Abstract
Habitat fragmentation can promote patches of small and isolated populations, gene flow disruption between those populations, and reduction of local and total genetic variation. As a consequence, these small populations may go extinct in the long-term. The ocelot (Leopardus pardalis), originally distributed from Texas to southern Brazil and northern Argentina, has been impacted by habitat fragmentation throughout much of its range. To test whether habitat fragmentation has already induced genetic differentiation in an area where this process has been documented for a larger felid (jaguars), we analyzed molecular variation in ocelots inhabiting two Atlantic Forest fragments, Morro do Diabo (MD) and Iguaçu Region (IR). Analyses using nine microsatellites revealed mean observed and expected heterozygosity of 0.68 and 0.70, respectively. The MD sampled population showed evidence of a genetic bottleneck under two mutational models (TPM = 0.03711 and SMM = 0.04883). Estimates of genetic structure (FST = 0.027; best fit of k = 1 with STRUCTURE) revealed no meaningful differentiation between these populations. Thus, our results indicate that the ocelot populations sampled in these fragments are still not significantly different genetically, a pattern that strongly contrasts with that previously observed in jaguars for the same comparisons. This observation is likely due to a combination of two factors: (i) larger effective population size of ocelots (relative to jaguars) in each fragment, implying a slower effect of drift-induced differentiation; and (ii) potentially some remaining permeability of the anthropogenic matrix for ocelots, as opposed to the observed lack of permeability for jaguars. The persistence of ocelot gene flow between these areas must be prioritized in long-term conservation planning on behalf of these felids.
1. Introduction
The long-term viability of a species is strongly influenced by its genetic variation. Habitat destruction and fragmentation caused by increasing deforestation, hunting, and other human interference have been promoting population declines in a number of tropical animal species, especially large mammals such as canids and felids [1,2,3]. Reduced genetic variation, inbreeding, and fitness decrease are expected genetic consequences of population decline [4,5], and often accompany population fragmentation [3]. Fragmented populations can become isolated, thus impairing gene flow, promoting genetic divergence between them [6,7] and reducing local effective population sizes and genetic diversity [8,9]. Nevertheless, dispersal capability across a human-dominated matrix can maintain the genetic equilibrium within the remaining populations [10,11,12]. For the effective management and conservation of an endangered species one needs to know how its genetic variation is distributed across the landscape, and if there is evidence of gene flow interruption induced by habitat fragmentation.
The ocelot (Leopardus pardalis) is a medium-sized felid that originally ranged from Texas, USA to southern Brazil and Argentina [13]. Until the 1980s, it was intensely hunted and suffered a decline due to the international interest in its pelts [14]. Currently, skin trade is prohibited and the main threats to this species are habitat loss and fragmentation [15]. In this context, recent analyses have indicated that the ocelots in the United States and Mexico suffer with habitat loss and population isolation, with detectable genetic depletion as a consequence [16,17,18].
Most of the habitats that ocelots occur in are currently highly fragmented, such as the Atlantic Forest in South America. About 80% of this ecosystem is composed by fragments of less than 50 hectares (ha), most of which are isolated by long distances [19]. The Interior Atlantic Forest, also known as Upper Parana Atlantic Forest (UPAF) ecoregion (471,204 km2) encompasses southwestern Brazil, northern Argentina, and eastern Paraguay. Most of this original area has undergone significant forest loss and fragmentation since the 1970s. In spite of the recent time frame of this fragmentation process, previous analyses using microsatellite loci have demonstrated that jaguars (Panthera onca) remaining in these forested patches have become significantly differentiated due to drift-induced loss of diversity [9]. This differentiation was strongest when the comparison involved the two most distant forest patches, Morro do Diabo (MD) in the north and Iguaçu region (IR) in the south. Those results indicated that jaguar effective population sizes were very small in the remaining patches, and that the intervening, human-dominated matrix was effectively preventing gene flow among them. Testing if a similar process is occurring with other predators in this region is a critical issue in the context of conservation planning, as different species may be affected differently by habitat loss and fragmentation.
