Biodiversity Dynamics on Islands: Explicitly Accounting for Causality in Mechanistic Models
Abstract
:1. Introduction
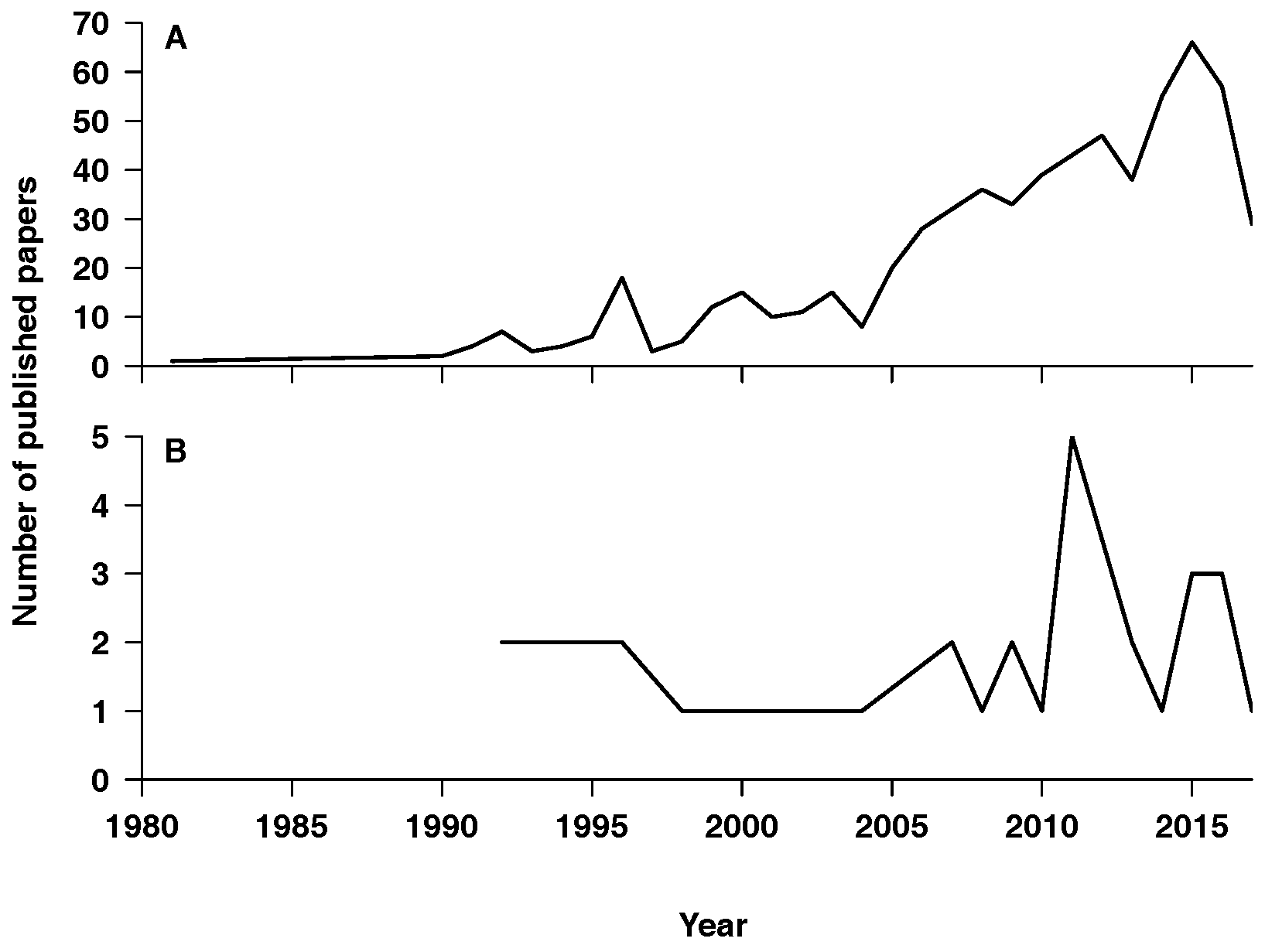
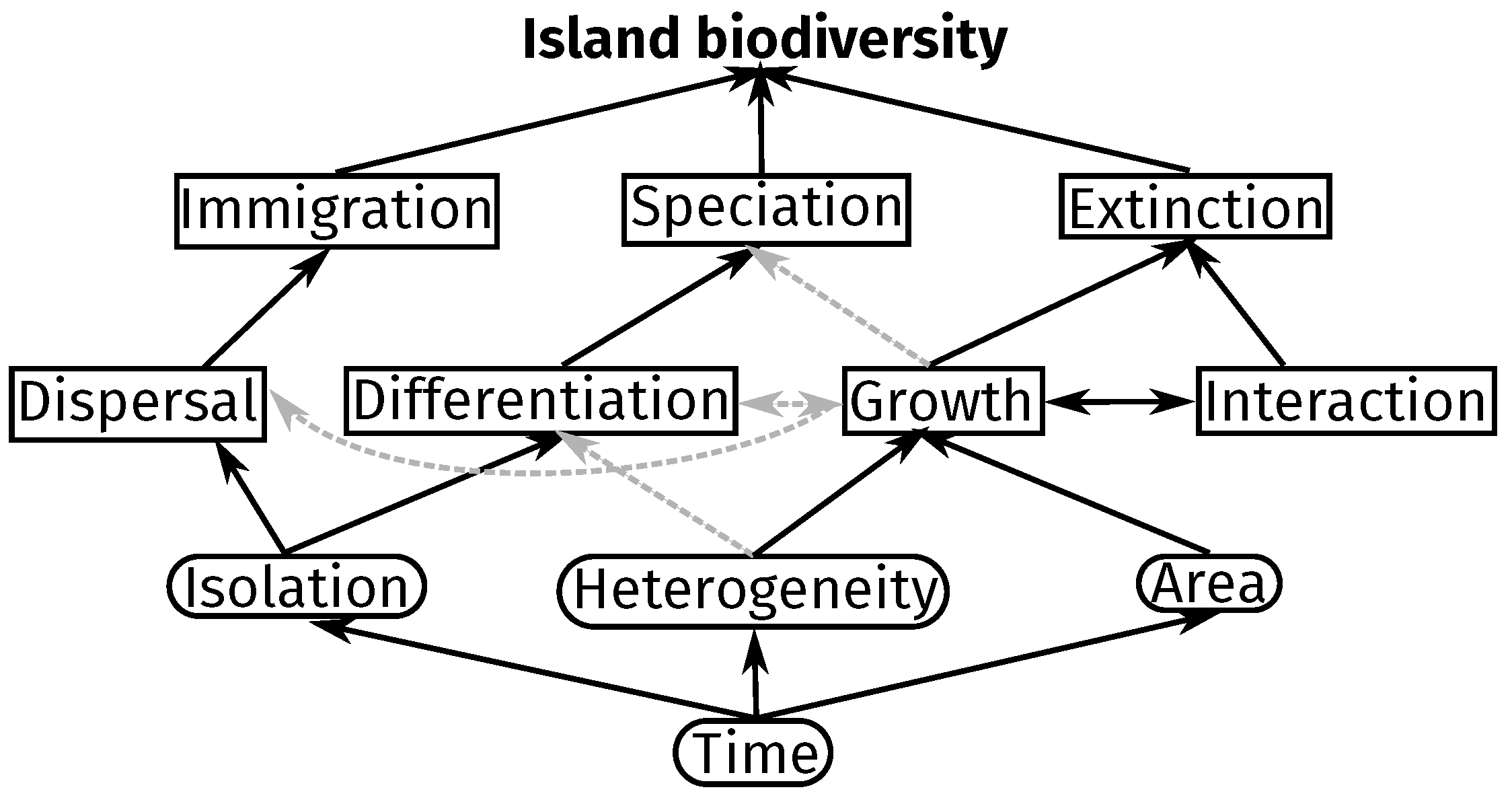
2. Results
2.1. Description
2.2. Properties
2.3. Findings
3. Discussion
3.1. General Modeling Trends
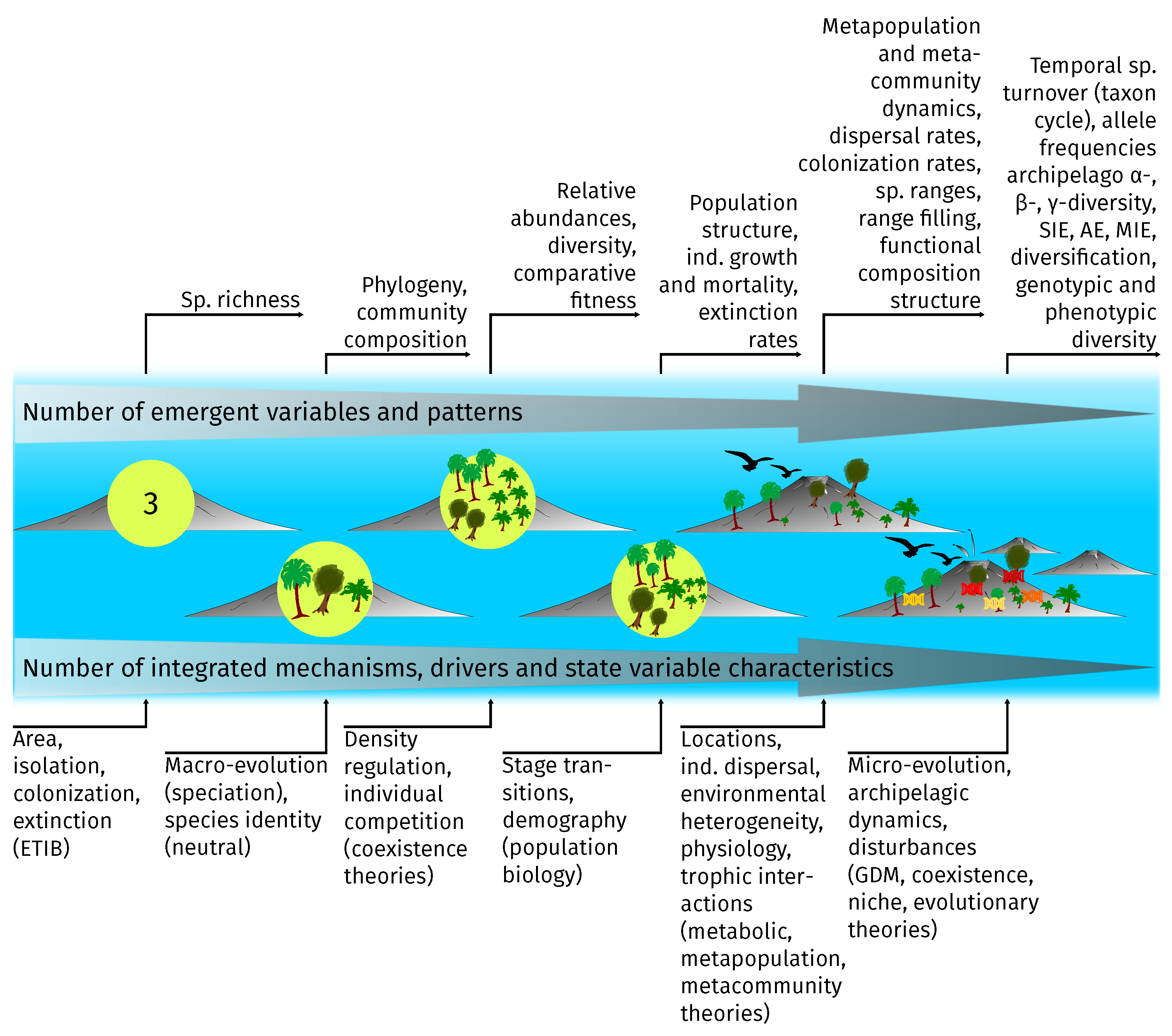
3.2. Integrated Processes and Emergent Patterns
3.3. Limitations and Modeling Agenda
3.4. Conclusions
4. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vitousek, P.M. Oceanic islands as model systems for ecological studies. J. Biogeogr. 2002, 29, 573–582. [Google Scholar] [CrossRef]
- Warren, B.H.; Simberloff, D.; Ricklefs, R.E.; Aguilée, R.; Condamine, F.L.; Gravel, D.; Morlon, H.; Mouquet, N.; Rosindell, J.; Casquet, J.; et al. Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol. Lett. 2015, 18, 200–217. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, P.; Kreft, H. Quantifying island isolation—Insights from global patterns of insular plant species richness. Ecography 2013, 36, 417–429. [Google Scholar] [CrossRef]
- Singer, A.; Johst, K.; Banitz, T.; Fowler, M.S.; Groeneveld, J.; Gutiérrez, A.G.; Hartig, F.; Krug, R.M.; Liess, M.; Matlack, G.; et al. Community dynamics under environmental change: How can next generation mechanistic models improve projections of species distributions? Ecol. Model. 2016, 326, 63–74. [Google Scholar] [CrossRef]
- Urban, M.; Bocedi, G.; Hendry, A.; Mihoub, J.B.; Pe’er, G.; Singer, A.; Bridle, J.; Crozier, L.; de Meester, L.; Godsoe, W.; et al. Improving the forecast for biodiversity under climate change. Science 2016, 353, aad8466. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.S.; Wiegand, K.; Kreft, H. Interactions between ecological, evolutionary, and environmental processes unveil complex dynamics of island biodiversity. bioRxiv 2017. [Google Scholar] [CrossRef]
- MacArthur, R.H.; Wilson, E.O. An equilibrium theory of insular zoogeography. Evolution 1963, 17, 373–387. [Google Scholar] [CrossRef]
- Whittaker, R.J.; Triantis, K.A.; Ladle, R.J. A general dynamic theory of oceanic island biogeography. J. Biogeogr. 2008, 35, 977–994. [Google Scholar] [CrossRef]
- Simberloff, D.S.; Wilson, E.O. Experimental zoogeography of islands: The colonization of empty islands. Ecology 1969, 50, 278–296. [Google Scholar] [CrossRef]
