Abstract
Whole-cell microbial biosensors are one of the newest molecular tools used in environmental monitoring. Such biosensors are constructed through fusing a reporter gene such as lux, gfp or lacZ, to a responsive promoter. There have been many reports of the applications of biosensors, particularly their use in assaying pollutant toxicity and bioavailability. This paper reviews the basic concepts behind the construction of whole-cell microbial biosensors for pollutant monitoring, and describes the applications of two such biosensors for detecting the bioavailability and biodegradation of Polychlorinated Biphenyls (PCBs).
1. Introduction
Environmental risk assessment is an essential tool in the investigation of polluted sites. Monitoring practices for assessing these risks usually involve the determination of the total concentration of pollutants using sophisticated chemical analytical techniques such as Gas Chromatography-Mass Spectroscopy (GC-MS) or High Performance Liquid Chromatography (HPLC) assays. The use of the total concentration is likely to overestimate the risk as only a fraction of the total amount of the pollutant, the bioavailable fraction, will actually have an impact on living organisms; this inability to differentiate between the two represents a major disadvantage of traditional analytical methods. This discrepancy between the total and the bioavailable fractions is particularly significant in the case of contaminants with poor aqueous solubility (e.g., PCBs, Poly Aromatic Hydrocarbons [PAHs]) [1]. The ability to monitor the bioavailability of a pollutant is essential, as it not only gives more accurate information regarding the risk that the contaminated site poses to human health, but also determines the effectiveness of potential bioremediation processes. Nowadays, increasing attention has been given to bioavailability assays that better predict the real exposure risks [2]. One such alternative is the use of biosensors which are highly selective and sensitive to a particular pollutant.
Whole-cell microbial biosensors have become one of the newest dimensions of molecular tools in environmental monitoring [3–5]. Microorganisms, due to their low cost, lifespan, and range of suitable pH and temperatures, have been widely employed as the biosensing elements in the construction of biosensors [6].
In the past decade, their applications were mainly focused in three areas:
- Monitoring survival and competition ability of bacteria [7–11].
- Monitoring plant root colonization of pollutant degrading bacteria in complex environmental samples [10,12–14].
- Monitoring the level of specific environmental pollutants [13,15–20].
In recent years, one of the most interesting areas utilising biosensor technology is the detection of environmental pollutant bioavailability, bioremediation, and toxicity. These biosensors are constructed by fusing a pollutant-responsive promoter to a reporter gene coding for a protein that can be easily quantified, and such constructs can be located on plasmids or on the chromosome (Figure 1). The efficacy of such biosensors was demonstrated by Willardson et al. [21]. The results they obtained showed that their toluene sensing, luciferase based whole-cell biosensor accurately reported toluene concentrations that were within the ±3% range as measured by standard GC-MS.
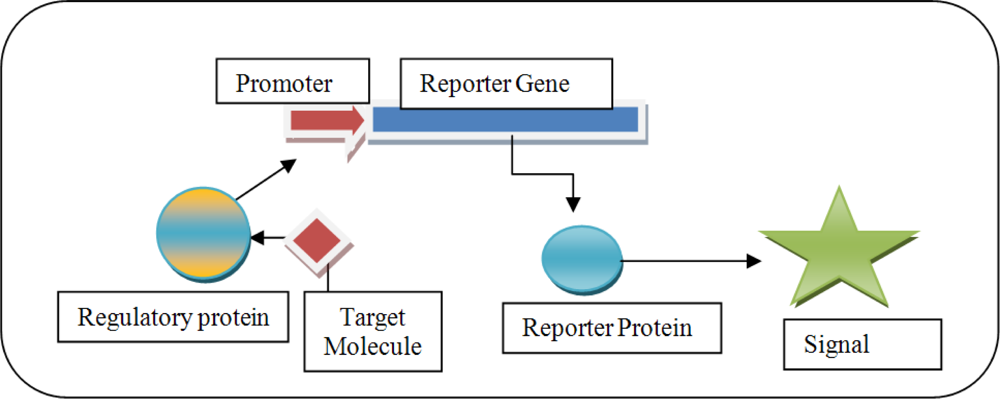
Figure 1.
Graphic illustrating the concept of a whole-cell transcriptional biosensor.
The biosensors rely on analysis of gene expression, typically by creating transcriptional fusions between a promoter of interest and the reporter gene. The extent of reporter gene expression may serve as a measure of the availability of specific pollutants in complex environments. Novel areas for applying these biosensors have been previously documented and include the construction of whole-cell biosensors as specific and sensitive devices for measuring biologically relevant concentrations of pollutants [4,15–18,21–27].
Previous applications of whole-cell microbial biosensors for environmental studies mainly concentrated on their use as biomarkers to investigate survival and competition ability [7–11] and as biosensors to detect the bioavailability or toxicity of environmental pollutants [15,16,28–33]. Layton et al. [34] reported a bioluminescent biosensor strain, Ralstonia eutropha ENV307 (pUTK60), detecting the bioavailability of PCBs by inserting the biphenyl promoter upstream of the bioluminescence genes. In the presence of biphenyl, bioluminescence was generated in a concentration-dependent manner. Kohler et al. [35] used an immobilized recombinant E. coli reporter to detect the bioavailability of 4-chlorobenzoate.
Compared with traditional detection methods for monitoring environmental pollutants, whole-cell biosensors provide the following advantages [36]:
- Biosensors determine only the bioavailable fraction of compounds, thus giving a more accurate response on the toxicity of a sample. Bioavailability is also important in bioremediation. If substances are bioavailable, they are potentially biodegradable.
- Biosensors provide an inexpensive and simple way of determining contaminants.
- As they are living organisms, they provide information on toxicology of different compounds.
- Some stress-induced biosensors report the mutagenic effects of samples with great sensitivity.
- Biosensors are unsurpassed in studying gene expression and physiology of bacteria in complex environments.
1.1. Commonly Used Reporter Genes
The reporter gene usually encodes an enzyme catalyzing a reaction that can be easily monitored. It determines the sensitivity and detection limits of the biosensor. Specific characteristics are needed for the reporter gene to be used in a biosensor. The gene must have an expression or activity that can be measured using a simple assay and it must reflect the level of chemical or physical change. Also, the biosensor must be free of any gene expression or activity similar to the desired gene expression or activity that is being measured to prevent misinterpretation of the response [37]. Several reporter genes meet the necessary requirements and are frequently used including gfp, lacZ, lucFF, luxAB, and luxCDABE with gfp and luxCDABE [29,38–40] being the most commonly used.
The gfp gene encoding Green Fluorescent Protein (GFP), originated from the jellyfish Aequorea victoria and its chromophore is assembled by the self-catalyzed covalent modification of amino acids Ser-Tyr-Gly at positions 65–67 to form a p-hydroxybenzylidene-imidazolidinone species [41,42]. The wild-type chromophore is excited with UV or blue light at 396 nm or 475 nm and emits green fluorescence at 508 nm [41]. The fluorescence of GFP can be monitored without the destruction of the biological sample [42,43]. A large collection of GFP derivatives have been constructed by the optimization of codon usage to alter the spectral properties of GFP for use in different organisms [41,44,45]. There are many examples of using different derivatives of GFPs in the construction of microbial biosensors for detecting environmental pollutants [46–50].
The bioluminescence gene lux cloned from Vibrio fischeri, Photorhabdus luminescens and others [51], coding for the enzyme luciferase, is another reporter gene regularly used for the construction of biosensors to monitor environmental pollutants. The light emitted by the labeled strain can be proportional to the concentration of the target pollutant. Bioluminescence has been used very successfully as a reporter for pollutant detection using sensitive instrumentation including fiber optic probes and integrated circuit chips detecting light production [52,53]. A comprehensive review of the application of bioluminescent genes and bacteria from 2000–2007 was reported by Girotti et al. [54].
Li et al. [26] constructed toluene bacterial biosensors which comprised of two reporters, gfp and luxCDABE. The bacterial luminescence biosensor allowed faster and more sensitive detection of toluene, while the fluorescence biosensor strain was much more stable and thus more applicable for long-term exposure.
1.2. Promoters and Regulatory Elements for the Construction of Biosensors
The selection of the promoter portion of the biosensor construct is dependent on the target molecule being monitored. A selected promoter sequence is normally placed at the 5′-region of the reporter system where it can be switched on in the presence of the target pollutant, thus turning on the expression of the reporter. The key factors when choosing promoters are sensitivity and specificity. Promoters often respond to groups of compounds rather than to a specific compound, and may also behave differently in different microorganisms. e.g., Winther-Larsen et al. [55] stated that the expression of the pm promoter is substrate-dependent and host-specific (more details on this promoter are described in 2.1).
A variety of well-characterized promoters are available for the construction of pollutant-reporting biosensors. These promoters include those for hydrocarbons and organic solvents [56–60], various heavy metals [17,18,61–63], pesticides [64,65], salicylates [66], various organo-phosphorous nerve agents [67,68], and mutagens and genotoxins [69,70]. Promoters are also available for the evaluation of general toxicity [71–74].
One of the greatest limitations of whole-cell biosensor development is the availability of strong promoters that respond only to relevant stimuli. To circumvent this obstacle, more knowledge on gene regulatory networks in bacteria is needed. Linking metagenome information with the meta-transcriptome analysis of microbial communities using microarray technology could provide an immense source of new regulatory elements in the future [75]. Another option is to synthesize ‘super promoters’ based on consensus sequences obtained from comparative studies of different promoters in known regulatory networks [75].