The ocelot’s ability to cross rivers and to use a natural corridor connecting different forest fragments has been detected by radio-telemetry in the Atlantic Forest [20,21]. On the other hand, the species was found to be incapable of crossing the human-dominated matrix between protected areas in Texas, and ~30 km were enough to hinder their dispersal and to induce significant genetic differentiation [17].
In this context, the goal of the present study was to analyze the genetic structure of ocelot populations inhabiting the IR and MD fragments of the Upper Parana Atlantic Forest ecoregion. We tested if habitat fragmentation has produced detectable genetic discontinuity between the studied populations, and if any loss of diversity can be observed in these forest remnants. By comparing our findings to previous analyses targeting jaguars in the same areas, and ocelots in different areas, we draw conclusions regarding the history of genetic differentiation due to landscape fragmentation in this region and its conservation implications.
2. Materials and Methods
2.1. Study Area and Sampling
We used samples from two protected areas located in the Brazilian Interior Atlantic Forest, within the Upper Paraná Atlantic Forest (UPAF) ecoregion. The Morro do Diabo (MD, n = 14) State Park spans ca. 34,000 ha and is located at the northern end of this ecoregion, in western São Paulo state (Figure 1). The landscape surrounding this park is a mosaic of farmlands, roads, and remaining forest fragments (totaling an area of about 10,000 ha). The other sampling site, referred to here as Iguaçu Region (IR, n = 18), comprised the Iguaçu National Park (Brazil) and the Iguazú National Park (Argentina), with a total area of ca. 255,000 ha. These areas are located at the southern end of the UPAF and are connected to each other and also to the forest remnants comprising the Green Corridor of Misiones, in Argentina. The MD and IR are located ca. 400 km from each other, and despite the presence of other protected areas in between, there is evidence indicating that they have been isolated from each other by human-induced fragmentation for 30–40 years [14].
2.2. Laboratory Protocols
The samples (n = 32) were derived from animals previously captured for independent ecological studies in these areas. Fourteen blood and tissue samples were collected in MD in 1999–2000, and 18 blood samples in the IR from 1993 to 2000. We extracted the DNA from all samples using a protocol modified from Sambrook et al. [22], and assessed genetic variation using 12 microsatellite loci initially developed for Felis catus [23]: FCA008, F42, FCA053, FCA077, FCA096, F98, FCA124, FCA146, FCA391, FCA424, FCA441 and FCA453. We used the fluorescent labeling protocol of Schuelke [24] and synthesized forward primers adding an M13 (-21) tail (5′-TGTAAAACGACGGCCAGT-3′) to their 5′ ends. The polymerase chain reaction (PCR) was performed in a final volume of 10 µL, containing 20 ng of DNA, 0.2 mM dNTPs, 1× PCR buffer, 8 pmol of the reverse primer, 2 pmol of the forward primer, 8 pmol of a M13 (-21) primer labeled with one of four dyes (6-FAM, NED, TET and HEX), 1.5 mM MgCl2 and 1 unit of Platinum Taq DNA Polymerase (Invitrogen). The PCR profile for all loci consisted of an initial denaturation step at 95 °C for 5 min, 35 cycles of 94 °C (1 min), 55–62 °C (1 min) and 72 °C (1 min), followed by 15 cycles of 94 °C (30 s), 55 °C (45 s), 72 °C (30 s) and a final extension at 72°C (10 min). We analyzed the results in a MegaBACE 1000 automated sequencer, and scored the allele sizes using the software Genetic Profiler (GE Healthcare).