- Borregaard, M.K.; Amorim, I.R.; Borges, P.A.; Cabral, J.S.; Fernández-Palacios, J.M.; Field, R.; Heaney, L.R.; Kreft, H.; Matthews, T.J.; Olesen, J.M.; et al. Oceanic island biogeography through the lens of the general dynamic model: Assessment and prospect. Biol. Rev. 2017, 92, 830–853. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.J.; Fernández-Palacios, J.M. Island Biogeography: Ecology, Evolution, and Conservation; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Dormann, C.F.; Schymanski, S.J.; Cabral, J.; Chuine, I.; Graham, C.; Hartig, F.; Kearney, M.; Morin, X.; Römermann, C.; Schröder, B.; et al. Correlation and process in species distribution models: Bridging a dichotomy. J. Biogeogr. 2012, 39, 2119–2131. [Google Scholar] [CrossRef]
- Weigelt, P.; Jetz, W.; Kreft, H. Bioclimatic and physical characterization of the world’s islands. Proc. Natl. Acad. Sci. USA 2013, 110, 15307–15312. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.S.; Weigelt, P.; Kissling, W.D.; Kreft, H. Biogeographic, climatic and spatial drivers differentially affect α-, β- and γ-diversities on oceanic archipelagos. Proc. R. Soc. Lond. B Biol. Sci. 2014, 281, 20133246. [Google Scholar] [CrossRef] [PubMed]
- Borges, P.A.; Brown, V.K. Effect of island geological age on the arthropod species richness of Azorean pastures. Biol. J. Linn. Soc. 1999, 66, 373–410. [Google Scholar] [CrossRef]
- Buskirk, R.E. Zoogeographic patterns and tectonic history of Jamaica and the northern Caribbean. J. Biogeogr. 1985, 12, 445–461. [Google Scholar] [CrossRef]
- Meyer, K.M.; Leveau, J.H. Microbiology of the phyllosphere: A playground for testing ecological concepts. Oecologia 2012, 168, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Maignien, L.; DeForce, E.A.; Chafee, M.E.; Eren, A.M.; Simmons, S.L. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 2014, 5, e00682-13. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, F.; Moloney, K.A.; Schurr, F.M.; Köchy, M.; Schwager, M. The state of plant population modelling in light of environmental change. Perspect. Plant Ecol. Evol. Syst. 2008, 9, 171–189. [Google Scholar] [CrossRef]
- Evans, M.R.; Grimm, V.; Johst, K.; Knuuttila, T.; De Langhe, R.; Lessells, C.M.; Merz, M.; O’Malley, M.A.; Orzack, S.H.; Weisberg, M.; et al. Do simple models lead to generality in ecology? Trends Ecol. Evol. 2013, 28, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.S.; Valente, L.; Hartig, F. Mechanistic simulation models in macroecology and biogeography: State-of-art and prospects. Ecography 2017, 40, 267–280. [Google Scholar] [CrossRef]