2. Development of Biosensors to Detect PCB Biodegradation
PCBs were detected in the environment for the first time in 1966 by Jensen [76], and they have since been found all over the world including in Arctic and Antarctic regions [77]. The production of PCBs was banned in 1970 in the USA and in the Czech Republic in 1984 [78]. However, several hundred million kilograms has been released into the environment. Wiegel and Wu [79] documented that one-third of all US produced PCBs currently reside in the natural environment.
One of the major threats to public health from PCBs is that they accumulate within the food chain [80,81]. Contaminated fish consumption is a major route of PCB bioaccumulation in humans [82]. The bioaccumulation capability of PCBs in salmon has increased to a much higher extent than in other foods [83]. Traditional methods applied in the remediation of PCB contamination include incineration, vitrification, solidification/stabilization, solvent extraction, thermal desorption and land filling [84]. In the last decade, microbial-mediated degradation has been considered as one of the main processes in the alleviation of PCB pollution from contaminated environments [85].
Microorganisms which are capable of growing on biphenyl as sole carbon source were first isolated in 1970 [86,87]. In 1973, Ahmed and Focht [88] reported that Achromobacter degrades a few lightly chlorinated PCBs. Since then, numerous PCB-degrading bacterial strains have been isolated from PCB contaminated sites [89–95]. Nearly all of these isolates are able to degrade only two bi-chlorinated PCBs and very few bacteria have been found with the ability to degrade more highly chlorinated congeners [91,96]. These microorganisms belong to both Gram-negative and Gram-positive genera including Pseudomonas, Burkholderia, Achromobacter, Comamonas, Ralstonia, Acinetobacter, Rhodococcus and Bacillus [97–99].
PCBs are broken down by the catabolic “biphenyl upper pathway” or the “bph pathway” [96,100] which involves four enzymes: biphenyl 2,3-dioxygenase (BphA), cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase (dihydrodiol dehydrogenase, BphB), 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) and 2-hydroxy-6-phenylhexa-2,4-dienoate hydrolase (HOPDA Hydrolase, BphD). The biphenyl upper pathway breaks down biphenyl into benzoic acid and 2-hydroxy-penta-2, 4-dienoic acid [101] as shown in Figure 2. The aliphatic acid is metabolized via acetyl-CoA through the tricarboxylic acid cycle ultimately leading to CO2. The chlorobenzoic acids can be mineralized in co-culture with bacterial strains which can use chlorobenzoic acid as carbon source.
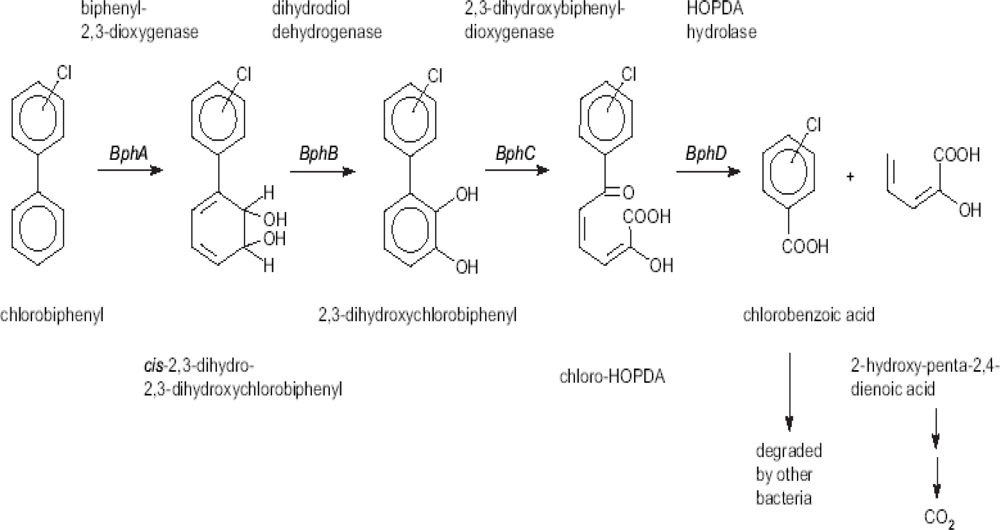
Figure 2.
Biphenyl (bph) upper-pathway of aerobic PCB degradation [89,100]. Intermediates such as hydroxylated chlorobiphenyls [102] and chlorobenzoic acids [13,19,103] been used as targets in developing biosensors for monitoring PCB biodegradation.
2.1. Biosensors Based on Monitoring Chlorobenzoic Acids
The pm promoter is derived from the toluene degrading TOL plasmid and regulates the meta-cleavage pathway of aromatic hydrocarbon degradation [104]. The meta-pathway is induced by various benzoic acid derivatives and this induction is mediated by the substrate-activated XylS protein. The pm promoter and its activator protein have previously been inserted into various vector systems and shown to be a useful expression system for controlled expression of recombinant proteins in several Gram-negative bacteria [105–111].
The promoter-reporter fusion KmR-xylS-pm-gfpmut3* in plasmid pJBA26 (Figure 3) [12] has been used to construct a whole-cell biosensor to report PCB degradation. The gfpmut3b gene in plasmid pJBA26 is a variant of the wild-type gfp gene in which two amino acids have been substituted (S65G, S72A). These substitutions result in up to eight-fold enhanced fluorescent signal [112]. Gfp-mut3*, a mutant of gfpmut3b, was inserted in to a pUC18-NotI based cloning vector by introducing a SphI site in the start codon of gfp-mut3b during PCR amplification. The sequence was also changed during PCR so that the gfp-mut3* contained an arginine residue instead of serine at position 2. The construct gfp-mut3* maintained its intensively fluorescent signal, and with an estimated half life of one day in vivo [113].
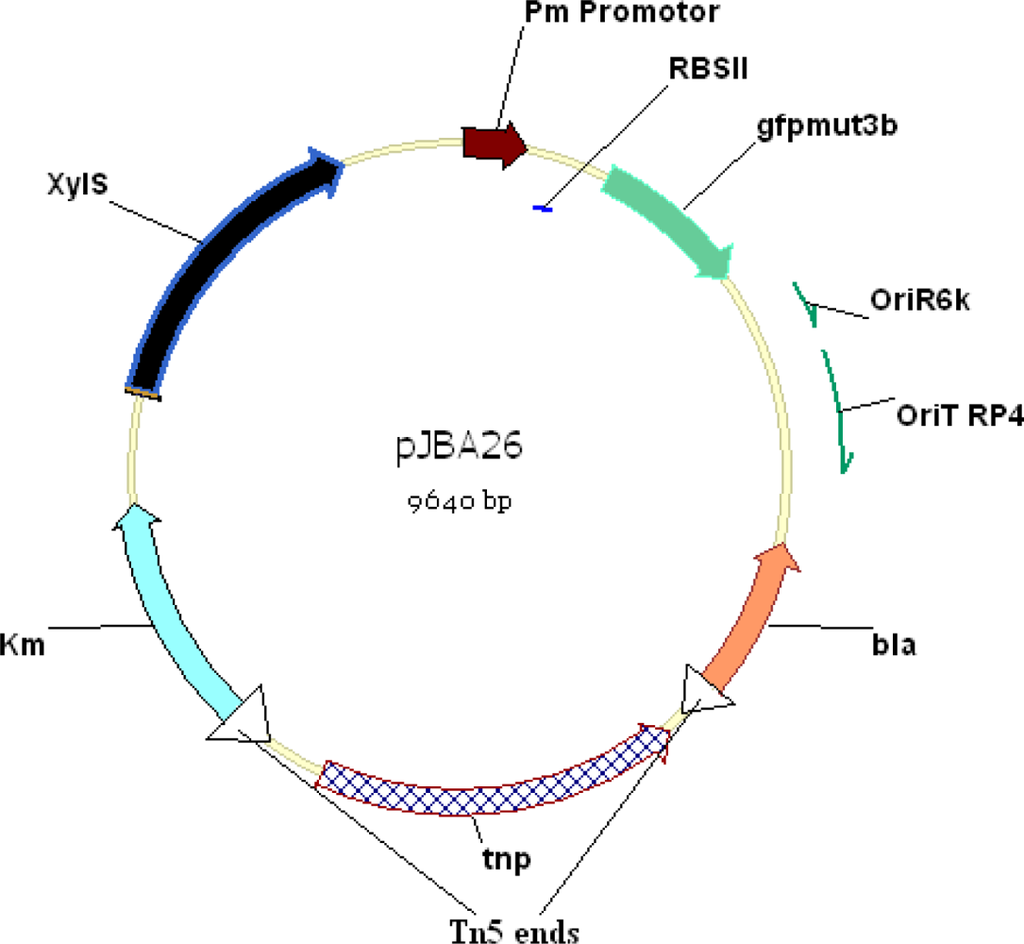
Figure 3.
Genetic Map of pJBA26 vector [12], the region between the Tn5 ends from the kanamycin resistance gene (Km) to the β-lactamase gene (bla) can be transposed into the chromosome of suitable recipient cells such as Pseudomonas fluorescens.
Boldt et al. [13] constructed a number of gfp-based P. fluorescens F113PCB biosensors using fusions of the Escherichia coli rrnBP1 ribosomal promoter and the pm-xylS system introduced by pJBA26 with gfp genes. P. fluorescens F113 was originally isolated from the rhizosphere of sugar beet [114] and was found to be an excellent root colonizer. The genes responsible for PCB degradation from Burkholderia xenovorans LB400, termed the bph operon [115], were chromosomally inserted into a spontaneous rifampicin-resistant mutant of F113 creating P. fluorescens F113rifpcb [116]. The biosensors constructed by Boldt et al. [13] were shown to be able to monitor the single-cell localization and activity of P. fluorescens F113 colonizing alfalfa roots. The monitoring systems permitted non-destructive in situ detection of cells on the entire root system grown in both the presence and absence of 3-chlorobiphenyl [13].