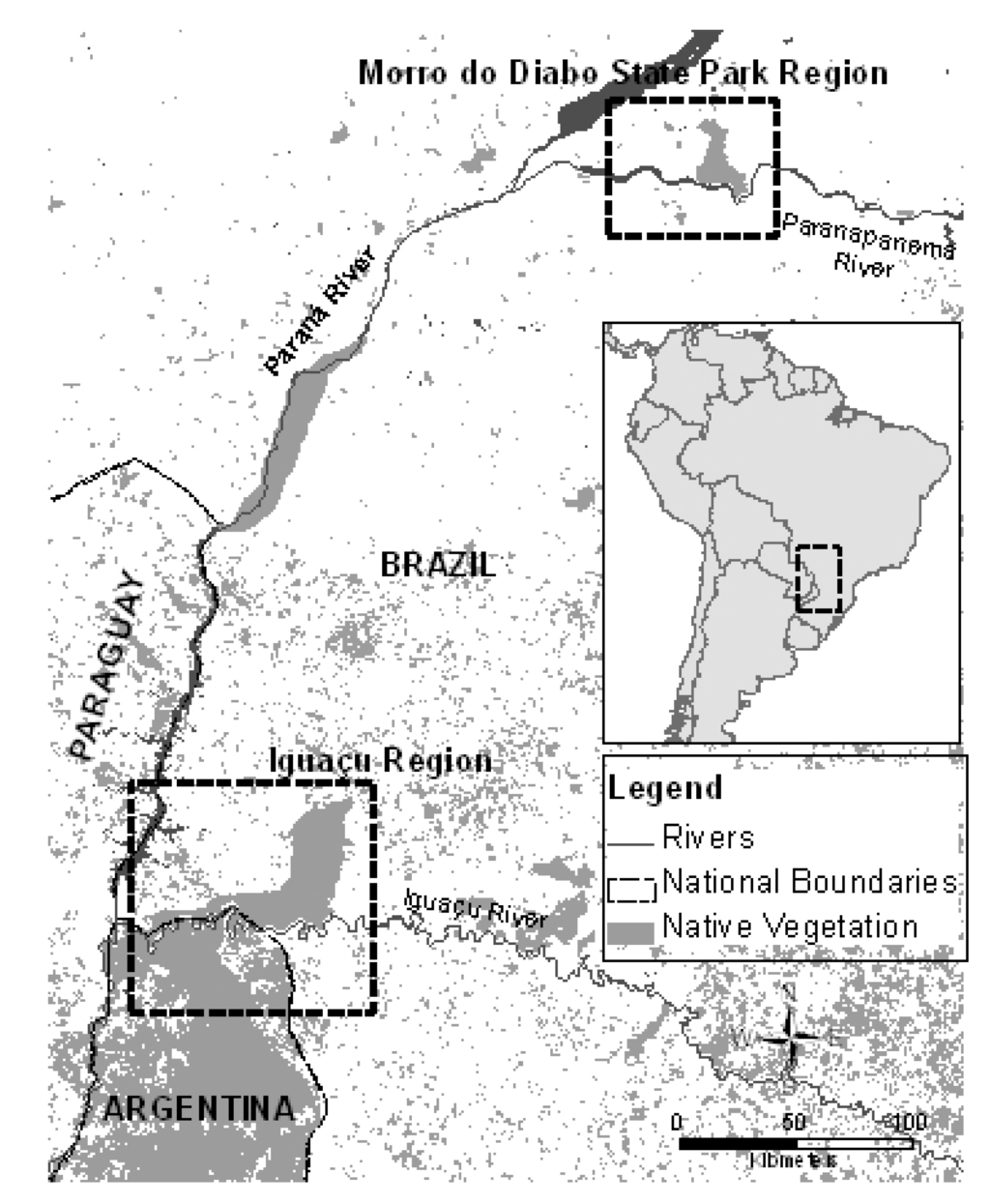
Figure 1.
Upper Parana ecoregion and the sampled areas. Each square represents the origin of the samples collected. The gray areas represent the remaining native vegetation.
2.3. Data Analysis
We calculated deviations from linkage equilibrium (LE) and Hardy-Weinberg equilibrium (HWE) with the software GENEPOP'007 [25] using the Markov chain method (10,000 iterations). We also corrected for multiple tests using the Benjamin and Yekutieli method [26], following the suggestions of Narum [27]. To detect the occurrence of null alleles or large allele dropout, we used the software MICRO-CHECKER 2.2.3 [28] and the Brookfield [29] null allele estimator.