- Villa, F.; Rossi, O.; Sartore, F. Understanding the role of chronic environmental disturbance in the context of island biogeographic theory. Environ. Manag. 1992, 16, 653–666. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Maher, S.P.; Brown, R.M. Species interactions during diversification and community assembly in an island radiation of shrews. PLoS ONE 2011, 6, e21885. [Google Scholar] [CrossRef] [PubMed]
- Cantrell, R.; Cosner, C.; Hutson, V. Spatially explicit models for the population dynamics of a species colonizing an island. Math. Biosci. 1996, 136, 65–107. [Google Scholar] [CrossRef]
- Chen, X.; Jiao, J.; Tong, X. A generalized model of island biogeography. Sci. China Life Sci. 2011, 54, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Borregaard, M.K.; Matthews, T.J.; Whittaker, R.J. The general dynamic model: Towards a unified theory of island biogeography? Glob. Ecol. Biogeogr. 2015, 25, 805–816. [Google Scholar] [CrossRef]
- Durrett, R.; Levin, S. Spatial models for species-area curves. J. Theor. Biol. 1996, 179, 119–127. [Google Scholar] [CrossRef]
- Rosindell, J.; Harmon, L.J. A unified model of species immigration, extinction and abundance on islands. J. Biogeogr. 2013, 40, 1107–1118. [Google Scholar] [CrossRef]
- Valente, L.M.; Phillimore, A.B.; Etienne, R.S. Equilibrium and non-equilibrium dynamics simultaneously operate in the Galápagos islands. Ecol. Lett. 2015, 18, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, S. The Unified Neutral Theory of Species Abundance and Diversity; Hubbell, SP (2004) Quarterly Review of Biology; Princeton University Press: Princeton, NJ, USA, 2001; Volume 79, pp. 96–97. [Google Scholar]
- Gavrilets, S.; Vose, A. Case studies and mathematical models of ecological speciation. 2. Palms on an oceanic island. Mol. Ecol. 2007, 16, 2910–2921. [Google Scholar] [CrossRef] [PubMed]
- Rosindell, J.; Cornell, S.J.; Hubbell, S.P.; Etienne, R.S. Protracted speciation revitalizes the neutral theory of biodiversity. Ecol. Lett. 2010, 13, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Kadmon, R.; Allouche, O. Integrating the effects of area, isolation, and habitat heterogeneity on species diversity: A unification of island biogeography and niche theory. Am. Nat. 2007, 170, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Avery, J.D.; Fonseca, D.M.; Campagne, P.; Lockwood, J.L. Cryptic introductions and the interpretation of island biodiversity. Mol. Ecol. 2013, 22, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Iwasa, Y. Parapatric speciation in three islands: dynamics of geographical configuration of allele sharing. Open Sci. 2017, 4, 160819. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.A.; Shnerb, N.M. Generalized model of island biodiversity. Phys. Rev. E 2015, 91, 042705. [Google Scholar] [CrossRef] [PubMed]
