J, Astakhova K. Revealing Nucleic Acid Mutations Using Förster Resonance Energy Transfer-Based Probes. Sensors. 2016; 16(8):1173. https://doi.org/10.3390/s16081173
Chicago/Turabian StyleJunager, Nina P. L., Jacob Kongsted, and Kira Astakhova. 2016. "Revealing Nucleic Acid Mutations Using Förster Resonance Energy Transfer-Based Probes" Sensors 16, no. 8: 1173. https://doi.org/10.3390/s16081173
APA StyleJunager, N. P. L., Kongsted, J., & Astakhova, K. (2016). Revealing Nucleic Acid Mutations Using Förster Resonance Energy Transfer-Based Probes. Sensors, 16(8), 1173. https://doi.org/10.3390/s16081173