We estimated the expected (HE) and observed (HO) heterozygosity across all loci at each sampling site using GENEPOP'007, and used the FSTAT 2.9.3.2 software [30] to calculate allelic richness (with the rarefaction method to correct for differences in sample size) and to estimate FIS. We used GenAlEx v. 6.5 [31,32] to survey the total number of private alleles in each population, and also used the rarefaction method correction with the HP-RARE software [33].
We inferred the possible occurrence of population bottlenecks using the software BOTTLENECK [34], which compares observed and expected heterozygosities under different mutation models, including the stepwise mutation model (SMM) and the two-phase model (TPM). The TPM included a 95% proportion of alleles attributed to SMM and significance testing with a Wilcoxon test. We also assessed the mean ratio (M) between the total number of alleles and the allele size range, following Garza and Williamson [35], with the AGARst population genetic package [36].
To investigate population differentiation, we calculated FST following Weir [37] and using the FREENA software [38], an efficient method to correct for null allele bias and FST overestimation (so-called ENA correction method). We also used an AMOVA to calculate the pairwise FST with the GenAlEx v. 6.5 software, without correcting for null alleles, and tested for significance with 9999 permutations. We performed an assignment test with GenAlEx 6.5 to test for individuals that were allocated in groups distinct from the one in which they had been collected.
We examined the optimal number of genetic clusters present in the sample with the Bayesian approach implemented in the STRUCTURE 2.1 software [39]. This method assumes HWE and LE within populations, and assesses if genetic subdivision may account for any detected deviation from either expectation. We calculated the most likely number of clusters using a Markov Chain Monte Carlo approach, given the multilocus genotypic data independently of locality information. We evaluated ln likelihoods for k = 1–3 by averaging the results from 30 runs for each value of k, each of which contained a sampled length of 1,000,000 iterations after a burn-in period of 100,000 steps. For these analyses, we assumed the admixture model with correlated allele frequencies.
Finally, we estimated the total effective population size (Ne) of ocelots in these fragments by including all the samples into a single assumed population. We used the linkage disequilibrium method implemented in LDNE 1.31 software [40] with the random mating model using the jackknife approach to assess significance (Pcrit = 0.05).
3. Results
Two of the 12 microsatellites that were initially tested were excluded from the analyses because they failed to amplify (FCA098) or to produce reliable genotypes (FCA146) despite the wide range of PCR conditions we explored. In addition, we were able to genotype locus FCA96 for only about 70% of the samples, most of which presented homozygous genotypes, including double negative amplifications suggesting the occurrence of null alleles. Therefore, this microsatellite locus was also excluded from the analyses. We thus employed the remaining nine, successfully genotyped polymorphic loci to investigate inter and intra-population levels of variation. The independence among these loci was assumed since we found no statistically significant linkage disequilibrium.
The two sampled populations exhibited very similar levels of genetic diversity (Table 1). When the mean between the two populations was computed, the expected heterozygosity was 0.71, the mean number of alleles per locus was 6.1, and the mean allelic richness was 5.7. The MD population exhibited a significant HWE deviation at locus FCA453, whereas the IR population showed a significant excess of homozygotes at locus FCA042, both after implementing the B-Y FDR correction (p < 0.0177). The locus FCA441 showed presence of null alleles only in the IR population (12.5%). No false allele or allelic dropout was detected for the others markers. The total FIS estimates were positive in the two populations (IR: 0.078; MD: 0.001), and significantly higher than zero for IR (p < 0.05). We found 15 private alleles in IR and 10 in MD. However, using the rarefaction estimation, the IR and MD values were lower (1.44 and 1.62, respectively).