- Valente, L.M.; Etienne, R.S.; Phillimore, A.B. The effects of island ontogeny on species diversity and phylogeny. Proc. R. Soc. Lond. B Biol. Sci. 2014, 281, 20133227. [Google Scholar] [CrossRef] [PubMed]
- Birand, A.; Howard, D.J. The relationship between proportion of endemics and species diversity on islands: Expectations from a null model. Ecography 2008, 31, 286–288. [Google Scholar] [CrossRef]
- Rosindell, J.; Phillimore, A.B. A unified model of island biogeography sheds light on the zone of radiation. Ecol. Lett. 2011, 14, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Gascuel, F.; Laroche, F.; Bonnet-Lebrun, A.S.; Rodrigues, A.S. The effects of archipelago spatial structure on island diversity and endemism: predictions from a spatially-structured neutral model. Evolution 2016, 70, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- De Blasio, F. Diversity variation in isolated environments: Species-area effects from a stochastic model. Ecol. Model. 1998, 111, 93–98. [Google Scholar]
- Wissel, C.; Maier, B. A stochastic model for the species-area relationship. J. Biogeogr. 1992, 19, 355–361. [Google Scholar] [CrossRef]
- Johnson, K.P.; Adler, F.R.; Cherry, J.L. Genetic and phylogenetic consequences of island biogeography. Evolution 2000, 54, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, R.E.; Bermingham, E. Application of Johnson et al.’s speciation threshold model to apparent colonization times of island biotas. Evolution 2004, 58, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Emerson, B.C.; Kolm, N. Species diversity can drive speciation. Nature 2005, 434, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; He, F. Speciation and endemism under the model of island biogeography. Ecology 2009, 90, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Hortal, J.; Triantis, K.A.; Meiri, S.; Thébault, E.; Sfenthourakis, S. Island species richness increases with habitat diversity. Am. Nat. 2009, 174, E205–E217. [Google Scholar] [CrossRef] [PubMed]
- Warren, P.B. Biodiversity on island chains: Neutral model simulations. Phys. Rev. E 2010, 82, 051922. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A. Species abundance distributions in neutral models with immigration or mutation and general lifetimes. J. Math. Biol. 2011, 63, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Gravel, D.; Massol, F.; Canard, E.; Mouillot, D.; Mouquet, N. Trophic theory of island biogeography. Ecol. Lett. 2011, 14, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. An island biogeography model for beta diversity and endemism: The roles of speciation, extinction and dispersal. Int. J. Biomath. 2015, 8, 1550011. [Google Scholar] [CrossRef]