Liu et al. [19] constructed two further biosensors based on this system using a modified F113PCB strain (F113L::1180) (Figure 4) where the bph pathway was under the regulation of a strong constitutive promoter (Nod D1) (see Villacieros et al. [117] for details) with a corresponding higher level of bph gene expression and PCB transformation. A second biosensor (F113rifgfp), which cannot degrade PCBs, only responds to external chlorobenzoic acid derivatives, and by using these two biosensors PCB degradation could be detected in vitro and in soil (Figure 5). The sensitivity of this system towards 3-chlorobenzoic acid, 2,3-dichlorobenzoic acid and 3,5-dichlorobenzoic acid was found to be 0.1ppm using epi-fluorescent microscopy.
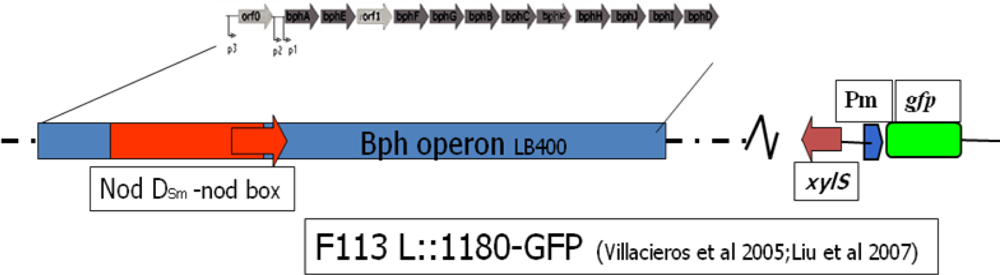
Figure 4.
Genetic organization of whole cell rhizosphere biosensor Pseudomonas fluorescens F113L::1180-GFP. This bacterium can utilize biphenyl as a carbon source and biodegrade a range of PCBs. The bph operon originated from the PCB degrader Burkholderia xenovorans LB400 and it is constitutively expressed by the NodD-nod box regulatory unit from the symbiotic bacterium Sinorhizobium meliloti. The XylS-pm-gfp cassette originated from plasmid pJBA26. The host bacterium P. fluorescens F113 is an active plant root colonizer and biocontrol strain.
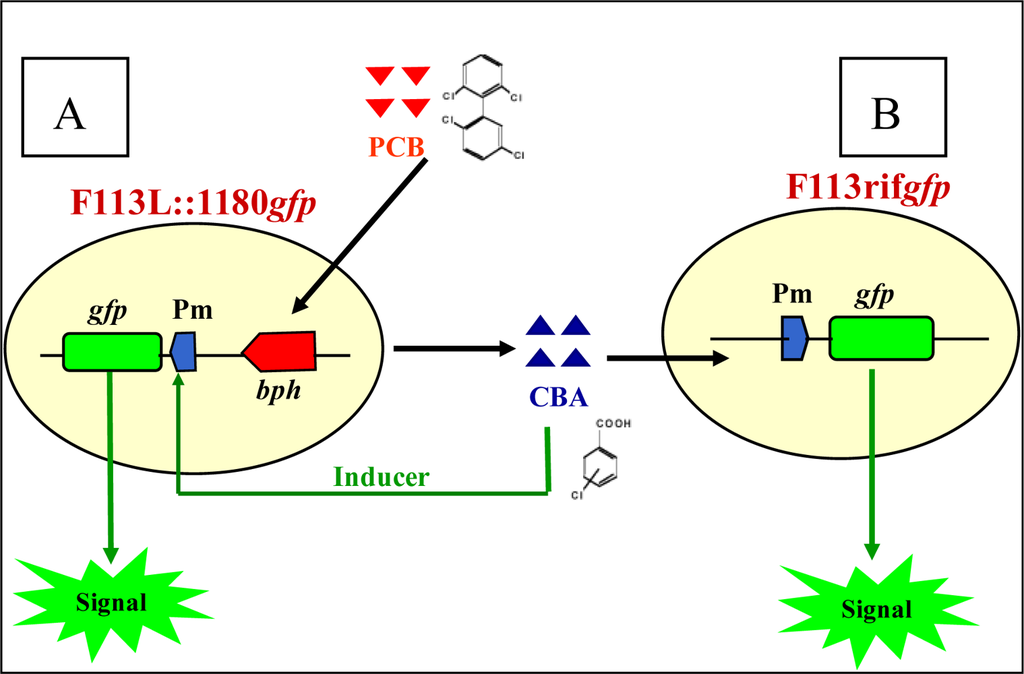
Figure 5.
The use of two PCB biosensors. (A) F113L::1180gfp is able to report its own biodegradation of PCBs by switching on the signal, and the biodegradation of chlorobenzoic acid (CBA) by other CBA degraders by switching off the signal. (B) F113rifgfp is able to sense the biodegradation of PCBs by other PCB degraders by switching on the signal and the biodegradation of CBA by other CBA degraders by switching off the signal [13,19,103].
Expression of the pm promoter is substrate-dependent and host-specific [55]. In a pure culture study [19], the pm promoter was induced by 3-CBA in all of the biosensor strains and was greatly induced by 2,3-DiCBA, 3,5-DiCBA and to a lesser extent by 3,4-DiCBA as determined by spectrofluoremetry and epifluorescent microscopy. In addition, a linear relationship was observed between the fluorescent intensity and the concentration of 3-CBA, 2,3-DiCBA and 3,5-DiCBA ranging from 4 to 400 ppm. Previous studies used an immobilized recombinant E. coli reporter to detect the bioavailability of 4-chlorobenzoate [35]. This work shows that the biosensors have the potential to detect the bioavailability of other CBA derivatives, which are produced by the intrinsic biodegradation of PCBs in the environment.
3. Encapsulation of Biosensors for Environmental Use
Whole-cell biosensors provide effective tools for detecting environmental pollution and toxicity. Important aspects of biosensing environmental pollutants are the ability to monitor in situ and preferably on-line. One of the main problems is the difficulty of maintaining constant sensing activity and variability for extended periods at room temperature. To meet these demands, various conservation techniques have been reported including freeze drying, vacuum drying, continuous cultivation, and immobilization in biocompatible polymers of organic or inorganic origin [72,118,119].
Encapsulating whole-cell biosensors in natural or synthetic polymers has been shown to be useful for the detection of environmental pollutants [120–123]. Polymeric matrices can provide a hydrated environment containing the nutrients and cofactors needed for cellular activity and growth. In addition, encapsulated cells are protected from toxic substances in their environment and maintain increased plasmid stability [124].
The following encapsulation parameters show the potential usefulness for developing an in situ application of whole-cell biosensors [53]:
- Agar/agarose: competent cells can be added to molten agar or agarose (1–5%). Gelation occurs as the agar or agarose cools to room temperature [125].
- Carrageenan: a 2% solution of carrageenan is warmed to 70–80 °C to initiate dissolution and then maintained at 35–50 °C. The cell culture is also warmed and added to the carrageenan solution. Gel formation occurs through the addition of cold 0.1 M potassium chloride [124].
- Alginate: cells are added to a 1–8% solution of alginate; addition of 0.5 M calcium chloride or 0.1 M strontium chloride causes polymerization [124]. The bioluminescent bioreporter P. fluorescens HK44 has been immobilized on the end of liquid light guides using this method [126–127].
- Polyurethane–polycarbomyl sulfonate (PCS): polyurethane or PCS at a polymer content of 30–50% is mixed with a 1% calcium-chloride solution, the pH is adjusted to approximately 6.5 and the cell mass is added. This mixture is sprayed into 0.75% calcium alginate, resulting in bead formation. After one hour, the beads are removed, washed and introduced into a 2% sodium-tripolyphosphate buffer, which dissolves the alginate layer leaving only a layer of polyurethane–PCS surrounding the cells [126].
- Polyacrylamide: cells are mixed in a solution of acrylamide and bisacrylamide. Ammonium persulfate and N,N,N′,N′-tetramethylethylenediamine (TEMED) are then added to initiate polymerization [128].
- Polyvinyl alcohol: the cell suspension is mixed with a 13% polyvinyl alcohol, 0.02% sodium-alginate mixture. Gel formation occurs on contact with a solution of saturated boric acid and 2% calcium chloride [129].
- Sol–gel: cells are combined with 0.1 M Tris-Cl and tetramethylorthosilicate, tetraethoxysilane, methyltrimethoxysilane, ethyltrimethoxysilane, propyltrimeth
- Oxysilane or polydimethylsiloxane. Solidification times vary depending on the concentrations used [130].
- Polyethylleneimine [131].
Alginate had been previously applied as a delivery system for Pseudomonas fluorescens F113lacZY in sugar-beet root colonization experiments [132]. It was shown that cells encapsulated in alginate polymers displayed more efficient root colonization and had a longer shelf life (up to eight weeks of storage), regardless of the conditions of incubation. Another F113 derivative F113rifPCB had also been alginate encapsulated and up to 100% of the encapsulated cells were found to be viable after 250 d storage [133].
3.1. Monitoring PCB Degradation in Vitro and in Soil Using Encapsulated PCB Biosensors
Liu [103] encapsulated two PCB/CBA biosensors (F113L::1180gfp or F113rifgfp) in alginate beads and examined the response of the biosensor cells to various PCBs and chlorinated benzoates in liquid cultures and in soil.