We detected significant evidence for a population bottleneck with the Wilcoxon sign-rank test for the MD population, and heterozygote excess under the two mutational models TPM and SMM (p = 0.03711 and 0.04883, respectively). Nevertheless, the mean ratio between the total number of alleles and the allele size range (M, following Garza and Williamson [36]) was 0.750 for MD and 0.776 for IR, in both cases higher than the expected M for a bottlenecked population (M < 0.68).
We detected no significant genotypic differences using the ENA correction method (which corrects for null alleles), with FST = 0.027, (p = 0.077). However, we found a larger divergence without this correction, with FST = 0.038, which was highly significant (p = 0.002). The most likely number of genetic clusters inferred using STRUCTURE was one (K = 1 (mean Ln P (D) = −971.6); K = 2 (mean Ln P (D) = −1060.0); K = 3 (mean Ln P (D) = −1259.5). In addition, when results from k = 2 and k = 3 runs were assessed, all individuals from both sampling sites were equally assigned to all possible clusters, supporting the interpretation that our data set can be best modeled by a single genetic population. Using the GenAlEx assignment test, 27 of 32 (84%) individuals were self-assigned to the population from which they had been sampled. Three individuals from IR and two from MD were assigned to the other population.
Since we found no evidence for genetic differentiation between these populations, we calculated a total Ne including all samples (total 32 individuals). This resulted in an estimate of Ne = 124.9 (CI 46.6–341.1) for the two combined sampling sites.

Table 1.
Genetic diversity at nine microsatellite loci for Leopardus pardalis sampled at the two areas investigated in this study.
| FCA008 | FCA042 ‡ | FCA053 | FCA077 | FCA124 | FCA391 | FCA424 | FCA441 | FCA453 † | Average | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Morro do Diabo (MD) (n = 14) | N | 7 | 9 | 3 | 9 | 2 | 7 | 5 | 3 | 7 | 5.8 |
| R | 6.77 | 8.69 | 3.00 | 8.92 | 2.00 | 6.97 | 5.00 | 3.00 | 6.84 | 5.8 | |
| HE | 0.78 | 0.88 | 0.56 | 0.89 | 0.30 | 0.84 | 0.76 | 0.54 | 0.83 | 0.71 | |
| HO | 0.92 | 1.00 | 0.33 | 0.79 | 0.36 | 0.64 | 0.79 | 0.64 | 0.92 | 0.71 | |
| Fis | −0.190 | −0.147 | 0.417 | 0.123 | −0.182 | 0.243 | −0.036 | −0.200 | −0.099 | 0.001 | |
| Iguaçu Region (IR) (n = 18) | N | 6 | 12 | 5 | 9 | 2 | 8 | 4 | 4 | 7 | 6.3 |
| R | 5.30 | 10.32 | 4.33 | 8.20 | 2.00 | 6.86 | 3.67 | 3.89 | 6.51 | 5.7 | |
| HE | 0.68 | 0.91 | 0.62 | 0.84 | 0.43 | 0.80 | 0.67 | 0.60 | 0.79 | 0.70 | |
| HO | 0.67 | 0.78 | 0.59 | 0.83 | 0.47 | 0.61 | 0.67 | 0.33 | 0.89 | 0.65 | |
| Fis | 0.012 | 0.141 | 0.048 | 0.012 | −0.103 | 0.243 | −0.002 | 0.450 | −0.136 | 0.078 |
N is the number of alleles in each population, R the allelic richness, HE expected heterozygosity, HO observed heterozygosity, † and ‡ indicate loci deviating from Hardy–Weinberg expectations (after B-Y FDR correction for multiple comparisons, p < 0.0177) for the Morro do Diabo population (†) and Iguaçu Region population (‡).
4. Discussion
The average expected heterozygosity (He = 0.71) and number of alleles per locus (6.1) were intermediate relative to values reported previously. A higher diversity was observed for Amazonian ocelots [41], whereas lower values were observed in the remaining ocelot populations from the United States [16,17,18]. It is noteworthy that the microsatellite loci employed in these previous studies were not identical to ours, and therefore this comparison should not be interpreted strictly.