- Jacquet, C.; Mouillot, D.; Kulbicki, M.; Gravel, D. Extensions of Island Biogeography Theory predict the scaling of functional trait composition with habitat area and isolation. Ecol. Lett. 2017, 20, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.H.; Kodric-Brown, A. Turnover rates in insular biogeography: Effect of immigration on extinction. Ecology 1977, 58, 445–449. [Google Scholar] [CrossRef]
- Lomolino, M.V. The Target Area Hypothesis: The Influence of Island Area on Immigration Rates of Non-Volant Mammals. Oikos 1990, 57, 297–300. [Google Scholar] [CrossRef]
- Keil, P.; Herben, T.; Rosindell, J.; Storch, D. Predictions of Taylor’s power law, density dependence and pink noise from a neutrally modeled time series. J. Theor. Biol. 2010, 265, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Lawton, J.H. Are There General Laws in Ecology? Oikos 1999, 84, 177–192. [Google Scholar] [CrossRef]
- Valente, L.; Etienne, R.S.; Dávalos, L.M. Recent extinctions disturb path to equilibrium diversity in Caribbean bats. Nat. Ecol. Evol. 2017, 1, 0026. [Google Scholar] [CrossRef]
- Terborgh, J.W. Toward a trophic theory of species diversity. Proc. Nat. Acad. Sci. USA 2015, 112, 11415–11422. [Google Scholar] [CrossRef] [PubMed]
- Barnes, N. Publish your computer code: It is good enough. Nature 2010, 467, 753. [Google Scholar] [CrossRef] [PubMed]
- Mislan, K.; Heer, J.M.; White, E.P. Elevating the status of code in ecology. Trends Ecol. Evol. 2016, 31, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Darwin, C. On the Origin of the Species by Natural Selection; J. Murray: London, UK, 1859. [Google Scholar]
- Schiffers, K.; Bourne, E.C.; Lavergne, S.; Thuiller, W.; Travis, J.M. Limited evolutionary rescue of locally adapted populations facing climate change. Philos. Trans. R. Soc. B 2013, 368, 20120083. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M. Species diversity and genetic diversity: Parallel processes and correlated patterns. Am. Nat. 2005, 166, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Laroche, F.; Jarne, P.; Lamy, T.; David, P.; Massol, F. A neutral theory for interpreting correlations between species and genetic diversity in communities. Am. Nat. 2014, 185, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Aguilée, R.; Claessen, D.; Lambert, A. Adaptive radiation driven by the interplay of eco-evolutionary and landscape dynamics. Evolution 2013, 67, 1291–1306. [Google Scholar] [CrossRef] [PubMed]