In liquid culture spiked with 3-chlorobiphenyl, 100% of the alginate encapsulated F113L::1180gfp biosensor cells were fluorescent after five days indicating that this strain was actively degrading the 3-chlorobiphenyl. In a similar soil based experiment, more than 30% of the alginate encapsulated F113L::1180gfp cells were visualized as gfp-expressing cells after 10 days. There was no other source of chlorobenzoates to induce this biosensor other than those derived from F113L::1180gfp own PCB degradation activity. This was demonstrated by the fact that when encapsulated F113rifgfp (a non PCB degrading chlorobenzoate biosensor) was inoculated under the same conditions no fluorescent cells were found.
When the encapsulated F113rifgfp biosensor was co-introduced into 3-chlorobiphenyl liquid culture with the natural PCB degrader Rhodococcus sp. ITCBP [134] about 80% of the biosensor cells were visualized as gfp-expressing cells after five days (Figure 6A). In soil, 50% of the encapsulated biosensor cells were fluorescent after 10 days when co- inoculated with Rhodococcus sp. ITCBP (Figure 6B). No fluorescent cells were visualized in the control experiment using a pure culture of either F113rifgfp or Rhodococcus sp. ITCBP, thus validating the premise that the encapsulated biosensor, F113rifgfp, could be used to detect PCB degradation by other bacteria, in this case Rhodococcus sp. ITCBP.
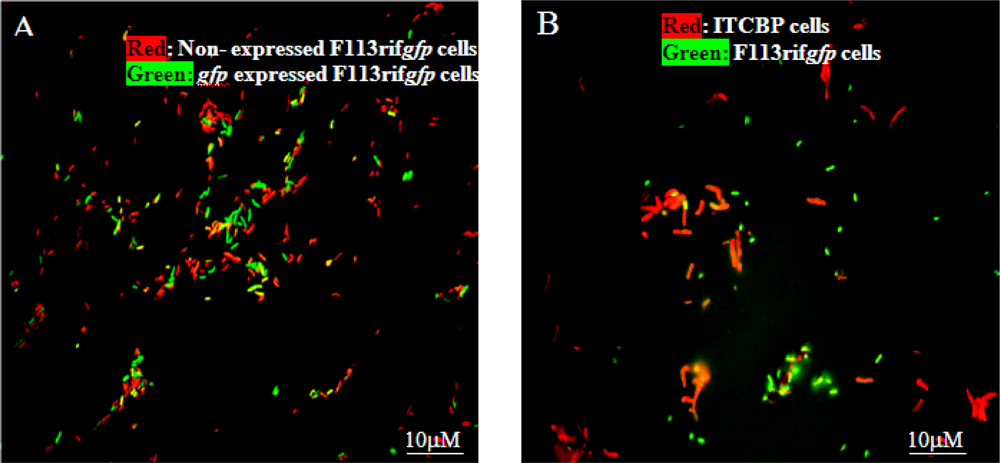
Figure 6.
Epifluorescent micrographs showing encapsulated biosensor F113rifgfp detecting 3-CBP degradation by Rhodococcus sp. ITCBP. (A) Biosensor F113rifgfp cells in liquid culture; (B) Rhodococcus sp. ITCBP cells and biosensor F113rifgfp cells in the soil.
3.2. Monitoring Chlorobenzoic Acids (CBA) Bioavailability and Biodegradation
When the encapsulated biosensors (F113L::1180gfp or F113rifgfp) were inoculated in minimal media broth or sterile soil supplemented with 0.1 mM 3-CBA or 2,3-CBA or 3,5-CBA, 100% of the cells were fluorescent after 24 hours. This result indicated that the encapsulation of the biosensors in alginate did not interfere with their CBA sensing ability either in liquid culture or in soil.
In 1 mM 3-CBA liquid culture, when either of the encapsulated biosensors (F113L::1180gfp or F113rifgfp) were co-inoculated with the chlorobenzoate degrader Pseudomonas sp. B13 [135] 80% of the biosensor cells were fluorescent after 10 hours. After 22 hours no fluorescent cells could be visualized (Figure 7). When the encapsulated biosensors were introduced into 3-CBA spiked soil previously inoculated with Pseudomonas sp. B13, the percentage of fluorescent cells dropped over time and after 10 days there were no fluorescent cells detected. These results demonstrated that the encapsulated biosensors could detect the biodegradation of chlorobenzoic acids by CBA degrading bacteria, in this case by Pseudomonas sp. B13.
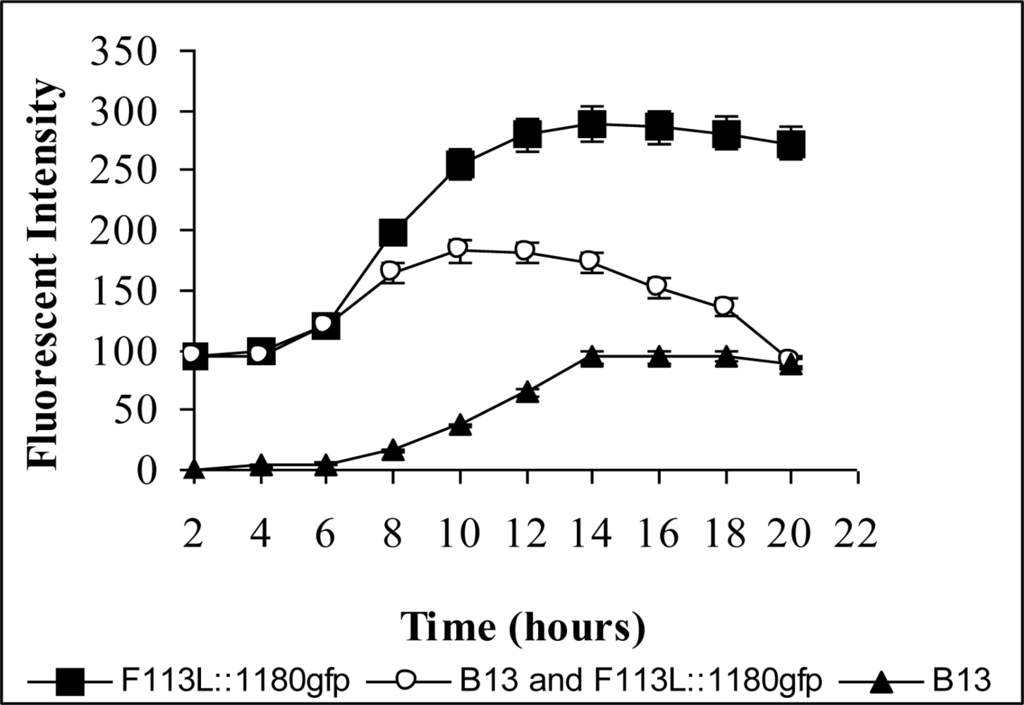
Figure 7.
Biosensor detecting 3-CBA degradation by Pseudomonas sp. B13. Gfp fluorescent signal detected using a spectrofluorimeter. Note in the mixed cultivation, fluorescence decreases after 10 hours due to removal/degradation of 3-CBA by Pseudomonas sp. B13.
4. Discussion and Conclusions
The majority of promoter-reporter biosensor systems currently being used are the result of cloning of a promoter upstream of a reporter gene cassette and the subsequent transfer of the plasmid construct into specific strains [30–32,34]. However, loss of these plasmids due to the starvation [136] and reduction in expression of the reporter gene due to multiple copies of the promoter binding region on the plasmid [137] poses problems when these biosensors are applied to in vivo situations. There are few reports of biosensors based on the chromosomal insertion of the promoter-reporter gene which produce more stable systems [16,128]. Liu [103] describes the construction of three chromosome-based biosensors P. fluorescens F113rifgfp, P. fluorescens F113rifPCBgfp and P. fluorescens F113L::1180gfp. The insertion of the gfp construct into the chromosome of these bacteria was confirmed by PCR and Southern blotting.
Studies have shown that the integration of the gfp reporter gene into the chromosome affected the growth ability of Ralstonia eutropha on 2, 4-dichlorophenoxyacetic acid [138] and on biphenyl [11]. It was important to ensure that the chromosomal insertion of the gfp construct did not affect any major catabolic pathway of these biosensor strains. Phenotypic characterization of F113rifgfp, F113rifPCBgfp and F113L::1180gfp by comparing the growth curve and Biolog® Gram Negative (GN) profiles of the three biosensor strains to their parental strains showed there was no detectable disruption of major metabolic pathways by the insertion of this gfp construct. Rhizosphere colonization ability was not affected in the biosensors as green fluorescent cells colonizing pea (Pisum sativum) root was observed by Epifluorescent microscopy. Also the bph operon was not affected by insertion of the gfp construct, as documented by Southern blotting using a Digoxigenin (DIG)-labeling bphC probe [19]. While random chromosomal insertion offers many advantages over plasmid-based construction of sensors, the ideal approach for commercial applications would be the use of targeted insertion to known chromosomal sites.
Environmental contamination by chlorobenzoic acids has occurred as a result of decades of excessive use of herbicides (e.g., 2,3,6- trichlorobenzoic acid) and through partial degradation of other xenobiotic compounds, such as PCBs [139]. One of the clear impacts of chlorobenzoates on the environment is that they inhibit PCB metabolism [140,141]. It is essential to develop an easy and cost effective method to detect and remove these compounds from contaminated environments.