We did not detect clear evidence of population subdivision in our sample, despite previously reported genetic differentiation and lack of migrants in ocelot populations separated by a mere 30 km in North America [17]. Bayesian analyses revealed no meaningful subdivision between our sampled sites, while the FST analyses indicated a low (FST = 0.027–0.038) but statistically significant results. These FST results are low in comparison with the ocelot populations in North America (FST = 0.146–0.367). Knutsen et al. [42] argued that it is difficult to interpret if a low but statistically significant level of genetic differentiation reflects a real substructure, or if it is caused by confounding factors such as retention of shared ancestral polymorphism, high gene flow, or selective sweeps.
However, a previous study on jaguars conducted in the same region reported a significant genetic differentiation between these sampling sites [9]. Since the timing of the fragmentation process (30–40 years) is identical for the two species, and they can be assumed to have similar generation times (ca. 5 years; implying ca. 6–8 generations of isolation in both cases), the contrasting results between them should be accounted for by other factors. One likely explanation would be a difference in their local effective population sizes, which would influence the pace of drift-induced genetic differentiation, and the retention of shared ancestral polymorphism. This is supported by the larger effective population size estimated for ocelots in this study (125 individuals) than that reported for jaguars (ca. 55 individuals [9]) in the same area.
In addition, it is reasonable to expect that ocelots might have better ability than jaguars to move across the highly modified habitat matrix without being detected (and killed) by farmers and ranchers. Such a difference would also lead to ocelots retaining population connectivity for a longer time than jaguars under this scenario of habitat fragmentation. However, previous telemetry studies demonstrate a strong habitat selection by ocelots for areas with closed wood cover [43], which may impose difficulties for dispersal in such a fragmented area, as is the case for jaguars [9]. Therefore, even if some gene flow still remains between the two ocelot populations studied here, it is predicted to decline or be completely abolished as habitat discontinuities continue to increase in the future within the UPAF ecoregion.
In conclusion, our data indicate that the ocelot populations sampled in these UPAF fragments must be viewed as a single genetic population. However, in the face of current levels of habitat loss and fragmentation, it is likely that anthropogenic barriers may lead (or have already led) to the complete severing of the connection between these fragments. Despite the low number of samples obtained in this study, the bottleneck inference for the MD population may already represent results of human-induced fragmentation and consequently an in-progress isolation of these populations. Considering that these results are based on samples collected between 1993 and 2000, and that habitat alteration in the region has continued to increase in the last decades, it may be reasonable to assume that this trend of isolation has already been enhanced, and may pose an important threat to these populations in the near future. It is therefore critical to foster immediate landscape management actions that allow the persistence of ocelot gene flow between these areas to preserve its original population structure and maintain its evolutionary potential in the face of continued disturbances. Such actions include the conservation and restoration of intervening habitats (e.g., riparian forests and native woodland patches within private ranches and farms), and the designation of additional protected areas in the region. Taken together, these actions should help ensure the long-term viability of this species in the region, and also benefit the persistence of the UPAF ecosystem as a whole.
Acknowledgments
The authors thank Jan K. Mähler Jr., Alessandra Nava, Cibele B. Indrusiak, Valeria Conforti and Anael A. Jacob for the samples used in this study. We also thank Rodrigo Begotti for help in the map production, Renata Miotto for help with laboratory procedures, and the Brazilian agencies Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for financial support.