- Thibert-Plante, X.; Gavrilets, S. Evolution of mate choice and the so-called magic traits in ecological speciation. Ecol. Lett. 2013, 16, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Gascuel, F.; Ferrière, R.; Aguilée, R.; Lambert, A. How ecology and landscape dynamics shape phylogenetic trees. Syst. Biol. 2015, 64, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Grimm, V.; Frank, K.; Jeltsch, F.; Brandl, R.; Uchmański, J.; Wissel, C. Pattern-oriented modelling in population ecology. Sci. Total Environ. 1996, 183, 151–166. [Google Scholar] [CrossRef]
- Grimm, V.; Revilla, E.; Berger, U.; Jeltsch, F.; Mooij, W.M.; Railsback, S.F.; Thulke, H.H.; Weiner, J.; Wiegand, T.; DeAngelis, D.L. Pattern-oriented modeling of agent-based complex systems: Lessons from ecology. Science 2005, 310, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Haydon, D.; Radtkey, R.R.; Pianka, E.R. Experimental Biogeography: Interactions between Stochastic, Historical, and Ecological Processes in a Model Archipelago. In Species Diversity in Ecological Communities: Historical and Geographical Perspectives; The University of Chicago Press: Chicago, IL, USA, 1993; pp. 117–130. [Google Scholar]
- Weigelt, P.; Steinbauer, M.J.; Cabral, J.S.; Kreft, H. Late Quaternary climate change shapes island biodiversity. Nature 2016, 532, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Palacios, J.M.; Rijsdijk, K.F.; Norder, S.J.; Otto, R.; Nascimento, L.; Fernández-Lugo, S.; Tjørve, E.; Whittaker, R.J. Towards a glacial-sensitive model of island biogeography. Glob. Ecol. Biogeogr. 2016, 25, 817–830. [Google Scholar] [CrossRef]
- Helmus, M.R.; Mahler, D.L.; Losos, J.B. Island biogeography of the Anthropocene. Nature 2014, 513, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Kubisch, A.; Winter, A.M.; Fronhofer, E.A. The downward spiral: Eco-evolutionary feedback loops lead to the emergence of ‘elastic’ranges. Ecography 2015, 39, 261–269. [Google Scholar] [CrossRef]
- Prevedello, J.A.; Gotelli, N.J.; Metzger, J.P. A stochastic model for landscape patterns of biodiversity. Ecol. Monogr. 2016, 86, 462–479. [Google Scholar] [CrossRef]
- Stier, A.C.; Hanson, K.M.; Holbrook, S.J.; Schmitt, R.J.; Brooks, A.J. Predation and landscape characteristics independently affect reef fish community organization. Ecology 2014, 95, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.4-1. 2016.