Whole-cell biosensor technology has been applied to detect the bioavailability or toxicity of environmental pollutants [4,36,142]. However, to our knowledge there are no studies to show the in vivo application of whole-cell biosensors to detect the biodegradation and decontamination of an environmental pollutant. Through the introduction of the biosensor F113L::1180gfp strain into PCB amended soil, PCB degradation could be monitored by observing gfp fluorescence in the cells. Another biosensor strain F113rifgfp could detect PCB degradation by other PCB degrader strains e.g., Rhodococcus sp. ITCBP. Potentially, this biosensor can be used to evaluate the PCB degrading ability in situ of intrinsic PCB degrading strains.
Previous studies had shown that the co-culture of PCB and CBA degrading strains led to the complete mineralization of PCB contamination from the environment [143–145]. The work detailed here showed that the PCB mineralization process can be monitored using the constructed biosensor strain F113L::1180gfp. When both the biosensor P. fluorescens F113L::1180gfp and Pseudomonas sp. B13 were co-inoculated into 3-CBP media, fluorescent cells were observed indicating that this biosensor detected its own biodegradation of PCBs. The percentage of fluorescent cells dropped to below detectable levels at the end of experiment, indicating that this biosensor also detected the mineralization of CBA by Pseudomonas sp. B13. Therefore, it is possible to monitor the complete mineralization process of PCB in situ using this biosensor.
It was also observed that the gfp gene was unevenly expressed within the biosensor populations. This is probably due to the spatial and temporal distribution of CBA compounds within the samples leading to the uneven expression of the pm promoter within the populations [30,55].
Previous studies have shown that using immobilized biosensor cells to easily and accurately detect the availability of pollutants based on induction and de novo synthesis of reporter proteins is possible [15,52,146,147]. Boldt et al. [13] reported using F113rifpcbgfp to detect the biodegradation of 3-CBP and root colonization in 3-CBP spiked soil. However, only 1% of the bacterial cells were shown to be Gfp fluorescent. In this study, it was suggested that the presence of PCB or CBA degraders would affect the accuracy of the biosensor. Thus, an alginate bead immobilized system for the delivery and examination of the biosensors was developed. The evaluation of alginate as a carrier was studied previously and the results indicated a long survival of the genetically modified strain F113lacZY [132].
In addition to the longer survival and easy release advantages of encapsulating the cells in an alginate polymer, there is also some evidence in the study carried out by Liu [103] to show that higher levels of gfp expression could be achieved by encapsulating the biosensor strains which in turn lead to more accurate results. This study showed that:
In 3-CBP media co-inoculated with F113L::1180gfp and Pseudomonas B13, more fluorescent cells were observed using the encapsulated cells than when using the free cells. This indicated that the alginate provided a protective barrier to maintain the CBA breakdown products from PCB degradation by F113L::1180gfp within the beads. These increased levels of CBAs then induced the biosensor so that it could detect the occurrence of this PCB breakdown process accurately. When F113L::1180gfp was applied as free cells to PCB contaminated environmental samples, only 1% of the cells showed fluorescence compared to 10%–43% cells that showed fluorescence when the encapsulated method was used.
It should be remembered that in nearly all cases whole-cell biosensors are not applied in situ to soil and water but rather samples are taken from polluted sites and incubated in in vitro assays. This may not precisely reflect the real conditions in nature with respect to prevailing environmental conditions. The development of robust permeable sealed immobilised biosensor systems could be useful to overcome this challenge. The presence of auto-fluorescence within the sample matrix can also be a problem and attempts to address this issue have included the use of reporters such as urophorphyrinogen II methyl transferase (UMT), using the CobA gene as a reporter, which converts UMT to two fluorescent compounds. A whole-cell biosensor for arsenate detection was developed using this approach (148). An alternate approach is to counter-stain with methyl violet to limit the interference of auto-fluorescence as reported by Germaine et al. (14).
The presence of toxic compounds (both target and non-target) in the sample matrix may have detrimental effects on the host cell with respect to viability, physiological state and survival may have negative impact on the function of the sensor. In many cases these issues can be addressed by the appropriate choice and detailed evaluation of the host strain under different polluted soils.
Whole-cell microbial biosensors offer excellent possibilities for assaying the complex nature of bioavailable and bioaccessible fractions in thousands of cases of severe and toxic pollution, which currently cannot be easily addressed [2] and in general there is good correlation between such sensors and standard quantitative methods. In the case of the PCB biosensor described here, preliminary data indicated a strong positive correlation between the number of fluorescent cells and concentration of PCBs in soils or sediments as measured by standard analytical methods. The work detailed in this review suggests that encapsulated biosensors have real potential to detect the bioavailability and biodegradation of PCBs in the environment.
Acknowledgments
This work was funded in part by EU contract QLRT-2001-00101, DAF Stimulus Programme P-Solve and the TSRIII programme “Agribiotics” sponsored by Technological Sector Research programme under the HEA.
References and Notes
- Harmsen, J. Measuring bioavailability: From a scientific approach to standard methods. J. Environ. Qual 2007, 5, 1420–1428. [Google Scholar]
- Tecon, R.; van der Meer, J.R. Bacterial biosensors for measuring availability of environmental pollutants. Sensors 2008, 8, 4062–4080. [Google Scholar]
- Purohit, H.J. Biosensors as molecular tools for use in bioremediation. J. Cleaner. Prod 2003, 11, 293–301. [Google Scholar]
- Ron, E. Biosensening environmental pollution. Curr. Opin. Biotechnol 2007, 18, 252–256. [Google Scholar]
- Yagi, K. Applications of whole-cell bacterial biosensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol 2007, 73, 1251–1258. [Google Scholar]
- Mulchandani, A.; Rogers, K.R. (Eds.) Enzyme and Micriobial Biosensors: Techniques and Protocols; Humana Press: Totowa, NJ, USA, 1998.
- Tresse, O.; Errampalli, D.; Kostrzynska, M.; Leung, K.T.; Lee, H.; Trevors, J.T.; Van Elsas, J.D. Green fluorescent protein as a visual marker in a p-nitrophenol degrading Moraxella sp. FEMS Microbiol. Lett 1998, 164, 187–193. [Google Scholar]
- Errampalli, D.; Okamura, H.; Lee, H.; Trevors, J.T.; Van Elsas, J.D. Green fluorescent protein as a marker to monitor survival of phenanthrene-mineralising Pseudomonas sp. UG14Gr in creosote-contaminated soil. FEMS Microbiol. Ecol 1998, 26, 181–191. [Google Scholar]
- Errampalli, D.; Tress, O.; Lee, H.; Trevors, J.T. Bacterial survival and mineralization of p-nitrophenol in soil by green fluorescent protein-marked Moraxella sp. G21 encapsulated cells. FEMS Microbiol. Ecol 1999, 30, 229–236. [Google Scholar]
- Cassidy, M.B.; Leung, K.T.; Lee, H.; Trevors, J.T. A comparison of enumeration methods for culturable Pseudomonas fluorescens cells marked with green fluorescent protein. J. Microbiol. Meth 2000, 40, 135–145. [Google Scholar]
- Abby, A.M.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Polychlorinated biphenyl (PCB) degradation and persistence of a gfp-marked Ralstonia eutropha H850 in PCB-contaminated soil. Appl. Microbiol. Biotechnol 2003, 63, 222–230. [Google Scholar]
- Moller, S.; Sternberg, C.; Andersen, J.B.; Christensen, B.B.; Ramos, J.L.; Givskow; Molin, S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol 1998, 64, 721–732. [Google Scholar]
- Boldt, T.S.; Sorensen, J.; Karlson, U.; Molin, S.; Ramos, C. Combined use of different Gfp reporters for monitoring single-cell activity of a genetically modified PCB degrader in the rhizosphere of Alfalfa. FEMS Microbiol. Ecol 2004, 48, 139–148. [Google Scholar]
- Germaine, K.; Keogh, E.; Garcia-Cabellos, G.; Borremans, B.; van der Lelie, D; Barac, T.; Oeyen, L.; Vangronsveld, J.; Porteous Moore, F.; Moore, E.R.B.; Campbell, C.D.; Ryan, D.; Dowling, D.N. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol. Ecol 2004, 48, 109–118. [Google Scholar]