Author Contributions
Fernando C. C. Azevedo, Laury Cullen Jr. and Peter G. Crawshaw Jr. collected the field data; Fernando P. Rodrigues and Eduardo Eizirik conceived the project; Marina G. Figueiredo, Marcelo Cervini and Pedro M. Galetti Jr conducted theexperiments and analyzed the data; and all authors wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marino, J. Threatened Ethiopian wolves persist in small isolated Afroalpine enclaves. Oryx 2003, 37, 62–71. [Google Scholar] [CrossRef]
- Galetti, M.; Eizirik, E.; Beisiegel, B.; Ferraz, K.; Cavalcanti, S.; Srbek-Araujo, A.C.; Crawshaw, P.; Paviolo, A.; Galetti, P.M., Jr.; Jorge, M.L.; et al. Atlantic Rainforest’s Jaguars in Decline. Science 2013, 342, 930. [Google Scholar] [CrossRef] [PubMed]
- Dirzo, R.; Young, H.S.; Galetti, M.; Ceballos, G.; Isaac, N.J.B.; Collen, B. Defaunation in the Anthropocene. Science 2014, 345, 401–406. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Roelke, M.E.; Marker, L.; Newman, A.; Winkler, C.A.; Meltzer, D.; Colly, L.; Evermann, J.F.; Bush, M.; Wildt, D.E. Genetic basis for species vulnerability in the cheetah. Science 1985, 227, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W.; Kalinowski, S.T. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Evol. Syst. 2000, 31, 139–162. [Google Scholar] [CrossRef]
- Coulon, A.; Guillot, G.; Cosson, J.-F.; Angibault, J.M.A.; Aulagnier, S.; Cargnelutti, B.; Galan, M.; Hewison, A.J.M. Genetic structure is influenced by landscape features: Empirical evidence from a roe deer population. Mol. Ecol. 2006, 15, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Ruell, E.W.; Boydston, E.E.; Lyren, L.M.; Alonso, R.S.; Troyer, J.L.; Crooks, K.R.; van de Woude, S. Gene flow and pathogen transmission among bobcats (Lynx rufus) in a fragmented urban landscape. Mol. Ecol. 2012, 21, 1617–1631. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.S.; Knight, M.E.; Darvill, B.; Goulson, D. Extremely low effective population sizes, genetic structuring and reduced genetic diversity in a threatened bumblebee species, Bombus sylvarum (Hymenoptera: Apidae). Mol. Ecol. 2006, 15, 4375–4386. [Google Scholar] [CrossRef] [PubMed]
- Haag, T.; Santos, A.S.; Sana, D.A.; Morato, R.G.; Cullen, R., Jr.; Crawshaw, P., Jr.; de Angelo, C.; di Bitetti, M.; Salzano, F.M.; Eizirik, E. The effect of habitat fragmentation on the genetic structure of a top predator: Loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol. Ecol. 2010, 19, 4906–4921. [Google Scholar] [CrossRef] [PubMed]
- Galbusera, P.; Githiru, M.; Lens, L.; Matthysen, E. Genetic equilibrium despite habitat fragmentation in an Afrotropical bird. Mol. Ecol. 2004, 13, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Purrenhage, J.L.; Niewiarowski, P.H.; Moore, F.B.-G. Population structure of spotted salamanders (Ambystoma maculatum) in a fragmented landscape. Mol. Ecol. 2009, 18, 235–247. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, E.S.; Tello, J.S.; Whitehead, A.; Rolón-Mendoza, C.M.J.; Maldonado-Rodríguez, M.C.D.; Stevens, R.D. Fragmentation of Atlantic Forest has not affected gene flow of a widespread seed-dispersing bat. Mol. Ecol. 2013, 22, 4619–4633. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; Gardner, G.L. Leopardus pardalis. Mammal. Spec. 1997, 548, 1–10. [CrossRef]
- Di Bitetti, M.S.; Placci, G.; Dietz, L. Uma Visão de Biodiversidade Para a Ecorregião Florestas do Alto Paraná—Bioma Mata Atlântica: Planejando a Paisagem de Conservação da Biodiversidade e Estabelecendo Prioridades Para Ações de Conservação; World Wildlife Fund Press: Washington, DC, USA, 2003. (In Portuguese) [Google Scholar]
- Di Bitetti, M.S.; Paviolo, A.; de Angelo, C. Density, habitat use and activity patterns of ocelots (Leopardus pardalis) in the Atlantic Forest of Misiones, Argentina. J. Zool. 2006, 270, 153–163. [Google Scholar] [CrossRef]