- De Leeuw, J.; Mair, P. Multidimensional Scaling Using Majorization: SMACOF in R. J. Stat. Softw. 2009, 31, 1–30. [Google Scholar] [CrossRef]


| Study | Year | System (h/r) | Theoretical Background | Processes (st/dn) * | No. par. | Stoc. * | Spt. Exp. * | Ev. * | Nt. * | No. nn. * | Agent Level * | Focal Level | Investigated Patterns |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wissel and Maier [42] | 1992 | MI (h) | ETIB, target effect | disp., repr., (st) | 7 | yes | no | no | yes | 0 | pop. | com. | SAR |
| Villa et al. [22] | 1992 | MA (h) | ETIB | disp., repr., dist., (st) | 9 | yes | yes | no | no | 4 | ind. | com. | SAR, eq. |
| Durrett and Levin [27] | 1996 | A (h) | ETIB | disp., comp., spec., (st) | 4 | yes | yes | yes | yes | 0 | sp. | com. | SAR |
| Cantrell et al. [24] | 1996 | MI (h) | ETIB, Allee effects | disp., repr., comp., (st) | 6 | no | yes | no | no | 2 | pop. | sp. | trait-dep. colonization |
| De Blasio [41] | 1998 | MI (h) | ETIB | disp., comp., (st) | 5 | yes | yes | no | yes | 0 | pop. | com. | SAR |
| Johnson et al. [43] | 2000 | MI (h) | ETIB, genetic divergence | disp., spec., ext., divergence, (st) | 7 | no | no | yes | yes | 0 | pop. | pop. | AID of diverging lineages over time |
| Ricklefs and Bermingham [44] | 2004 | MI (h) | [43] | mut., spec., (st) | 6 | yes | no | yes | yes | 0 | sp. | sp. | SIR, divergence-dep. no. of lineages |
| Gavrilets and Vose [31] | 2007 | MI (h) | sympatric spec. | disp., sex. repr., mut., (st) | 6 | yes | yes | yes | no | 5 | ind. | com. | time- & niche-dep. trait distribution |
| Kadmon and Allouche [33] | 2007 | MI (h) | ETIB, UNTB, niche | disp., repr., ext., (st) | 5 | no | no | no | no | 1 | ind. | com. | SAR, SHR |
| Birand and Howard [38] | 2008 | A (h) | [45] | disp., spec., ext., (st) | 4 | yes | no | yes | yes | 0 | sp. | com. | sp.-div.-dep.E |
| Chen and He [46] | 2009 | MI (h) | ETIB | disp., spec., ext., (st) | 4 | no | no | yes | yes | 0 | sp. | com. | rate-dep. E |
| Hortal et al. [47] | 2009 | MI (r) | [33] | disp, repr., (st) | 5 | yes | no | no | no | 2 | ind. | com. | SAR, SHR |
| Warren [48] | 2010 | MA (h) | UNTB | disp, spec., (st) | 6 | yes | no | yes | yes | 0 | ind. | com. | SIR, rate-dep. -div. |
| Rosindell and Phillimore [39] | 2011 | MI (r) | UNTB | disp., repr., spec., (st) | 5 | yes | no | yes | yes | 0 | ind. | com. | SAD, SAR, SIR |
| Lambert [49] | 2011 | MI (h) | ETIB, UNTB | disp., repr., mut., (st) | 6 | yes | no | yes | yes | 0 | pop. | com. | rate-dep. SAD |
| Esselstyn et al. [23] | 2011 | A (r) | not explicit | disp., (st) | 1 | yes | yes | yes | yes | 0 | sp. | sp. | radiation history |