- Schreiter, P.P.; Gillor, O.; Post, A.; Belkin, S.; Schmid, R.D.; Bachmann, T.T. Monitoring of phosphorus bioavailability in water by an immobilized luminescent cyanobacterial reporter strain Biosens. Bioelectron 2001, 16, 811–818. [Google Scholar]
- Taylor, C.J.; Bain, L.A.; Richardson, D.J.; Spiro, S.; Russell, D.A. Construction of whole-cell gene reporter for the fluorescent bioassay of nitrate. Anal. Biochem 2004, 328, 60–66. [Google Scholar]
- Brandt, K.K.; Holm, P.E.; Nybroe, O. Bioavailability and toxicity of soil particle-associated copper as determined by two bioluminescent Pseudomonas fluorescens biosensor strains. Environ. Toxicol. Chem 2006, 25, 1738–1741. [Google Scholar]
- Fujimoto, H.; Wakabayashi, M.; Yamashiro, H.; Maeda, I.; Isoda, K.; Kondoh, M.; Kawase, M.; Miyasaka, H.; Yagi, K. Whole-cell arsenite biosensor using photosynthetic bacterium Rhodovulum sulfidophilum: Rhodovulum sulfidophilum as an arsenite biosensor. Appl. Microbiol. Biotechnol 2006, 73, 332–338. [Google Scholar]
- Liu, X.; Germaine, K.; Ryan, D.; Dowling, D.N. Development of a GFP-based biosensor for detecting the bioavailability and biodegradation of polychlorinated biphenyls (PCBs). J. Environ. Eng. Landsc. Manag 2007, 15, 261–268. [Google Scholar]
- Lejon, D.; Martins, J.; Leveque, J.; Spanini, L.; Pascault, N.; Landry, M.; Milloux, M.; Nowak, V.; Chaussod, R.; Ranjard, L. Copper dynamics and impacts on microbial communities in soils of variable organic status. Environ. Sci. Technol 2008, 42, 2819–2825. [Google Scholar]
- Willardson, M.B.; Wilkins, F.J.; Rand, A.T.; Schupp, M.J.; Hill, K.K.; Keim, P.; Jackson, J.P. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microbiol 1998, 64, 1006–1012. [Google Scholar]
- Diesel, E.; Schreiber, M.; van der Meer, J. Development of bactyeria-based bioassays for arsenic detection in natural waters. Anal. Bioanal. Chem 2009, 394, 687–693. [Google Scholar]
- Fu, Y.; Chen, W.; Huang, Q. Construction of two lux-tagged Hg2+ specific biosensors and their luminescence performance. Appl. Microbiol. Biotechnol 2008, 79, 363–370. [Google Scholar]
- Keane, A.; Lau, P.; Ghoshal, S. Use of a whole cell biosensor to assess the bioavailability enhancement of aromatic hydrocarbon compounds bu nonionic surfactants. Biotechnol. Bioeng 2008, 99, 86–98. [Google Scholar]
- Kohlmeier, S.; Mancuso, M.; Deepthike, U.; Tecon, R.; van der Meer, J.; Harms, H.; Wells, M. Comparison of naphthalene bioavailability determined by whole cell biosensing and availability determined by extraction with Tenax. Environ. Pollut 2008, 156, 803–808. [Google Scholar]
- Li, Y.; Li, F.; Ho, C.; Liao, V.H. Construction and comparison of fluorescence and bioluminescence bacterial biosensors for the detection of bioavailable toluene and related compounds. Environ. Pollut 2008, 152, 123–129. [Google Scholar]
- Fiorentino, G.; Ronca, R.; Bartolucci, S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl. Microbiol. Biotechnol 2009, 82, 67–77. [Google Scholar]
- Applegate, M.B.; Kehrmeyer, R.S.; Sayler, S.G. A chromosomally based tod luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl. Environ. Microbiol 1998, 64, 2730–2735. [Google Scholar]
- Roberto, F.; Barnes, J.; Bruhn, D. Evaluation of a GFP reporter gene construct for environmental arsenic detection. Talanta 2002, 58, 181–188. [Google Scholar]
- Stiner, L.; Halverson, L.J. Development and characterization of a green fluorescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl. Environ. Microbiol 2002, 68, 1962–1971. [Google Scholar]
- Ikeno, S.; Ogino, C.; Ito, T.; Shimizu, N. Detection of benzene derivatives by recombinant E. coli with Ps promoter and GFP as a reporter protein. J. Biochem. Eng 2003, 15, 193–197. [Google Scholar]
- Chang, S.T.; Lee, H.J.; Gu, M.B. Enhancement in the sensitivity of an immobilized cell-based soil biosensor for monitoring PAH toxicity. Sens. Actuat 2004, 97, 272–276. [Google Scholar]
- Peca, L.; Kos, P.; Mate, Z.; Farsang, A.; Vass, I. Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol. Lett 2008, 289, 258–64. [Google Scholar]
- Layton, A.C.; Muccini, M.; Ghosh, M.M.; Sayler, G.S. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl. Environ. Microbiol 1998, 64, 5023–5026. [Google Scholar]
- Kohler, S.; Bachmann, T.T.; Schmitt, J.; Belkin, S.; Schmid, R.D. Detection of 4-chlorobenzoate using immobilized recombinant Escherichia coli reporter strains. Sens. Actuators 2000, 70, 139–144. [Google Scholar]
- Hansen, L.H.; Sorensen, S.J. The use of whole-cell biosensors to detect and quantify compounds or conditions affecting biological systems. Microb. Ecol 2001, 42, 483–494. [Google Scholar]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smith-Spencer, W. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev 2000, 100, 2705–2738. [Google Scholar]
- Turpeinen, R.; Virta, M.; Haggblom, M. Analysis of arsenic bioavailability in contaminated soils. Environ. Toxicol. Chem 2003, 22, 1–6. [Google Scholar]
- Tauriainen, S.; Virta, M.; Karp, M. Detecting bioavailable toxic metals and metalloids from natural water samples using luminescent sensor bacteria. Wat. Res 2000, 34, 2661–2666. [Google Scholar]
- Petanen, T.; Virta, M.; Karp, M.; Romantschuk, M. Construction and use broad host range mercury and arsenite sensor plasmids in the soil bacterium Pseudomonas fluorescens OS8. Microb. Ecol 2001, 41, 360–368. [Google Scholar]
- Heim, R.; Prasher, D.; Tsien, R. Wavelength mutations and post-translational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci USA 1994, 91, 12501–12504. [Google Scholar]
- Cubitt, A.B.; Heim, R.; Adams, S.R.; Boyd, A.E.; Gross, L.A.; Tsien, R.Y. Understanding, improving and using green fluorescent proteins. TIBS 1995, 20, 448–455. [Google Scholar]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.; Prasher, D. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar]
- Heim, R.; Cubitt, A.; Tsien, R. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar]
- Heim, R.; Tsien, R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol 1996, 6, 178–182. [Google Scholar]
- Cha, H.J.; Srivastava, R.; Vakharia, V.N.; Rao, G.; Bentley, W.E. Green fluorescent protein as a noninvasive stress probe in resting Escherichia coli cells. Appl. Environ. Microbiol 1999, 65, 409–414. [Google Scholar]
- Joyner, D.C.; Lindow, S.E. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 2000, 146, 2435–2445. [Google Scholar]
- Brandl, M.T.; Quinones, B.; Lindow, S.E. Heterogeneous transcription of an indoleacetic acid biosynthetic gene in Erwinia herbicola on plant surfaces. Proc. Natl. Acad. Sci. USA 2001, 98, 3454–3459. [Google Scholar]
- Leveau, J.H.; Lindow, S.E. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 2001, 98, 3446–3453. [Google Scholar]
- Miller, W.G.; Brandl, M.T.; Quinones, B.; Lindow, S.E. Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl. Environ. Microbiol 2001, 67, 1308–1317. [Google Scholar]
- Meighen, E.A. Molecular biology of bacterial bioluminescence. Microbiol. Rev 1991, 55, 123–142. [Google Scholar]
- Heitzer, A.; Malachowsky, K.; Thonnard, J.E.; Bienkowski, P.R.; White, D.C.; Sayler, G.S. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol 1994, 60, 1487–1494. [Google Scholar]
- Simpson, M.L.; Sayler, G.S.; Applegate, B.M.; Ripp, S.; Nivens, D.E.; Paulus, M.J.; Jellison, G.E. Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol 1998, 16, 332–338. [Google Scholar]
- Girotti, S.; Ferri, E.N.; Fumo, M.G.; Maiolini, E. Monitoring of environmental pollutants by bioluminescent bacteria. Analytica Chimica. Acta 2008, 608, 2–29. [Google Scholar]
- Winther-Larsen, H.C.; Josefsen, K.D.; Brautaset, T.; Valla, S. Parameters Affecting Gene Expression from the Pm Promoter in Gram-Negative Bacteria. Metab. Eng 2000, 2, 79–91. [Google Scholar]
- Phoenix, P.; Keane, A.; Patel, A.; Bergeron, H.; Ghoshal, S.; Lau, P.C. Characterization of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ. Microbiol 2003, 5, 1309–1327. [Google Scholar]
- Campbell, D.W.; Muller, C.; Reardon, K.F. Development of a fiber optic enzymatic biosensor for 1,2-dichloroethane. Biotechnol. Lett 2006, 28, 883–887. [Google Scholar]
- Leedjarv, A.; Ivask, A.; Virta, M.; Kahru, A. Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere 2006, 64, 1910–1919. [Google Scholar]
- Tizzard, A.C.; Bergsma, J.H.; Lloyd-Jones, G. A resazurin-based biosensor for organic pollutants. Biosens. Bioelectron 2006, 22, 759–763. [Google Scholar]