- Janečka, J.E.; Tewes, M.E.; Laack, L.L.; Grassman, L.I., Jr.; Haines, A.M.; Honeycutt, R.L. Small effective population sizes of two remnant ocelot populations (Leopardus pardalis albescens) in the United States. Conserv. Genet. 2008, 9, 869–878. [Google Scholar] [CrossRef]
- Janečka, J.E.; Tewes, M.E.; Laack, L.L.; Caso, A.; Grassman, L.I., Jr.; Haines, A.M.; Shindle, D.B.; Davis, B.W.; Murphy, W.J.; Honeycutt, R.L. Reduced genetic diversity and isolation of remnant ocelot populations occupying a severely fragmented landscape in southern Texas. Anim. Conserv. 2011, 14, 608–619. [Google Scholar] [CrossRef]
- Janečka, J.E.; Tewes, M.E.; Laack, L.; Caso, A.; Grassman, L.; Honeycutt, R.L. Loss of Genetic Diversity among Ocelots in the United States during the 20th Century Linked to Human Induced Population Reductions. PLOS 2014, 9, e89384. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.C.; Metzger, J.-P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Crawshaw, P.G., Jr. Comparative Ecology of Ocelot (Leopardus pardalis) and Jaguar (Panthera onca) in a Protected Subtropical Forest in Brazil and Argentina. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 1995. [Google Scholar]
- Jacob, A.A. Ecologia e Conservação da Jaguatirica (Leopardus pardalis) no Parque Estadual Morro do Diabo, Pontal do Paranapanema, SP. Master’s Thesis, University of Brasília, Brasília, Brazil, 2002. [Google Scholar]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Press: New York, NY, USA, 1989. [Google Scholar]
- Menotti-Raymond, M.; David, V.A.; Lyons, L.A.; Schäffer, A.A.; Tomlin, J.F.; Hutton, M.K.; O’Brien, S.J. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 1999, 57, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, Y.; Yekutieli, D. The control of false discovery rate under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar]
- Narum, S. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv. Genet. 2006, 7, 783–787. [Google Scholar] [CrossRef]
- Van Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Brookfield, I.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996, 5, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT, Version 2.9.3.2. Institute of Ecology: Lausanne, 2002; Available online: http://www2.unil.ch/izea/softwares/fstat.html (accessed on 30 October 2009).
- Peakall, R.; Smouse, P.E. GENEALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [PubMed]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Harley, E.H. AGARST (version 3.3): A Program for Calculating Allele Frequencies, Gst and Rst from Microsatellite Data Plus a Number of Other Population Genetic Estimates and Outputting Files Formatted for Various Other Population Genetic Programs; Department of Chemical Pathology. University of Cape Town: Rondebosch, South Africa, 2003. [Google Scholar]
- Weir, B.S. Genetic Data Analysis II; Sinauer & Associates: Sunderland, MA, USA, 1996. [Google Scholar]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Waples, R.S.; Do, C. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, M.; Murillo, A.; Corrales, C.; Romero-Aleán, N.; Alvarez-Prada, D. Genética de poblaciones amazónicas: La historia evolutiva del jaguar, ocelote, delfín rosado, mono lanudo y piurí, reconstruida a partir de sus genes. Anim. Biodivers. Conserv. 2007, 30, 115–130. (In Spanish) [Google Scholar]
- Knutsen, H.; Olsen, E.M.; Jorde, P.E.; Espeland, S.H.; Andre’, C.; Stenseth, N.C. Are low but statistically significant levels of genetic differentiation in marine fishes “biologically meaningful”? A case study of coastal Atlantic cod. Mol. Ecol. 2011, 20, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Horne, J.S.; Haines, A.M.; Tewes, M.E.; Laack, L.L. Habitat partitioning by sympatric ocelots and bobcats: Implications for recovery of ocelots in southern Texas. Southwest. Nat. 2009, 54, 119–126. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).