| Gravel et al. [50] | 2011 | MI (r) | ETIB, trophic th. | disp, ext., (st) | 2 | yes | no | no | no | 1 | sp. | com. | SAR |
| Chen et al. [25] | 2011 | MI (h) | GDM, ETIB | disp, spec., ext., (dn) | 12 | no | no | yes | yes | 0 | sp. | com. | time-dep. E & sp. richness |
| Avery et al. [34] | 2013 | MI (r) | not explicit | disp., repr., mut., (dn) | 6 | yes | no | yes | yes | 0 | ind. | pop. | colonization history |
| Rosindell and Harmon [28] | 2013 | MI (h) | UNTB, ETIB | disp., death, (st) | 6 | yes | no | no | yes | 0 | ind. | com. | rate-dep. SAD |
| Valente et al. [37] | 2014 | MI (h) | GDM, ETIB | disp., spec., ext., (dn) | 4 | yes | no | yes | yes | 0 | sp. | com. | rate- & time-dep. sp. richness |
| Chen [51] | 2015 | MA (h) | [45,46] | disp., ext., spec., (st) | 3 | no | no | yes | yes | 0 | sp. | sp. | rate-dep. E & -div. |
| Kessler and Shnerb [36] | 2015 | MI (h) | ETIB, UNTB, Lotka-Volterra | disp., repr., comp., (st) | 4 | yes | no | no | no | 1 | pop. | com. | comp.-dep.com. assembly |
| Valente et al. [29] | 2015 | MI (r) | UNTB, ETIB | disp., spec., ext., (st) | 6 | yes | no | yes | yes | 0 | sp. | com. | time-dep. phyl. rates |
| Borregaard et al. [26] | 2016 | I (h) | GDM | disp., spec., ext., (dn) | 13 | no | no | yes | yes | 0 | sp. | com. | time-dep. SAR, SHR |
| Gascuel et al. [40] | 2016 | MA (h) | UNTB, [39] | repr., disp., spec., (st) | 6 | yes | yes | yes | yes | 0 | ind. | com. | SIR, E in A |
| Jacquet et al. [52] | 2017 | MI (h) | ETIB, trophic th. | disp., ext., (st) | 14 | yes | no | no | no | 1 | sp. | com. | AID of traits |
| Yamaguchi and Iwasa [35] | 2017 | A (h) | pop. genetics | disp., mut., (st) | 4 | yes | no | yes | yes | 0 | pop. | pop. | time-dep. SIR |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leidinger, L.; Cabral, J.S. Biodiversity Dynamics on Islands: Explicitly Accounting for Causality in Mechanistic Models. Diversity 2017, 9, 30. https://doi.org/10.3390/d9030030
Leidinger L, Cabral JS. Biodiversity Dynamics on Islands: Explicitly Accounting for Causality in Mechanistic Models. Diversity. 2017; 9(3):30. https://doi.org/10.3390/d9030030
Chicago/Turabian StyleLeidinger, Ludwig, and Juliano Sarmento Cabral. 2017. "Biodiversity Dynamics on Islands: Explicitly Accounting for Causality in Mechanistic Models" Diversity 9, no. 3: 30. https://doi.org/10.3390/d9030030
APA StyleLeidinger, L., & Cabral, J. S. (2017). Biodiversity Dynamics on Islands: Explicitly Accounting for Causality in Mechanistic Models. Diversity, 9(3), 30. https://doi.org/10.3390/d9030030