- Dawson, J.J.C.; Iroegbu, C.O.; Maciel, H.; Paton, G.I. Application of luminescent biosensors for monitoring the degradation and toxicity of BTEX compounds in soils. J. Appl. Microbiol 2008, 1, 141–151. [Google Scholar]
- Bontidean, I.; Mortari, A.; Leth, S.; Brown, N.L.; Karlson, U.; Larsen, M.M.; Vangvonsveld, J.; Corbisier, P.; Csoregi, E. Biosensors for detection of mercury in contaminated soils. Environ. Pollut 2004, 131, 255–262. [Google Scholar]
- Shetty, R.S.; Deo, S.K.; Liu, Y.; Daunert, S. Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol. Bioeng 2004, 88, 664–670. [Google Scholar]
- Magrisso, S.; Erel, Y.; Belkin, S. Microbial reporters of metal bioavailability. Microb. Biotechnol 2008, 4, 320–330. [Google Scholar]
- Farre, M.; Goncalves, C.; Lacorte, S.; Barcelo, D.; Alpendurada, M.F. Pesticide toxicity assessment using an electrochemical biosensor with Pseudomonas putida and a bioluminescence inhibition assay with Vibrio fischeri. Anal. Bioanal. Chem 2002, 373, 696–703. [Google Scholar]
- Chinalia, F.A.; Paton, G.I.; Killham. Physiological and toxicological characterisation of an engineered whole-cell biosensor. Bioresour. Technol 2008, 99, 714–721. [Google Scholar]
- Huang, W.E.; Wang, H.; Zheng, H.; Huang, L.; Singer, A.C.; Thompson, I.; Whiteley, A.S. Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ. Microbiol 2005, 7, 1339–1348. [Google Scholar]
- Lei, Y.; Mulchandani, P.; Wang, J.; Chen, W.; Mulchandani, A. Highly sensitive and selective amperometric microbial biosensor for direct determination of p-nitrophenyl-substituted organophosphate nerve agents. Environ. Sci. Technol 2005, 39, 8853–8857. [Google Scholar]
- Mulchandani, P.; Chen, W.; Mulchandani, A. Direct determination of p-nitrophenyl substituent organophosphorus nerve agents using a recombinant Pseudomonas putida js444-modified clark oxygen electrode. J. Agric. Food. Chem 2005, 53, 524–527. [Google Scholar]
- Norman, A.; Hansen, L.H.; Sorensen, S.J. A flow cytometry-optimized assay using an SOS-green fluorescent protein (SOS-GFP) whole-cell biosensor for the detection of genotoxins in complex environments. Mutat. Res 2006, 603, 164–172. [Google Scholar]
- Matsui, N.; Kaya, T.; Nagamine, K.; Yasukawa, T.; Shiku, H.; Matsue, T. Electrochemical mutagen screening using microbial chip. Biosens. Bioelectron 2006, 21, 1202–1209. [Google Scholar]
- Belkin, S.; Smulski, D.R.; Vollmer, A.C.; Van Dyk, T.K.; LaRossa, R.A. Oxidative stress detection with Escherichia coli harboring a katG’:lux fusion. Appl. Environ. Microbiol 1996, 62, 2252–2256. [Google Scholar]
- Choi, J.W.; Park, K.W.; Lee, D.B.; Lee, W.; Lee, W.H. Cell immobilization using self-assembled synthetic oligopeptide and its application to biological toxicity detection using surface plasmon resonance. Biosens. Bioelectron 2005, 20, 2300–2305. [Google Scholar]
- Neufeld, T.; Biran, D.; Popovtzer, R.; Erez, T.; Ron, E.Z.; Rishpon, J. Genetically engineered pfabA pfabR bacteria: an electrochemical whole cell biosensor for detection of water toxicity. Anal. Chem 2006, 78, 4952–4956. [Google Scholar]
- Sorensen, S.J.; Burmolle, M.; Hansen, L.H. Making bio-sense of toxicity: new developments in whole-cell biosensors. Curr. Opin. Biotechnol 2006, 17, 11–16. [Google Scholar]
- Cases, I.; de Lorenzo, V. Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol 2005, 3, 105–118. [Google Scholar]
- Jensen, S. Report of a new chemical hazard. New. Sci 1966, 32, 612. [Google Scholar]
- Risebrough, R.W.; Walker, W.; Schmidt, T.T.; deLappe, B.W.; Connors, C.W. Transfer of chlorinated biphenyls to Antarctica. Nature 1976, 264, 738–739. [Google Scholar]
- Triska, J.; Kuncova, G.; Mackova, M.; Novakova, H.; Paasivirta, J.; Lahtipera, M.; Vrchotova, N. Isolation and identification of intermediates from biodegradation of low chlorinated biphenyls (Delor 103). Chemosphere 2004, 54, 725–733. [Google Scholar]
- Weigel, W.; Wu, Q. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol 2000, 32, 1–15. [Google Scholar]
- Arnett, C.M.; Parales, J.V.; Haddock, J.D. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp. strain LB400. Appl. Environ. Microbiol 2000, 66, 2928–2933. [Google Scholar]
- Abraham, W.R.; Nogales, B.; Golyshin, P.N.; Pieper, D.H.; Timmis, K.N. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol 2002, 5, 246–253. [Google Scholar]
- Johnson, B.L.; Hicks, H.E.; Cibulas, W.; Faroon, O.; Ashizawa, A.E.; De Rosa, C.T.; Cogliano, V.J.; Clark, M. Public Health implications of exposure to polychlorinated biphenyls (PCBs). Agency for Toxic Substances and Disease Registry: Atlanta, GA, 1998; Available online: www.atsdr.cdc.gov/DT/pcb007.html/ (accessed on 24 January 2010).
- EWG. PCBs in Farmed Salmon. Internet Commun. 2005. Available online: http://www.ewg.org/reports/farmedPCBs/ (accessed on 1 February 2010).
- ATSDR. Proceedings of the expert panel workshop to evaluate the public health implications for the treatment and disposal of polychlorinated biphenyls contaminated waste. Internet Commun. 1993. Available online: http://www.atsdr.cdc.gov/HAC/PCB/b_pcb_c1.html/ (accessed on 1 February 2010).
- Kolar, A.B.; Hrsak, D.; Fingler, S.; Cetkovic, H.; Petric, I.; Kolic, N.U. PCB degrading potential of aerobic bacteria enriched from marine sediments International. Int. Biodeterior. Biodegrad 2007, 60, 16–24. [Google Scholar]
- Catelani, D.; Mosselmans, G.; Nienhaus, J.; Sorlini, C.; Treccani, V. Microbial degradation of aromatic hydrocarbons used as reator coolants. Experientia 1970, 26, 922–923. [Google Scholar]
- Lunt, D.; Evans, W.C. The microbial metabolism of biphenyl. J. Biochem 1970, 118, 54–55. [Google Scholar]
- Ahmed, M.; Focht, D.D. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can. J. Microbiol 1973, 19, 47–52. [Google Scholar]
- Furukawa, K.; Matsumura, F. Microbial metabolism of polychlorinated biphenyls-studies on relative degradability of polychlorinated biphenyl components by Alcaligenes sp. J. Agric. Food Chem 1976, 24, 251–256. [Google Scholar]
- Sylvestre, M.; Fauteux, J. A new facultative anaerobe capable of growth on 4-chlorobiphenyl. J. Gen. Appl. Microbiol 1982, 28, 61–72. [Google Scholar]
- Bedard, D.L.; Unterman, R.; Ropp, L.H.; Brennan, M.I.; Haber, M.L.; Johnson, C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol 1986, 51, 761–768. [Google Scholar]
- Asturias, J.A.; Timmis, K.N. Three different 2,3-dihydroxybiphenyl- 1,2 dioxygenase genes in the grampositive polychlorobiphenyl degrading bacterium Rhodococcus globerulus P6. J. Bacteriol 1993, 175, 4631–4640. [Google Scholar]
- Seto, M.; Kimbara, K.; Shimura, M.; Hatta, T.; Fukuda, M. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Applied Appl. Environ. Microbiol 1995, 61, 3353–3358. [Google Scholar]
- Sakai, M.; Masai, E.; Asami, H.; Sugiyama, K.; Kimbara, K.; Fukuda, M. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1. J. Biosci. Bioeng 2002, 93, 421–427. [Google Scholar]
- Sierra, A.; Valera, J.L.; Marina, M.L.; Laborda, F. Study of the biodegradation process of polychlorinated biphenyls in liquid medium and soil by a new isolated aerobic bacterium (Janibacter sp). Chemosphere 2003, 53, 609–618. [Google Scholar]
- Abramowicz, D.A. Aerobic and anaerobic biodegradation of PCBs: a review. Crit. Rev. Biotechnol 1990, 10, 241–251. [Google Scholar]
- Furukawa, K. Engineering dioxygenases for efficient degradation of environmental pollutants. Curr. Opin. Biotechnol 2000, 11, 244–249. [Google Scholar]
- Borja, J.; Taleon, D.M.; Auresenia, K.; Gallardo, S. Polychlorinated biphenyls and their biodegradation. Process Biochem 2005, 40, 1999–2013. [Google Scholar]
- Pieper, D.H. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol 2005, 67, 170–191. [Google Scholar]
- Sylvestre, M. Genetically modified organisms to remediate polychlorinated biphenyls. Where Do We Stand? Int. Biodeterior. Biodegrad 2004, 54, 153–162. [Google Scholar]
- Bedard, D.L. Polychlorinated biphenyls in aquatic sediments: environmental fate and outlook for biological treatment. In Dehalogenation: Microbial Processes and Environmental Applications; Haggblom, M.M., Bossert, I.D., Eds.; Kluwer Academic Publishers: Boston, MA, USA, 2003; pp. 443–465. [Google Scholar]
- Turner, K.; Xu, S.; Pasini, P.; Deo, S.; Bachas, L.; Daunert, S. Hydroxylated polychlorinated biphenyls detection based on a genetically engineering bioluminescent whole-cell sensing system. Anal. Chem 2007, 79, 5740–5745. [Google Scholar]
- Liu, X. Development and application of biosensor technologies for the biodegradation of environmental pollutants. PhD. Thesis. Institute of Technology, Carlow, Ireland. 2008. [Google Scholar]
- Franklin, F.C.; Bagdasarian, M.; Bagdasarian, M.M.; Timmis, K.N. Molecular andfunctional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 1981, 78, 7458–7462. [Google Scholar]
- Ramos, J.L.; Gonzalez-Carrero, M.; Timmis, K.N. Broad-host range expression vector containing manipulated meta-cleavagepathway regulatory elements of the TOL plasmid. FEBS Lett 1988, 226, 241–246. [Google Scholar]
- de Lorenzo, V.; Fernandez, M.H.; Jakubzik, U.; Timmis, K.N. Engineering of alkyl-and haloaromatic-responsive gene expression mini-transposon containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 1993, 130, 41–46. [Google Scholar]
- Herrero, M.; de Lorenzo, V.; Ensley, B.; Timmis, K.N. A T7 RNA polymerase-based system for the construction of Pseudomonas strain with phenotypes dependent on TOL-meta pathway effectors. Gene 1993, 134, 103–106. [Google Scholar]
- Cebolla, A.; Guzman, C.; de Lorenzo, V. Nondisruptive detection of activity of catabolic promoters of Pseudomonas putida with an antigenic surface reporter system. Appl. Environ. Microbiol 1996, 62, 214–220. [Google Scholar]
- Mermod, N.; Ramos, J.L.; Lehrbach, P.R.; Timmis, K.N. Vectors for regulated expression of cloned genes in a wide range of gram-negative bacteria. J. Bacteriol 1986, 167, 447–454. [Google Scholar]
- Blatny, J.M.; Brautaset, T.; Winther-Larsen, H.C.; Haugan, K.; Valla, S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol 1997, 63, 370–379. [Google Scholar]
- Blatny, J.M.; Brautaset, T.; Winther-Larsen, H.C.; Karunakaran, P.; Valla, S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 1997, 38, 35–51. [Google Scholar]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjorn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol 1998, 64, 2240–2246. [Google Scholar]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2, 4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol 1992, 58, 353–358. [Google Scholar]
- Goris, J.; Vos, P.D.; Caballero-Mellado, J.; Park, J.; Falsen, E.; Quensen, J.F., III; Tiedje, J.M.; Vandamme, P. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp nov. Int. J. Syst. Evol. Microbiol 2004, 54, 1677–168. [Google Scholar]
- Brazil, G.M.; Kenefick, L.; Callanan, M.; Haro, A.; De Lorenzo, V.; Dowling, D.N.; O'Gara, F. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol 1995, 61, 1946–1952. [Google Scholar]
- Villacieros, M.; Whelan, C.; Mackova, M.; Molgaard, J.; Sánchez-Contreras, M.; Lloret, J.; Aguirre de Cárcer, D.; Oruezábal, R.I.; Bolanos, L.; Macek, T.; Karlson, U.; Dowling, D.N.; Martín, M.; Rivilla, R. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol 2005, 71, 2687–2694. [Google Scholar]
- Bjerketorp, J.; Hakansson, S.; Belkin, S.; Jansson, J.K. Advances in preservation methods: keeping biosensor microorganisms alive and active. Curr. Opin. Biotechnol 2006, 17, 43–49. [Google Scholar]
- Marques, S.; Aranda-Olmedo, I.; Ramos, J.L. Controlling bacterial physiology for optimal expression of gene reporter constructs. Curr. Opin. Biotechnol 2006, 17, 50–56. [Google Scholar]
- Sticher, P.; Jaspers, M.C.; Stemmler, K.; Harms, H.; Zehnder, A.J.; van der Meer, J.R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol 1997, 63, 4053–4060. [Google Scholar]
- Premkumar, J.R.; Lev, O.; Rosen, R.; Belkin, S. Encapsulation of Luminous Recombinant E. coli in Sol-Gel Silicate Films. Adv. Mater 2001, 13, 1773–1775. [Google Scholar]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol 2003, 6, 206–212. [Google Scholar]
- Trogl, J.; Ripp, S.; Kuncova, G.; Sayler, G.S.; Churava, A.; Parik, P.; Demnerova, K.; Halova, J.; Kubicova, L. Selectivity of whole cell optical biosensor with immobilized bioreporter Pseudomonas fluorescens HK44. Sen. Actuat 2005, 107, 98–103. [Google Scholar]
- Cassidy, M.B.; Lee, H.; Trevors, J.T. Environmental applications of immobilized microbial cells: A review. J. Ind. Microbiol 1996, 16, 79–101. [Google Scholar]
- Mamo, G.; Gessesse, A. Immobilization of alkaliphilic Bacillus sp. cells for xylanase production using batch and continuous culture. Appl. Biochem. Biotechnol 2000, 87, 95–101. [Google Scholar]
- Kanasawud, P.; Hjorleifsdottir, S.; Holst, O.; Mattiasson, B. Studies on immobilization of the thermophilic bacterium Thermus aquaticus YT-1 by entrapment in various matrices. Appl. Microbiol. Biotechnol 1989, 31, 228–233. [Google Scholar]
- Heitzer, A.; Webb, O.F.; Thonnard, J.E.; Sayler, G.S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl. Envion. Microbiol 1992, 58, 1839–1846. [Google Scholar]
- Vorlop, K.; Muscat, A.; Beyersdorf, J. Entrapment of microbial cells withinpolyurethane hydrogel beads with the advantage of low toxicity. Biotechnol. Tech 1992, 6, 483–488. [Google Scholar]
- Wu, K.Y.A.; Wisecarver, K.D. Cell immobilization using PVA crosslinked with boric acid. Biotechnol. Bioeng 1992, 39, 447–449. [Google Scholar]
- Rietti-Shati, M.; Ronen, D.; Mandelbaum, R.T.J. Atrazine degradation by Pseudomonas strain ADP entrapped in sol-gel glass. J. Sol-Gel Sci. Technol 1996, 7, 77–79. [Google Scholar]
- Chu, Y.F.; Hsu, C.; Soma, P.; Lo, Y. Immobilization of bioluminescent Escherishia coli cells using natural and artificial fibers treated with polyethyleneimine. Bioresour. Technol 2009, 100, 3167–3174. [Google Scholar]
- Russo, A.; Moënne-Loccoz, Y.; Fedi, S.; Higgins, P.; Fenton, A.; Dowling, D.N.; O'Regan, M.; O'Gara, F. Improved delivery of biocontrol Pseudomonas and their antifungal metabolites using alginate polymers. Appl. Microbiol. Biotechnol. 1996, 44, 740–745. [Google Scholar]
- Power, B.; Dowling, D.N. Alginate studies on Pseudomonas fluorescens F113rifPCB. (Unpublished).
- Gilmartin, N. Biodegradation of PCBs: Characterisation of a Rhodococcus strain and possible function of a glutathione S-transferase (BphK) from Burkholderia LB400. PhD thesis. Institute of Technology Carlow, Carlow, Ireland. 2004. [Google Scholar]
- Dorn, E.; Hellwig, M.; Reineke, W.; Knackmuss, H.J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol 1974, 99, 61–70. [Google Scholar]
- Leff, L.G.; Leff, A.A. Use of green fluorescent protein to monitor survival of genetically engineered bacteria in aquatic environments. Appl. Environ. Microbiol 1996, 62, 3486–3488. [Google Scholar]
- Wang, Y.; Rawlings, M.; Gibson, D.T.; Labbe, D.; Bergeron, H.; Brousseau, R.; Lau, P.C. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol. Gen. Genet 1995, 246, 570–579. [Google Scholar]
- Fuchslin, H.P.; Ruegg, I.; van der Meer, J.R.; Egli, T. Effect of integration of a GFP reporter gene on fitness of Ralstonia eutropha during growth with 2,4-dichlorophenoxyacetic acid. Environ. Microbiol 2003, 5, 878–887. [Google Scholar]
- Marks, T.S.; Smith, A.R.; Quirk, A.V. Degradation of 4-Chlorobenzoic Acid by Arthrobacter sp. Appl._Environ. Microbiol 1984, 48, 1020–1025. [Google Scholar]
- Nawaz, M.S.; Chapatawala, K.D. Simultaneous degradation of acetonitrile and biphenyl by Pseudomonas aenrginosa. Can. J. Microbiol 1991, 37, 411–418. [Google Scholar]
- Sondossi, M.; Sylvestre, M.; Ahmad, D. Effects of chlorobenzoate on the Pseudomonas testosterone biphenyl and chlorobiphenyl degradation pathway. Appl. Environ. Microbiol 1992, 58, 485–495. [Google Scholar]
- Harms, H.; Rime, J.; Leupin, O.; Hug, S.J.; van der Meer, J. Effect of groundwater composition on arsenic detection by bacterial biosensors. Microchimica Acta 2005, 151, 217–222. [Google Scholar]
- Pettigrew, C.A.; Breen, A.; Corcoran, C.; Sayler, G.S. Chlorinated biphenyl mineralization by individual populations and consortia of freshwater bacteria. Appl. Environ. Microbiol 1990, 56, 2036–2045. [Google Scholar]
- Hickey, W.J.; Searles, D.B.; Focht, D.D. Enhanced mineralization of polychlorinated biphenyls in soil inoculated with chlorobenzoate-degrading bacteria. Appl. Environ. Microbiol 1993, 59, 1194–1200. [Google Scholar]
- Di Gioia, D.; Bertin, L.; Zanaroli, G.; Marchetti, L.; Fava, F. Polychlorinated biphenyl degradation in aqueous wastes by employing continuous fixed-bed bioreactors. Process Biochemistry 2006, 41, 935–940. [Google Scholar]
- Ikariyama, Y.; Nishiguchi, S.; Koyama, T.; Kobatake, E.; Aizawa, M. Fiber-optic-based biomonitoring of benzene derivatives by recombinant E. coli bearing luciferase gene-fused TOL-plasmid immobilized on the fiber-optic end. Anal. Chem 1997, 69, 2600–2605. [Google Scholar]
- Gil, G.C.; Mitchell, R.J.; Chang, S.T.; Gu, M.B. A biosensor for the detection of gas toxicity using a recombinant bioluminescent bacterium. Biosens. Bioelectron 2000, 15, 23–30. [Google Scholar]
- Feliciano, J.; Liu, Y.; Daunert, S. Novel reporter gene in a fluorescent-based whole cell sensing system. Biotechnol. Bioeng 2006, 93, 989–997. [Google Scholar]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).